Abstract
Plant mitochondrial genomes show much more evolutionary plasticity than those of animals. We analysed the first mitochondrial DNA (mtDNA) of a lycophyte, the quillwort Isoetes engelmannii, which is separated from seed plants by more than 350 million years of evolution. The Isoetes mtDNA is particularly rich in recombination events, and chloroplast as well as nuclear DNA inserts document the incorporation of foreign sequences already in this most ancestral vascular plant lineage. On the other hand, particularly small group II introns and short intergenic regions reveal a tendency of evolution towards a compact mitochondrial genome. RNA editing reaches extreme levels exceeding 100 pyrimidine exchanges in individual mRNAs and, hitherto unobserved in such frequency, also in tRNAs with 18 C-to-U conversions in the tRNA for proline. In total, some 1500 sites of RNA editing can be expected for the Isoetes mitochondrial transcriptome. As a unique molecular novelty, the Isoetes cox1 gene requires trans-splicing via a discontinuous group I intron demonstrating disrupted, but functional, RNAs for yet another class of natural ribozymes.
INTRODUCTION
Mitochondrial DNAs (mtDNAs) trace back in evolution to the genome of an α-proteobacterial endosymbiont which gave rise to the mitochondria of eukaryotic cells (1). The mitochondrial genomes in most animal (metazoa) lineages are compact, circular DNAs of some 16 kb which encode a standard set of 37 or fewer tightly packed genes (2). The mtDNAs of other eukaryotes, however, are significantly more diversified, most notably between different protist lineages, which reflect most of the evolutionary history and diversity of eukaryotic cells (3). These, for example, include obvious evolutionary ancestral states such as the gene-rich 69-kb mtDNA of the jakobid protist Reclinomonas americana with nearly 100 mitochondrial genes (4) as well as the massively reduced 6-kb mtDNA of the malaria parasite Plasmodium falciparum with only five genes (5), reflecting a massive gene transfer into the nuclear genome.
Land plant (embryophyte) mtDNAs in contrast are significantly extended in size and may exceed 2000 kb in certain flowering plant (angiosperm) families (6). The embryophyte mtDNAs encode some of the genes for protein subunits of the respiratory chain complexes, for ribosomal proteins and for proteins involved in cytochrome c biogenesis which are found in protists but are generally absent from animal or fungal mitochondrial genomes. Many plant mitochondrial genes are interrupted by introns belonging to either of the two classes of ribozyme-type group I or group II introns, which are commonly encountered in fungal, algal and plant organelle genomes and occasionally also in bacteria, phages and exceptionally also in the mtDNAs of primitive metazoan lineages (2,7). Besides intron gains, size increases of plant mitochondrial genomes have occurred mainly through the extension of non-coding intergenic regions. This becomes immediately apparent when the available mtDNAs of land plants (Figure 1) are compared to those of the charophyte algae (8,9) phylogenetically related to the embryophyte lineage (10). Some of the additional sequences, at least in flowering plant (angiosperm) mtDNA, have been identified as copies of chloroplast or nuclear DNA (11,12) or even as gained via horizontal gene transfer (13–16).
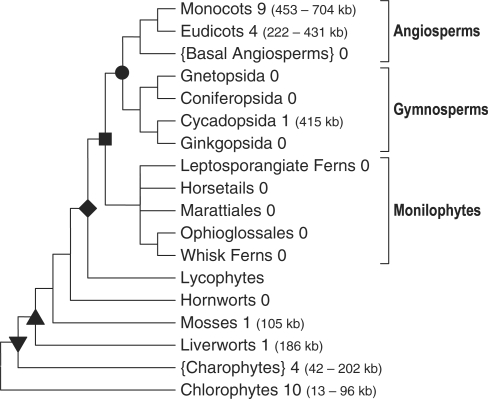
Figure 1.
Current view of a simplified phylogeny of extant Viridiplantae (green plants sensu lato). The cladogram shown summarizes insights from recent molecular studies of land plant phylogeny (e.g. Qiu et al., 2006). Numbers of completely sequenced mtDNAs (http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plants_tax.html) are indicated for each group. Brackets indicate paraphyletic grades, all other designations indicate reasonably well supported monophyletic groups. Further well supported monophyletic clades of higher order are the spermatophytes (seed plants, circle), the euphyllophytes (square), the tracheophytes (vascular plants, rhomb), the embryophytes (land plants, up triangle) and the streptophytes (down triangle) whereas the bryophytes (liverworts, mosses and hornworts) are paraphyletic.
Moreover, plant mitochondria have evolved complex features with respect to genome arrangements and gene expression that are contrary to the general evolutionary trend of compaction and streamlining of endosymbiotic genomes (17,18). The gain of RNA editing activity in the organelles (19,20) to correct gene sequences by pyrimidine exchanges (mainly cytidine to uridine) at the transcript level likewise appears to be gained with the earliest embryophytes, although this phenomenon is suspiciously absent in the subclass of marchantiid liverworts (21). Despite size increase to more than 100 kb in early embryophyte evolution (Figure 1), the two so far available completely sequenced mtDNAs of bryophytes—those of the liverwort Marchantia polymorpha (22) and the moss Physcomitrella patens (23)—are recognized as simple, circular-mapping genomes. However, linear DNAs may in fact contribute significantly to the population of mtDNA molecules actually present in the mitochondria (24). Flowering plant (angiosperm) mtDNAs are rich in active recombination resulting in co-existing alternative mitochondrial genome arrangements (e.g. 25–27), the stoichiometries of which are now understood to be regulated by nuclear-encoded protein factors related to bacterial rec proteins (28). A ‘master-circle’ representing the full mitochondrial genome complexity in a single circular DNA molecule may be entirely hypothetical in these cases (29).
Evidently coinciding with the rise of recombinational activity during the evolution of plant mitochondrial genomes is the appearance of trans-splicing group II introns producing peculiar arrangements of the affected genes with exons distributed across wide distances in the mtDNA. The origins of trans-splicing group II introns have been traced back through plant evolution (Figure 1) as having arisen through disruption of ancestral, conventional group II introns that can still be identified as their respective orthologues in ferns, hornworts and mosses (30,31).
In the absence of complete mtDNA information for ferns, horsetails, lycophytes or hornworts (Figure 1), there is currently a large phylogenetic gap remaining between the available mtDNA sequences of the liverwort Marchantia or the moss Physcomitrella and the first recently completed mtDNA of a gymnosperm, the cycad Cycas taitungensis (32). Accordingly, we have investigated the mtDNA of the quillwort Isoetes engelmannii. As a lycophyte, Isoetes represents the most ancestral lineage of recent vascular plants (tracheophytes). The mtDNA of I. engelmannii offers a plethora of surprising findings, which include particularly small group II introns, extreme frequencies of DNA recombination and RNA editing also in tRNAs, insertions of chloroplast and nuclear DNA and, most notably, a trans-splicing group I intron.
