Abstract
The CUG-binding protein 1 (CUG-BP1) is a member of the CUG-BP1 and ETR-like factors (CELF) family or the Bruno-like family and is involved in the control of splicing, translation and mRNA degradation. Several target RNA sequences of CUG-BP1 have been predicted, such as the CUG triplet repeat, the GU-rich sequences and the AU-rich element of nuclear pre-mRNAs and/or cytoplasmic mRNA. CUG-BP1 has three RNA-recognition motifs (RRMs), among which the third RRM (RRM3) can bind to the target RNAs on its own. In this study, we solved the solution structure of the CUG-BP1 RRM3 by hetero-nuclear NMR spectroscopy. The CUG-BP1 RRM3 exhibited a noncanonical RRM fold, with the four-stranded β-sheet surface tightly associated with the N-terminal extension. Furthermore, we determined the solution structure of the CUG-BP1 RRM3 in the complex with (UG)3 RNA, and discovered that the UGU trinucleotide is specifically recognized through extensive stacking interactions and hydrogen bonds within the pocket formed by the β-sheet surface and the N-terminal extension. This study revealed the unique mechanism that enables the CUG-BP1 RRM3 to discriminate the short RNA segment from other sequences, thus providing the molecular basis for the comprehension of the role of the RRM3s in the CELF/Bruno-like family.
INTRODUCTION
The CUG-binding protein 1 (CUG-BP1) was first identified as a protein that binds to the CUG triplet repeat sequence in the 3′-untranslated region (UTR) of the pre-mRNA encoding the mytonin protein kinase (Mt-PK), which was suggested to be associated with myotonic dystrophy (1). Subsequently, there have been a number of reports that CUG-BP1 and its homologs bind to specific RNA elements and play various important roles in the post-transcriptional processing of mRNA, such as alternative splicing, translational control and the regulation of mRNA decay.
Specifically, in the nucleus, CUG-BP1 binds to the CUG repeats in the cardiac troponin T (cTNT) pre-mRNA to regulate the alternative splicing of its pre-mRNA (2). The zebrafish homolog is involved in the alternative splicing of the α-actinin pre-mRNA upon binding to the Bruno responsive element, which is also referred to as the repeat of uridine and purine elements (UREs) (3). In the cytoplasm, CUG-BP1 binds to the CUG/CCG sequence in the 5′ region of the mRNA encoding the CCAAT/enhancer binding protein β (C/EBPβ) and regulates the translation to produce a low-molecular-weight isomer of C/EBPβ (4). CUG-BP1 also reportedly binds to the class III AU-rich element in the TNFα and c-jun mRNAs, as well as to the GC-rich sequence in the 5′ UTR of the p21 mRNA, and increases the expression levels of their gene-products in the cells (5). Moreover, several reports pointed out the importance of CUG-BP1 for the control of mRNA deadenylation and degradation. First, in the Xenopus oocyte, the embryo deadenylation element [EDEN; U(A/G) repeat in Xenopus laevis maternal mRNAs] was identified as the target sequence of the EDEN-binding protein (EDEN-BP), the X. laevis ortholog of CUG-BP1, and the binding of EDEN-BP to EDEN accelerates the deadenylation of the Eg5 mRNA (6). Further investigations revealed that CUG-BP1 mediates the deadenylation and the decay of the mRNAs, through interactions with the deadenylation enzyme upon binding to a GU-rich element (GRE) in the 3′ UTR of the TNFR 1B, c-jun, junB, TNFα and c-fos mRNAs (7–9).
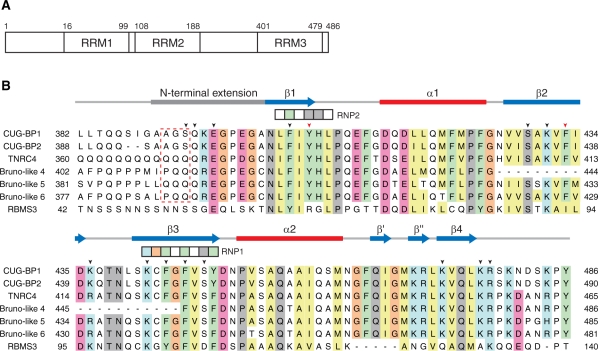
In the human genome, six proteins have been identified as homologs of CUG-BP1: CUG-BP1, CUG-BP2, CELF3, CELF4, CELF5 and CELF6. They form a protein group referred to as the CUG-BP1 and ETR-like factors (CELF)/Bruno-like family. All of the family members have three RNA recognition motifs (RRM1–3): two consecutive N-terminal RRMs and a single C-terminal RRM (Figure 1A) (5).