MATERIALS AND METHODS
Fosmid analyses
Isoetes engelmannii plant material originally collected in South Central Indiana (USA) by Jerry Gastony, and subsequently greenhouse cultivated, was kindly made available through Jeff Palmer and Erin Badenhop (Bloomington, IN). The non-green bulb tissue of plants was used to enrich for mitochondrial vs. chloroplast DNA. Total genomic DNA was isolated using a CTAB protocol. After size-fractionation into ∼38 kb fragments, DNA was blunt-ended and cloned into the fosmid vector pCC1FOS using the CopyControl Fosmid Library Production Kit (EPICENTRE, Madison, Wisconsin). A library of 11 700 fosmid clones was sorted and filter-spotted for successive rounds of hybridization initially using a mixture of PCR-derived gene probes of cox3, nad2, nad5 and nad7 and subsequently with probes derived from the sequence-verified mitochondrial fosmids. Identity of fosmid clones was initially verified through terminal insert sequencing and positive clones were used for sub-library production. Fosmid DNAs were isolated using NucleoBond Xtra Midi EF Kit (Macherey Nagel, Düren, Germany), sheared by Nebulizers (Invitrogen, Carlsbad, California), blunted using a End-It DNA End-Repair Kit (EPICENTRE, Madison, Wisconsin), A-tailed with Taq-Polymerase (Genaxxon, Biberach, Germany), and fractionated by preparative electrophoresis in 0.8% agarose. Fragments of 2–2.5 kb in size were recovered using the NucleoSpin Extract II Kit (Macherey Nagel, Düren, Germany) and cloned into pGEM-T Easy vector (Promega, Madison, Wisconsin). Minimally 400 plasmid clones were sequenced for each fosmid to reach ∼8-fold sequencing coverage. Five fosmid clones (11P20, 19N12, 26A6, 28M14 and 30K18) were validated as native mtDNA. Graphical maps of the fosmid clones created with OGDRAW v1.1 (33) are given in Supplementary Figure 1. The respective fosmid insert sequences were annotated and deposited in the database under accession numbers FJ010859, FJ536259, FJ390841, FJ176330 and FJ628360, respectively.
Sequence analyses
Sequence handling and analysis of final fosmid assemblies was essentially done using the alignment explorer of the MEGA software (34). Identification of loci was essentially done using similarity searches with Basic Local Alignment Search Tool (BLAST) service at the NCBI (35). Candidate sites of RNA editing in Isoetes were identified manually in alignments of deduced protein sequences with homologues in Chara (AY267353) and Marchantia (M68 929), species devoid of RNA editing.
Trancript analyses
Total I. engelmannii RNA was isolated using the NucleoSpin RNA Plant Kit (Macherey Nagel, Düren, Germany); cDNA was synthesized with RevertAid First Strand cDNA Synthesis Kit (Fermentas, Burlington, Ontario) in the presence of random hexamer primers as recommended by the manufacturer. Oligonucleotide pairs (all sequence information available from the authors upon request) were used for RT–PCR amplification according to the standard protocol of GoTaq DNA Polymerase (Promega, Madison, Wisconsin) in a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, California) with annealing temperatures between 50°C and 55°C. Amplicons were recovered from agarose gel and cloned into pGEM T Easy vector as described above. On average, 10 cDNA clones per locus were sequenced and analysed by comparison with the corresponding DNA sequences. RNA self ligation for cox1 transcript end mapping followed published procedures (36). Total I. engelmannii RNA was ligated by T4 RNA ligase (New England Biolabs, Ipswich, Massachusetts) and cDNA was synthesized with RevertAid First Strand cDNA Synthesis Kit (Fermentas, Burlington, Ontario) in the presence of 200 pmol of outward directed primers cox1leftdo1 and cox1rightup (1 and 2, respectively, in Figure 2). The same oligonucleotide pairs were used for first PCR amplification according to the standard protocol of BD Advantage 2 polymerase (BD Bioscience, Franklin Lakes, New Jersey) in a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, California) with annealing temperatures at 45°C. Amplification results in several molecules of different sizes which were recovered from an agarose gel using the NucleoSpin Extract II Kit (Macherey Nagel, Düren, Germany). These molecules served as templates for subsequent nested PCR in presence of primers cox1leftdo and cox1rightup (1 and 3, respectively, in Figure 2) with same preferences as initial PCR.
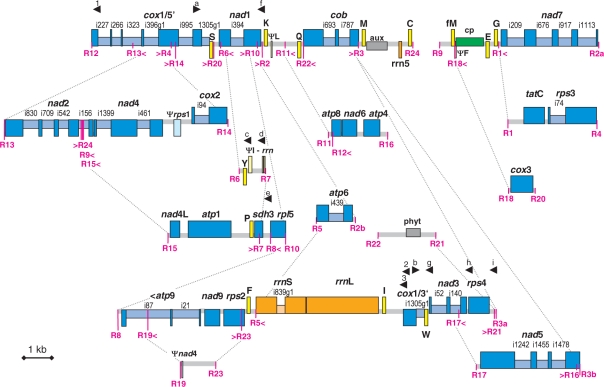
Figure 2.
The Isoetes engelmannii mtDNA with protein-coding genes shown in blue, tRNAs in yellow, rRNAs in orange, pseudogenes in the respective lighter colours and the cp and nuc DNA inserts indicated with green and grey boxes, respectively. Drawing is approximately to scale. Genes shown above or below the lines indicate directions of transcription to the right or to the left, respectively. Recombination points (R1–R24) are highlighted in magenta with arrows indicating recombination forks. Selected connections between islands of recombination are exemplarily shown with stippled grey lines. Unique net mtDNA sequences add up to 57 571 bp. Arrowheads indicate oligonucleotide primers (1–3 and a–i) anchoring in regions not affected by RNA editing to analyze the arrangement of cox1 and its transcript maturation.
Southern blotting
For Southern blotting, ∼10 µg of I. engelmannii total DNA was digested with combinations of restriction enzymes (either EcoRI and Cfr9I or EcoRI and EcoRV, see Figure 6) and separated on a 0.8% agarose gel prior to blotting following established procedures (37). Approximately 100 ng of PCR-derived probes (Figure 6) were radioactively labelled with 50 µCi of α-P32-dCTP. Hybridization of the nylon blotting membranes was overnight at 65°C in 50 mM sodium phosphate buffer containing 0.9 M NaCl, followed by washing in 2× SSC with 0.1% SDS at 65°C before exposure on a phosphor imager.
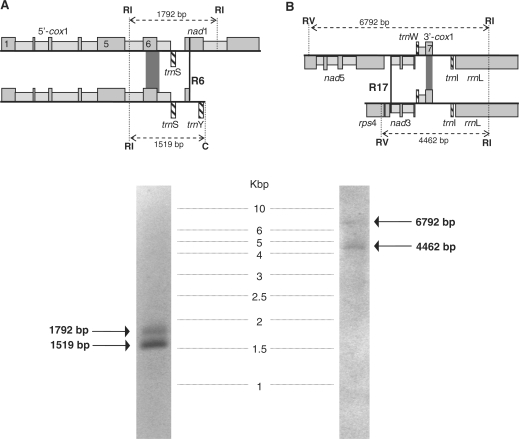
Figure 6.
Southern-blot hybridization to verify cox1 gene arrangements for the upstream (A) and downstream (B) part of the gene using probes covering cox1 exons 6 and 7, respectively (dark grey rectangles). Isoetes engelmannii total genomic DNA was digested with EcoRI (RI) and Cfr9I (C) or with EcoRI (RI) and EcoRV (RV) to include the nearest identified recombination sites R6 or R17, respectively, in each case. Only two restriction fragments of expected sizes for coexisting genomic arrangements across R6 and R17 were identified by hybridization in each case: the 5′ cox1 part followed by trnS and either nad1 or by Ψnad1-trnY (A) and the 3′ cox1 part preceded by either nad5, Ψnad3 and trnW or by rps4, nad3 and trnW (B).
RESULTS
Genomic features and gene complement of the Isoetes mtDNA
The I. engelmannii mtDNA sequence was assembled from fosmid clones, identified in an arrayed library by hybridization with mitochondrial gene probes and verified in their mitochondrial nature through complete sequencing of the inserts. As more fosmid sequences were analyzed in the course of our studies, it became apparent that the same genes were repeatedly identified. Mitochondrial genes were found in different genomic environments, indicating a particularly high frequency of recombination events resulting in co-existing alternative gene arrangements (Figure 2). A total of 24 recombination breakpoints were identified, making the physical existence of a potential mtDNA master-circle encompassing the full mtDNA complexity highly unlikely. Different fosmid inserts reflected different pathways through the recombination points and the resulting products of DNA recombination were exemplarily verified as co-existing (see below). The net mtDNA sequence complexity of the analyzed I. engelmannii fosmid clones is 57 571 bp, with an overall A+T content of 51.3% and a percentage of 46.2% coding sequences.
We identified a typical complement of plant mitochondrial genes (Table 1) encoding subunits of respiratory chain complex I (nad genes nad1, 2, 3, 4, 4L, 5, 6, 7 and 9), complex II (sdh3), complex III (cob), complex IV (cox1, 2 and 3) and of complex V, the ATP synthase (atp1, 4, 6, 8 and 9). Likewise present are the genes for the large, small and 5S rRNAs (rrnL, rrnS, rrn5), for four ribosomal proteins (rpl5, rps2, rps3 and rps4) as well as the tatC gene encoding a subunit of the sec-independent transport pathway, and thirteen intact tRNA genes. Hence, on the one hand, four ribosomal protein genes demonstrated to be frequently transferred to the nucleus in angiosperms (38) are present in the Isoetes mtDNA. On the other hand, genes encoding cytochrome biogenesis components (ccmB, ccmC, ccmF) are completely lacking as had previously been observed for the land plant lineage only in the mtDNA of the green alga Chaetosphaeridium (8). To exclude the possibility that the ccm genes were accidently missed through yet a further recombination event, we have used oligonucleotide primers directed against conserved ccmB, ccmC and ccmF sequences but were unable to retrieve them in PCR approaches using I. engelmannii DNA. In addition, we identified small pseudogene fragments of three tRNA genes, of the rrn genes and of the nad4 and rps1 genes.
Table 1.
The gene and intron complement of Isoetes engelmannii (Ie) mtDNA in comparison to the mtDNAs of the liverwort Marchantia polymorpha (Mp) the moss Physcomitrella patens (Pp), the gymnosperm Cycas taitungensis (Ct) and the angiosperm Arabidopsis thaliana (At)
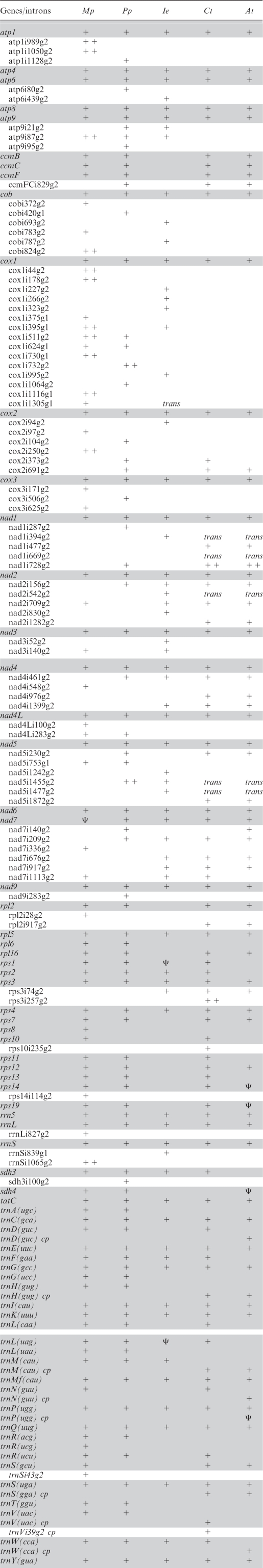 |
Plus signs indicate presence of a gene or intron, respectively and Ψ indicates recognizable pseudogene remnants (indicated only where not accompanied by a co-existing functional gene copy). Intron designations are according to the position of the preceding nucleotide in the uninterrupted coding sequence in the reading frames of M. polymorpha with the addendum g1 or g2 indicating group I or group II introns, respectively. Double plus signs (++) indicate presence of intron-borne ORFs.
Recombination points (Figure 2) were identified both in intergenic regions (R1, R2, R8, R11, R20 and R21) as well as in coding regions (of atp6: R5, atp8: R12, atp9: R19, cob: R3, R22, cox1: R12, R13, R4, R14, rpl5: R10, rps2: R23, rps3: R4, trnF: R18, nad1: R6, R10, nad2: R24, R9, R15, nad3: R17, nad5: R16, R3b and sdh3: R7, respectively), which accordingly result in fragmented pseudogene copies co-existing with the functional genes. In agreement with the observation of highly frequent recombination, only two evolutionary ancient gene linkages (trnP–sdh3 and nad4–nad2) are conserved as mere traces of much more extended syntenies that were identified when the liverwort Marchantia and the moss Physcomitrella mtDNAs were compared (23) to those of the streptophyte algae Chaetosphaeridium globosum (8) and Chara vulgaris (9). When not affected by recombination, intergenic regions between functional genes are small (only 17 bp between nad4 and nad2 and 8 bp between atp8 and nad6, respectively) with the exception of the spacer between trnK and trnQ carrying the pseudo-trnL fragment and the non-coding regions extended through the insertions of foreign DNA fragments.