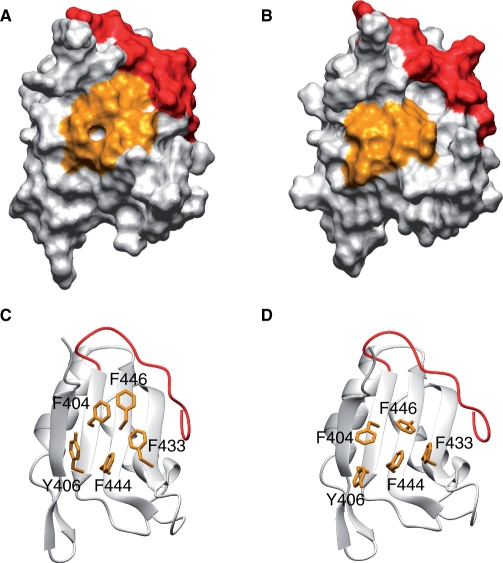
Figure 1.
Primary structures of the third RRMs of (CELF)/Bruno-like family members and the first RRM of RBMS3. (A) Schematic diagram of the human CUG-BP1 protein. CUG-BP1 possesses three RNA-recognition motifs (RRMs) (10,11). (B) Multiple sequence alignment of the CUG-BP1 and ETR-like factors (CELF)/Bruno-like family. The RRM3 domains of CUG-BP1 (Q92879), CUG-BP2 (NP_001020247), trinucleotide repeat containing 4 (TNRC4, NP_009116), Bruno-like 4 (NP_064565), Bruno-like 5 (NP_068757) and Bruno-like 6 (NP_443072) were aligned using ClustalX (48). Secondary structure elements are depicted with blue arrows (β-sheet) and red bars (α-helix) above the sequence alignment. The conserved signature sequences of RNP1 and RNP2 are indicated by the boxes. The arrowheads indicate the residues that play important roles in the RNA-binding. Two aromatic residues (Tyr406 on β1 and Phe433 on β2), which are characteristic of RRM3, are indicated by the red arrowheads. The red dashed box indicates the residues mutated for the ITC experiments. In addition, the amino acid sequence of the single-stranded-interacting protein (RBMS3) is compared with the CELF/Bruno-like family members.
RRMs have been found in many kinds of eukaryotic RNA-binding proteins. For instance, 901 RRM-containing human proteins are included in the Pfam release 23.0 database (10,11). RRMs play important roles in sequence-specific RNA binding (12–14). In some cases, a single RRM can bind to a structured or a single-stranded RNA in a sequence-specific manner, for example in U1A (15), U2B′′/U2A′ (16), Fox-1 (17) and SRp20 (18). On the other hand, two consecutive RRMs often function cooperatively for the recognition of the target RNA, such as in Sxl (19), polyadenylate-binding protein (PABP) (20), polypyrimidine tract binding protein (PTBP) (21) and U2AF65 (22). Furthermore, some RRM-containing proteins have multiple RNA-binding modules and recognize their target RNA molecules specifically by the combination of these RNA-binding modules. For instance, the HuC and HuD proteins, which are members of the Elav-type RRM protein family, have the same domain architecture as CUG-BP1. Their N-terminal consecutive RRMs bind cooperatively to the AU-rich elements in the 3′ UTR (23), and the C-terminal RRM of HuC reportedly binds to poly(A) (24).
The RRM consists of a four-stranded anti-parallel β-sheet packed against two α-helices (βαββαβ topology) and has two conserved motifs, referred to as RNP2 and RNP1, which correspond to the first and third β-strands, respectively. Most RRMs interact with their target RNA molecules on their four-stranded β-sheet surface. Specifically, two well-conserved aromatic amino acids, which are aligned next to each other on RNP1 and RNP2, stack with the bases of nucleic acids. Further examination of the RRM–RNA recognition also revealed that in a single RRM, the C-terminal extension of the RRM body frequently plays an important role in increasing the binding strength to RNA (25). On the other hand, in the case of the RNA recognition by tandem RRMs connected by a short interdomain linker, the two RRMs cooperatively provide a large RNA-binding surface for strong binding to the target RNA molecule (25). As described above, there have been several structural reports about RRM–RNA interactions. However, the presently available information was not sufficient to allow the prediction of the target RNA sequences for putative RRMs or to understand the diverse RNA recognition modes of the RRMs. Therefore, further structural information about the RRM–RNA complexes was desired.