Insertions of foreign DNA
Three ‘promiscuous’ DNA inserts of foreign origin were identified in the I. engelmannii mtDNA. A 1208 bp fragment of chloroplast DNA located between trnE and trnfM (Figure 2) covers parts of the chloroplast trnA and 23S rRNA genes. Highest similarity of this chloroplast sequence insert is found with the corresponding chloroplast sequence of another Isoetes species deposited in the database (I. malinverniana, DQ629281) indicating (recent) inter-organellar rather than horizontal gene transfer. Sequence deviations of the chloroplast insert in the I. engelmannii mtDNA from the native chloroplast homologue show striking pattern of degeneration with only two base changes within 1080 nt of 23S rRNA but indels of exclusively 5 or 6 bp (Supplementary Figure 2).
A 735 bp sequence stretch in the intergenic region between rrn5 and trnM bears strong similarity with nuclear encoded auxin-responsive transcription factors and a 533-bp sequence with similarity to phytochrome genes occurs between rps4 and trnQ (Figure 2). Like the chloroplast insertion, both nuclear sequence inserts are non-functional pseudogene fragments.
RNA editing in mRNAs
The protein-encoding genes in the I. engelmannii mtDNA show a very strong requirement for mRNA editing via pyrimidine exchanges to reconstitute evolutionary conserved codons. Altogether more than 1420 positions (over 1200 C-to-U and 220 U-to-C changes) in the Isoetes mitochondrial transcriptome appear to be subject to editing (Supplementary Table 1). This includes the reconstitution of appropriate AUG start codons from ACG threonine codons which is required in 12 cases and the introduction of stop codons which is required for nine reading frames, respectively. In fact, the introduction of both the start and stop codons at the same time to correctly define the reading frames is necessary in five genes: atp6, atp9, cox3, nad4 and nad4L. Reverse U-to-C editing is required in the majority of mRNAs to convert genomically encoded stop codons into conserved glutamine or arginine codons. Indeed, only three out of the 24 protein encoding genes in the Isoetes mtDNA (atp4, nad3, nad4L) are without any stop codons on DNA level. To confirm the expectations on RNA editing, we performed exemplary cDNA analyses (Supplementary Table 1). RNA editing was indeed found to affect more than one out of five amino acid identities in the atp1 reading frame (115 of 515 codons) through 105 C-to-U exchanges and 23 U-to-C exchanges in the mRNA, including a change of six codons in a row with the sequence motif H-C-STOP-T-P-S changed into Y-R-Q-M-S-L by four C-to-U and two U-to-C exchanges in the transcript (not shown). A full 14 stop codons are removed through U-to-C RNA editing in the case of the atp1 mRNA. Approximately one out of seven nucleotides is affected by RNA editing in the atp9 reading frame (Figure 3A) resulting in sense changes in one out of three codons exactly as predicted to reconstitute evolutionary conserved codons, including introduction of both the start and the stop codon. Typically, the steady state pool of flowering plant mitochondrial mRNAs contains transcripts edited to different degrees, reflecting only partial editing of some sites. To investigate this for I. engelmannii we examined 30 cDNA clones for the nad7 gene, for which we postulated RNA editing to correct 92 codon identities, including removal of nine stop codons. Complete editing of all the sites exactly as predicted was observed in 20 of the 30 cDNA clones, whereas four cDNAs lacked one editing to remove one of the stop codons (Figure 3B). The remaining six cDNA clones showed individual patterns lacking editing at this or another of one of five codons in total affected by partial editing (Figure 3B).
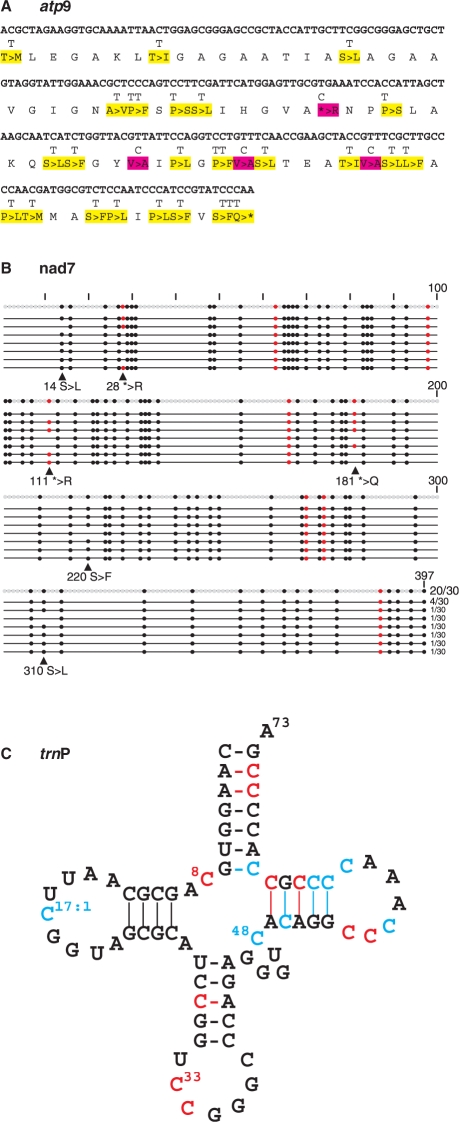
Figure 3.
Examples of cDNA analyses for RNA editing: atp9 (A), nad7 (B) and trnP (C). (A) Yellow and magenta shading indicate changes introduced through C-to-U or U-to-C editing events in the atp9 mRNA. (B) The degree of partial editing of nad7 transcripts was investigated with 30 cDNA clones covering the complete reading frame of 397 codons. Black dots indicate codon sense changes, red dots the removal of stop codons. Twenty of 30 cDNA clones reflected complete editing at all predicted sites, the remaining clones lacked editing at certain positions. (C) Ten expected (red) and eight additional, non-predicted RNA editing sites (cyan) were found in trnP. Numbering of tRNA positions follows the standard convention with numbers after the colons indicating optional nucleotides not present in all tRNAs.
RNA editing in tRNAs
Cloverleaf modelling of the 13 tRNAs present in the Isoetes mtDNA strongly suggested frequent RNA editing activity to act on tRNAs as well. Several base-pairings in the four conserved stems and unpaired conserved uridines need to be re-established through C-to-U RNA editing in eleven tRNAs. The number of sites with predicted RNA editing events varied from single positions each in tRNA-fM and tRNA-G to six in tRNA-Q and even ten in tRNA-P (Figure 3C), respectively. Assuming that tRNA editing may take place in a precursor-transcript before processing we targeted a likely co-transcript of trnP with sdh3 (Figure 2) by RT–PCR, one of the rare cases of an ancient, conserved gene arrangement. Sequencing the cDNA product revealed not only the ten postulated positions of C-to-U exchanges, but also eight additional sites of C-to-U editing (but no reverse U-to-C changes), i.e. a total of 18 RNA editing positions in the trnP coding sequence (Figure 3C).
Introns
A total of 27 group II introns were identified in the I. engelmannii mtDNA (Table 1, nomenclature according to ref. (39), all of which are located in protein coding genes and most of which are particularly small. In fact, the I. engelmannii intron cox1i266 has a size of only 327 bp—to our knowledge, the smallest known group II intron as yet identified in any organism. Despite the strong recombinational activity in I. engelmannii mtDNA, none of the group II introns is in a trans-splicing arrangement. On the contrary, four of the known trans-splicing group II introns in angiosperms have cis-arranged counterparts in I. engelmannii (nad1i394, nad2i542, nad5i1455 and nad5i1477), seed plant introns nad1i669 and nad1i728 (in e.g. Beta vulgaris and Oryza sativa) obviously only appear later in evolution and get disrupted into trans-arrangements. A total of nine group II introns appear at novel insertion sites not yet observed in green algae (Charophytes or Chlorophytes), bryophytes or seed plants: atp6i439, cobi693, cobi787, cox1i227, cox1i266, cox1i323, cox1i995, cox2i94 and nad2i830 (Table 1).
Three group I introns were found in the Isoetes mtDNA, one in rrnS and two in cox1 (Figure 2). Orthologues of group I intron cox1i395 had previously been identified the liverwort Marchantia and in the alga Chaetosphaeridium, both of which carry endonuclease ORFs typical for this intron class. The I. engelmannii counterpart now identified is a small group I intron of only 328 bp without an ORF and hence similarly size-reduced as the group II introns. Yet smaller with a size of only 237 bp is a group I intron (rrnSi839g1) in the small ribosomal RNA gene rrnS.
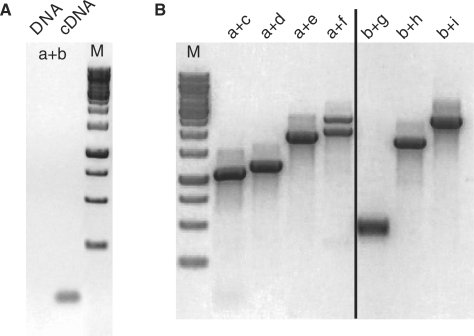
The most notable genomic peculiarity of the Isoetes mtDNA resides in the 3′ part of the cox1 gene (Figure 2). A group I intron (coxi1305) with a known homologue in the Marchantia mtDNA interrupts the cox1 coding region. However, intron homology breaks off sharply 210 bp after the splice donor site, 130 bp upstream of the trnS(uga) gene located downstream in inverted orientation. The seemingly missing terminal cox1 coding sequence was found elsewhere, preceded by the adequate splice acceptor site for joining the exons appropriately, 290 bp downstream of the trnW(cca) gene. Both parts of the cox1 coding regions have comparable similarities to other cox1 sequences in the database making independent, foreign origins, of the one or the other part of the gene, for example through horizontal gene transfer (15,16), unlikely.
To elucidate whether we had failed to identify a cox1 sequence continuity, we used primers anchoring in the directly flanking and also in distant cox1 exons, respectively, for PCR amplification assays on I. engelmannii DNA but failed to retrieve products (Figure 4A). To exclude potential malfunctions of the primers we used them individually in combinations with other primers anchoring in genomic distances ∼2 kb apart in each case, as predicted from the recombinational mtDNA map (Figure 2). Expected products were retrieved both for the upstream part of cox1 extending downstream across several other genes and recombination points (trnS-R6-trnY-trnI-R7-sdh3-rpl5-R10-nad1) as well as the downstream part of cox1 extending upstream across other genes (trnW-nad3-rps4), respectively (Figure 4B). Most notably, the different genomic routes downstream of cox1/5′ (Figure 2) identified through fosmid mapping were found to be faithfully reflected by two PCR products confirming the coexisting gene arrangements (Figure 4B).
Figure 4.
(A) Primers anchoring in the sub-terminal (a) and terminal (b) exons of cox1 fail to detect a genomic continuity across cox1i1305 or an alternative, intron-less cox1 copy with DNA but readily amplify the expected, spliced product with cDNA. (B) Each of the primers a and b combined with other primers anchoring in distant mtDNA regions (see Figure 2) downstream of trnY (a + c), downstream of ΨtrnI (a + d), downstream of sdh3 (a + e), downstream of nad1 (a + f) or upstream of trnW (b + g), downstream of nad3 (b + h) or downstream of rps4 (b + i), respectively, all reveal amplicon products as expected. Two PCR products obtained simultaneously with primer combination a–f (lane 4) faithfully reflect the co-existing alternative genomic arrangements downstream of cox1/5′ to nad1 either via the trnS-nad1 continuity directly or alternatively through the trnS-R6-trnY-ΨtrnI-R7-sdh3-rpl5-R10-nad1 pathway.
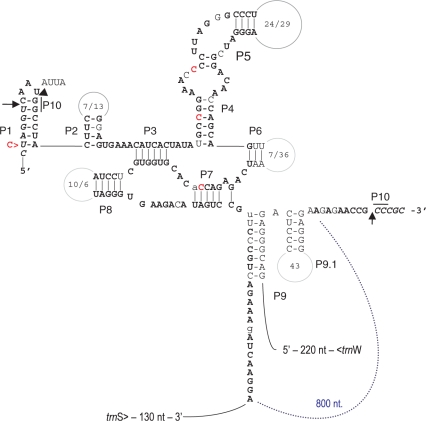
RT–PCR products across the cox1i1305 discontinuity were easily retrieved from cDNA (Figure 4A). Cloning and sequencing verified correct splicing of cox1i1305 and all five additional upstream cox1 introns and showed differences to the genomic sequence exclusively at 106 positions of RNA editing, exactly as expected. Modelling the discontinuous group I intron sequences of cox1i1305 flanking the distantly located terminal cox1 exons reveals that the two sequence halves can combine for a classic group I secondary structure (Figure 5), to our knowledge the first example of a trans-splicing group I intron identified in nature. The typical ribozymic intron core structure of group I introns (40–42) is well conserved in comparison to its conventionally cis-arranged homologue in Marchantia (43).
Figure 5.
The trans-splicing group I intron cox1i1305 in I. engelmannii. The ribozymic core in the 5′-half (paired regions P1–P8) is conserved with the cis-arranged orthologue in Marchantia polymorpha. Italic letters indicate flanking exon sequences, bold letters indicate nucleotide identities in Marchantia, non-bold letters indicate transitions, lower case letters indicate transversions and numbers indicate loop sizes L2, L5, L6 and L8 in Isoetes and Marchantia (after the oblique), respectively. The intron discontinuity in Isoetes coincides with rearrangements in P9 which embraces a large continuous L9 loop of 800-nt in Marchantia (dotted line, blue). The Marchantia orthologue carries only traces of a formerly functional intron-encoded ORF. A C-to-U RNA editing event in the upstream exon is shown, three further cytidines in the intron core may likewise be subject to editings which could improve conservation of base-pairings.
To complement the PCR approaches outlined above in targeting potential alternatively arranged cox1 loci, we used cox1 cDNA as well as a mixture of the cox1i1305 intron halves as new probes. Rehybridization into our fosmid library, however, identified only those fosmids that had been identified and sequenced before.
To independently investigate the cox1 gene arrangement in I. engelmannii as deduced from the mtDNA map (Figure 2), we have used probes covering the terminal and sub-terminal exons 6 and 7 of the cox1 gene in a Southern blot hybridization experiment (Figure 6). Restriction sites for digestion of total genomic Isoetes DNA were selected to include the proximal recombination points identified near the upstream (R6) and the downstream part of the cox1 gene (R17). With hybridizations using probes for the upstream (Figure 6A) and downstream part of the cox1 gene (Figure 6B), two hybridizing restriction fragments were indeed identified in each case, reflecting the co-existing genomic rearrangements exactly as predicted from the genomic map (Figure 2). No further, additional hybridization signals were identified, which could potentially represent a continuous cox1 gene copy (either intron-less or with a cis-splicing cox1i1305 counterpart) in full accord with the PCR experiments detailed above.
To determine cox1 transcript ends we used an approach of RNA circularization by self-ligation, followed by cDNA synthesis and RT–PCR with outward directed primers (oligonucleotides 1 and 2 in Figure 2). This revealed a 3′-UTR (untranslated region) extending 18 bp downstream of the stop codon and a 5′-UTR of 71 bp. The first seven cox1 codons are identical to those of the atp8 gene provided via recombination event R12 (Figure 2).
DISCUSSION
Lycophytes occupy a crucial position in the phylogeny of land plants (Figure 1), now unequivocally recognized as the sister group to euphyllophytes, which comprise the seed plants and the monilophytes with the latter encompassing the ferns, horsetails and whisk ferns (10,44,45). Comparatively poor in numbers of genera, families and with only three orders (Isoetales, Lycopodiales and Selaginellales) the recent lycophytes represent the most ancient lineage of vascular plants. As such, they could be expected to assume an intermediary position between the non-vascular bryophytes and the evolutionary advanced tracheophytes also with respect to the evolution of complexity in plant mtDNA. However, the I. engelmannii mitochondrial genome reported here as a first lycophyte mtDNA rather underlines the notion that plant mitochondria are ‘more unique than ever’ (46) by providing yet another example of unique pathways of organelle genome evolution.
Two main evolutionary trends have evidently shaped the I. engelmannii mtDNA. The gain and rise of recombinational activity seems to be the evolutionary force producing co-existing gene arrangements and the discontinuous group I intron now discovered in the cox1 gene. Likewise, highly active DNA recombination may be the ultimate prerequisite for the incorporation of DNA from the nuclear and chloroplast genomes, which has not been observed in bryophyte mtDNAs. After the recent report of chloroplast DNA inserts in the mtDNA of the gymnosperm Cycas taitungensis (47), the first occurrences of such ‘promiscuous’ inserts of foreign DNA are now pushed back yet way further in plant evolution. The peculiar disposition of plant mtDNA to incorporate foreign genetic material originating from the other two genomes in the plant cell may have evolved with the increase of recombinational activity in the earliest tracheophytes (Figure 1).
The small introns and the small intergenic regions in Isoetes mtDNA on the other hand seem to reflect a counter-acting trend for organelle genome compaction. This tendency is also reflected in the Isoetes mtDNA gene complement itself, which generally mirrors the observations made for independent nuclear gene transfer in a survey of 280 flowering plant genera (38). Ribosomal protein genes that were found frequently and independently lost from the angiosperm mtDNAs are similarly missing from the Isoetes mtDNA, whereas those found to be transferred to the nucleus more rarely are (still) present (notably rps2, rps3 and rps4). Obvious exceptions on the other hand are the ccm genes not present in the Isoetes mtDNA. The inability to identify ccm genes independently via PCR may either indicate the commonly observed significant sequence alteration after nuclear gene transfer or, as a more remote possibility, an evolutionary switch to an alternative pathway of cytochrome c maturation (48,49). Vice versa, the sdh3 gene was here identified in the Isoetes mtDNA but is frequently transferred to the nucleus in angiosperms.
Notably, despite high recombinational activity, none of the 27 group II introns in the I. engelmannii mtDNA was found in a trans-splicing arrangement. On the contrary, four of the conserved trans-splicing group II introns of angiosperms find their small orthologues as cis-arranged counterparts in Isoetes (Table 1). Hence, the trans-splicing group I intron reported here to occur in the cox1 gene may represent a mere chance product with recombination acting before size reduction towards a minimum ribozyme core had reduced the chances of creating a discontinuous, yet functional, intron. Trans-splicing group II introns are known for more than 20 yrs since their discoveries both in chloroplasts (50,51) of algae and land plants and briefly thereafter in plant mitochondria (52–54) and, more recently, also in the mtDNA of an alga (55).
The first example of a trans-splicing group I Intron in nature shows that discontinuous molecules exist in yet another class of ribozyme-type RNAs after a discontinuous hammerhead RNA had been reported very recently (56). For mitochondrial genomes, yet another type of gene discontinuity with ‘modules’ distributed over separate DNA molecules for which the mechanisms of RNA maturation still have to be determined had recently been described for the protist Diplonema (57). Other examples for unusual modes of RNA maturation have also been reported outside of mitochondria such as tRNAs encoded in separate genes for 5′ and 3′ halves in Nanoarchaeum (58,59) or circularly permuted tRNAs expressed via circular RNA intermediates in the red alga Cyanidioschyon (60).
Whereas several of the introns in the Isoetes mtDNA have clear homologues at identical positions in bryophyte or seed plant mitochondrial genomes (Table 1), nine of the group II insertion sites are so far unique in the quillwort. It will be highly interesting whether homologues of these introns can be identified in the remaining major land plant clades for which complete mitochondrial genomes are still missing (Figure 1): ferns, horsetails, whisk ferns and hornworts given that their gains and losses could add independent further data relevant to the backbone of land plant phylogeny.
The extreme requirement of RNA editing in I. engelmannii mitochondrial RNAs not only to re-establish conserved reading frames in mRNAs but also to reconstitute secondary structures of tRNAs exceeds what has been observed before including the recent estimates of some 1000 editing sites in Cycas taitungensis mitochondrial mRNAs (32). The C-to-U editings hitherto observed in tRNAs of plant organelles were considered functionally essential, yet rare, events, e.g. (61–63). Likewise, such events of C-to-U editing have also been reported in animal mitochondria, e.g. (64). Multiple sites of RNA editing in single mitochondrial tRNA species had been observed as different types of editing: the replacement of nucleotides in tRNA acceptor stems, e.g. in the protist Acanthamoeba castellanii (65), the chytridomycete fungus Spizellomyces punctatus (66), de novo synthesis of 3′ ends in a centipede (67) or the pyrimidine insertional type of editing in slime molds (68).
Given the extraordinary degree of recombination, the presence of chloroplast and nuclear sequence inserts, a trans-splicing group I intron, the extraordinary amounts of RNA editing in mRNAs and, most notably, in hitherto unseen amounts also in tRNAs the I. engelmannii mtDNA once more demonstrates that ‘anything goes’ in mitochondrial genome evolution (69).
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [DFG Kn411/6-1].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are highly grateful to Dr Jeffrey Palmer and co-workers (Bloomington, IN, USA.) for generously making fresh material of Isoetes engelmannii available to us, to Diana Kühn at the Max-Planck-Institut (MPI) für Züchtungsforschung, Cologne, for fosmid clone sorting and filter spotting. We also wish to thank Monika Polsakiewicz for her skillful technical assistance; and Julia Neuwirt, Yesim Kümetepe and Patrick Johner for their early work on lycophyte mtDNA cosmid cloning attempts. Finally, we are very grateful for helpful comments on this manuscript by two anonymous reviewers. Sequences obtained in this study were deposited in GenBank (accession numbers FJ010859, FJ536259, FJ390841, FJ176330, FJ628360).
REFERENCES
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Lavrov DV. Key transitions in animal evolution: a mitochondrial DNA perspective. Integr. Compar. Biol. 2007;47:734–743. doi: 10.1093/icb/icm045. [DOI] [PubMed] [Google Scholar]
- 3.Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu. Rev. Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 4.Lang BF, Burger G, O'Kelly CJ, Cedergren R, Golding GB, Lemieux C, Sankoff D, Turmel M, Gray MW. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 5.Vaidya AB, Akella R, Suplick K. Sequences similar to genes for two mitochondrial proteins and portions of ribosomal RNA in tandemly arrayed 6-kilobase-pair DNA of a malarial parasite. Mol. Biochem. Parasitol. 1989;35:97–107. doi: 10.1016/0166-6851(89)90112-6. [DOI] [PubMed] [Google Scholar]
- 6.Ward BL, Anderson RS, Bendich AJ. The mitochondrial genome is large and variable in a family of plants (Cucurbitaceae) Cell. 1981;25:793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]
- 7.Beagley CT, Okada NA, Wolstenholme DR. Two mitochondrial group I introns in a metazoan, the sea anemone Metridium senile: one intron contains genes for subunits 1 and 3 of NADH dehydrogenase. Proc. Natl Acad. Sci. USA. 1996;93:5619–5623. doi: 10.1073/pnas.93.11.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turmel M, Otis C, Lemieux C. The chloroplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proc. Natl Acad. Sci. USA. 2002;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turmel M, Otis C, Lemieux C. The mitochondrial genome of Chara vulgaris: insights into the mitochondrial DNA architecture of the last common ancestor of green algae and land plants. Plant Cell. 2003;15:1888–1903. doi: 10.1105/tpc.013169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl Acad. Sci. USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern DB, Lonsdale DM. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature. 1982;299:698–702. doi: 10.1038/299698a0. [DOI] [PubMed] [Google Scholar]
- 12.Knoop V, Unseld M, Marienfeld J, Brandt P, Sünkel S, Ullrich H, Brennicke A. copia-, gypsy- and LINE-like retrotransposon fragments in the mitochondrial genome of Arabidopsis thaliana. Genetics. 1996;142:579–585. doi: 10.1093/genetics/142.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaughn JC, Mason MT, Sper-Whitis GL, Kuhlman P, Palmer JD. Fungal origin by horizontal transfer of a plant mitochondrial group I intron in the chimeric CoxI gene of Peperomia. J. Mol. Evol. 1995;41:563–572. doi: 10.1007/BF00175814. [DOI] [PubMed] [Google Scholar]
- 14.Cho YR, Palmer JD. Multiple acquisitions via horizontal transfer of a group I intron in the mitochondrial cox1 gene during evolution of the Araceae family. Mol. Biol. Evol. 1999;16:1155–1165. doi: 10.1093/oxfordjournals.molbev.a026206. [DOI] [PubMed] [Google Scholar]
- 15.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 16.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J. Exp. Bot. 2006;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 17.Kubo T, Mikami T. Organization and variation of angiosperm mitochondrial genome. Phys. Plant. 2007;129:6–13. [Google Scholar]
- 18.Knoop V. The mitochondrial DNA of land plants: peculiarities in phylogenetic perspective. Curr. Genet. 2004;46:123–139. doi: 10.1007/s00294-004-0522-8. [DOI] [PubMed] [Google Scholar]
- 19.Takenaka M, Verbitskiy D, van der Merwe JA, Zehrmann A, Brennicke A. The process of RNA editing in plant mitochondria. Mitochondrion. 2008;8:35–46. doi: 10.1016/j.mito.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Maier RM, Zeltz P, Kössel H, Bonnard G, Gualberto JM, Grienenberger JM. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996;32:343–365. doi: 10.1007/BF00039390. [DOI] [PubMed] [Google Scholar]
- 21.Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V. Plant mitochondrial RNA editing: extreme in hornworts and dividing the liverworts? J. Mol. Evol. 1999;48:303–312. doi: 10.1007/pl00006473. [DOI] [PubMed] [Google Scholar]
- 22.Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J. Mol. Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-r. [DOI] [PubMed] [Google Scholar]
- 23.Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol. Biol. Evol. 2006;24:699–709. doi: 10.1093/molbev/msl198. [DOI] [PubMed] [Google Scholar]
- 24.Oldenburg DJ, Bendich AJ. Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J. Mol. Biol. 2001;310:549–562. doi: 10.1006/jmbi.2001.4783. [DOI] [PubMed] [Google Scholar]
- 25.Unseld M, Marienfeld JR, Brandt P, Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 26.Sugiyama Y, Watase Y, Nagase M, Makita N, Yagura S, Hirai A, Sugiura M. The complete nucleotide sequence and multipartite organization of the tobacco mitochondrial genome: comparative analysis of mitochondrial genomes in higher plants. Mol. Genet. Genom. 2005;272:603–615. doi: 10.1007/s00438-004-1075-8. [DOI] [PubMed] [Google Scholar]
- 27.Ogihara Y, Yamazaki Y, Murai K, Kanno A, Terachi T, Shiina T, Miyashita N, Nasuda S, Nakamura C, Mori N, et al. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005;33:6235–6250. doi: 10.1093/nar/gki925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shedge V, Arrieta-Montiel M, Christensen AC, Mackenzie SA. Plant mitochondrial recombination surveillance requires unusual RecA and MutS homologs. Plant Cell. 2007;19:1251–1264. doi: 10.1105/tpc.106.048355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bendich AJ. Reaching for the ring: the study of mitochondrial genome structure. Curr. Genet. 1993;24:279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- 30.Malek O, Knoop V. Trans-splicing group II introns in plant mitochondria: the complete set of cis-arranged homologs in ferns, fern allies, and a hornwort. RNA. 1998;4:1599–1609. doi: 10.1017/s1355838298981262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Groth-Malonek M, Pruchner D, Grewe F, Knoop V. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol. Biol. Evol. 2005;22:117–125. doi: 10.1093/molbev/msh259. [DOI] [PubMed] [Google Scholar]
- 32.Chaw SM, Chun-Chieh SA, Wang D, Wu YW, Liu SM, Chou TY. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 33.Lohse M, Drechsel O, Bock R. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007;52:267–274. doi: 10.1007/s00294-007-0161-y. [DOI] [PubMed] [Google Scholar]
- 34.Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kempken F, Bolle N, Forner J, Binder S. Transcript end mapping and analysis of RNA editing in plant mitochondria. In: Leister D, Herrmann JM, editors. Mitochondria: Practical Protocols, Vol. 372. Totowa, NJ: Humana Press; 2008. pp. 177–192. [Google Scholar]
- 37.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 38.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl Acad. Sci. USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dombrovska E, Qiu YL. Distribution of introns in the mitochondrial gene nad1 in land plants: phylogenetic and molecular evolutionary implications. Mol. Phylogenet. Evol. 2004;32:246–263. doi: 10.1016/j.ympev.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 41.Saldanha R, Mohr G, Belfort M, Lambowitz AM. Group I and group II introns. FASEB J. 1993;7:15–24. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 42.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 43.Ohta E, Oda K, Yamato K, Nakamura Y, Takemura M, Nozato N, Akashi K, Ohyama K, Michel F. Group I introns in the liverwort mitochondrial genome: the gene coding for subunit 1 of cytochrome oxidase shares five intron positions with its fungal counterparts. Nucleic Acids Res. 1993;21:1297–1305. doi: 10.1093/nar/21.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pryer KM, Schneider H, Smith AR, Cranfill R, Wolf PG, Hunt JS, Sipes SD. Horsetails and ferns are a monophyletic group and the closest living relatives to seed plants. Nature. 2001;409:618–622. doi: 10.1038/35054555. [DOI] [PubMed] [Google Scholar]
- 45.Qiu YL. Phylogeny and evolution of charophytic algae and land plants. J. Syst. Evol. 2008;46:287–306. [Google Scholar]
- 46.Rasmusson AG, Handa H, Moller IM. Plant mitochondria, more unique than ever. Mitochondrion. 2008;8:1–4. doi: 10.1016/j.mito.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Wang D, Wu YW, Shih AC, Wu CS, Wang YN, Chaw SM. Transfer of chloroplast genomic DNA to mitochondrial genome occurred at least 300 MYA. Mol. Biol. Evol. 2007;24:2040–2048. doi: 10.1093/molbev/msm133. [DOI] [PubMed] [Google Scholar]
- 48.Giegé P, Grienenberger JM, Bonnard G. Cytochrome c biogenesis in mitochondria. Mitochondrion. 2008;8:61–73. doi: 10.1016/j.mito.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Allen JW, Jackson AP, Rigden DJ, Willis AC, Ferguson SJ, Ginger ML. Order within a mosaic distribution of mitochondrial c-type cytochrome biogenesis systems? FEBS J. 2008;275:2385–2402. doi: 10.1111/j.1742-4658.2008.06380.x. [DOI] [PubMed] [Google Scholar]
- 50.Koller B, Fromm H, Galun E, Edelman M. Evidence for in vivo trans splicing of pre-mRNAs in tobacco chloroplasts. Cell. 1987;48:111–119. doi: 10.1016/0092-8674(87)90361-8. [DOI] [PubMed] [Google Scholar]
- 51.Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kuck U, Bennoun P, Rochaix JD. Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell. 1988;52:903–913. doi: 10.1016/0092-8674(88)90432-1. [DOI] [PubMed] [Google Scholar]
- 52.Knoop V, Schuster W, Wissinger B, Brennicke A. Trans splicing integrates an exon of 22 nucleotides into the nad5 mRNA in higher plant mitochondria. EMBO J. 1991;10:3483–3493. doi: 10.1002/j.1460-2075.1991.tb04912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wissinger B, Schuster W, Brennicke A. Trans splicing in Oenothera mitochondria: nad1 mRNAs are edited in exon and trans-splicing group II intron sequences. Cell. 1991;65:473–482. doi: 10.1016/0092-8674(91)90465-b. [DOI] [PubMed] [Google Scholar]
- 54.Chapdelaine Y, Bonen L. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a trans-splicing model for this gene-in-pieces. Cell. 1991;65:465–472. doi: 10.1016/0092-8674(91)90464-a. [DOI] [PubMed] [Google Scholar]
- 55.Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol. Biol. Evol. 2002;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- 56.Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marande W, Burger G. Mitochondrial DNA as a genomic jigsaw puzzle. Science. 2007;318:415. doi: 10.1126/science.1148033. [DOI] [PubMed] [Google Scholar]
- 58.Randau L, Münch R, Hohn MJ, Jahn D, Söll D. Nanoarchaeum equitans creates functional tRNAs from separate genes for their 5′- and 3′-halves. Nature. 2005;433:537–541. doi: 10.1038/nature03233. [DOI] [PubMed] [Google Scholar]
- 59.Randau L, Soll D. Transfer RNA genes in pieces. EMBO Rep. 2008;9:623–628. doi: 10.1038/embor.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science. 2007;318:450–453. doi: 10.1126/science.1145718. [DOI] [PubMed] [Google Scholar]
- 61.Binder S, Marchfelder A, Brennicke A. RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- 62.Maréchal-Drouard L, Ramamonjisoa D, Cosset A, Weil JH, Dietrich A. Editing corrects mispairing in the acceptor stem of bean and potato mitochondrial phenylalanine transfer RNAs. Nucleic Acids Res. 1993;21:4909–4914. doi: 10.1093/nar/21.21.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miyata Y, Sugita C, Maruyama K, Sugita M. RNA editing in the anticodon of tRNA(Leu) (CAA) occurs before group I intron splicing in plastids of a moss Takakia lepidozioides S. Hatt. & Inoue. Plant Biol (Stuttg) 2008;10:250–255. doi: 10.1111/j.1438-8677.2007.00027.x. [DOI] [PubMed] [Google Scholar]
- 64.Janke A, Pääbo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lonergan KM, Gray MW. Editing of transfer RNAs in Acanthamoeba castellanii mitochondria. Science. 1993;259:812–816. doi: 10.1126/science.8430334. [DOI] [PubMed] [Google Scholar]
- 66.Laforest MJ, Roewer I, Lang BF. Mitochondrial tRNAs in the lower fungus Spizellomyces punctatus: tRNA editing and UAG ‘stop’ codons recognized as leucine. Nucleic Acids Res. 1997;25:626–632. doi: 10.1093/nar/25.3.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lavrov DV, Brown WM, Boore JL. A novel type of RNA editing occurs in the mitochondrial tRNAs of the centipede Lithobius forficatus. Proc. Natl Acad. Sci USA. 2000;97:13738–13742. doi: 10.1073/pnas.250402997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antes T, Costandy H, Mahendran R, Spottswood M, Miller D. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell Biol. 1998;18:7521–7527. doi: 10.1128/mcb.18.12.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.