In the case of CUG-BP1, several types of RNA elements were predicted as the target RNA sequences, as described above. Taken together, the CUG, UG and UA repeats are considered as the fundamental RNA-binding elements of CUG-BP1 thus far. Among the three RRMs of CUG-BP1, the two consecutive N-terminal RRMs (RRM1 and RRM2) cooperatively bind to CUG-repeats (26,27) and those of the zebrafish CUG-BP1 homolog bind to URE-repeats (26,27). However, neither RRM1 nor RRM2 is able to bind to the RNA repeats on its own (26,27). On the other hand, the C-terminal RRM3 could bind to the UG repeat by itself (26); however, unlike RRM1 and RRM2, it does not target the CUG repeats (27). Thus, like the HuC protein, the CELF/Bruno-like family members have multiple RNA-binding modules (RRM1–2 and RRM3) that exhibit distinct preferences for the primary and tertiary structures of RNA molecules. In a variety of biological contexts, CUG-BP1 could recognize its respective target RNAs by utilizing various combinations of these binding modules.
The putative binding sequences for CUG-BP1 (the CUG, UG and UA repeats) are similar to each other. Therefore, in order to elucidate the versatility of CUG-BP1, more precise information about the RNA recognition mechanism of the RNA-binding modules in the CUG-BP1 protein is necessary. However, the mechanism by which CUG-BP1 discriminates between these RNA sequences has remained unclear. Among the three RRMs, RRM3 is the evolutionarily best conserved within the CELF/Bruno-like family members (Figure 1B) (5). Thus, it is conceivable that the RRM3 of the CELF/Bruno-like family members assumes the principal role for the function of the protein family. Therefore, the elucidation of the sequence preference and the molecular mechanism of the RNA binding of CUG-BP1 RRM3 will clarify the regulation of the CELF/Bruno-like family members.
Here, we determined solution structure of CUG-BP1 RRM3, and revealed by NMR chemical shift perturbation analysis that it prefers UG repeat sequences rather than CUG and UA repeat sequences. Furthermore, we determined the solution structure of CUG-BP1 RRM3 in complex with (UG)3 RNA, and elucidated the mechanism for the recognition of the (UG)3 RNA by CUG-BP1 RRM3. Unlike the canonical RNA recognition mode by a single RRM, the N-terminal extension plays an important role in the RNA recognition by the CUG-BP1 RRM3. Our study, therefore, provides significant insight into the sequence-specific RNA recognition mode of the CUG-BP1 and CELF/Bruno-like family proteins, as well as a comprehensive understanding of the RNA recognition mode mediated by RRM folds.
MATERIALS AND METHODS
Protein expression and purification
The DNA encoding the third RRM domain (Leu382–Lys480) of human CUG-BP1 (SwissProt accession no. Q92879) was subcloned by PCR from the human full-length cDNA clone. This DNA fragment was cloned into the expression vector pCR2.1 (Invitrogen), as a fusion with an N-terminal native His affinity tag and a Tobacco Etch Virus (TEV) protease cleavage site. The 13C, 15N labeled fusion protein was synthesized by a cell-free protein expression system (28–30). The lysate was clarified by centrifugation at 16 000 g for 20 min and filtration with a 0.45-mm membrane (Millipore). The clarified lysate was applied to a 5-ml His Trap 5ml column (GE Healthcare Biosciences), which was eluted with a 12–500-mM imidazole gradient, and the tag was removed by an incubation with TEV protease for 1 h at 30°C. The tag-free CUG-BP1 RRM3 was further purified by HiTrap Q and HiTrap SP column chromatography (GE Healthcare).
In order to purify the sample for the biochemical experiments, such as the isothermal titration calorimetry (ITC) measurements, we used an in vivo protein production system. For the system, the amplified DNA fragment was cloned into the expression vector pET-15b (Novagen), as a fusion with an N-terminal native His affinity tag and a TEV protease cleavage site. Mutant proteins, in which the residues Ala391-Gly-Ser393 in the His-tagged CUGBP-1 RRM3 (384–480) were replaced with Pro-Gln-Gln or Gln-Gln-Gln, were generated by PCR, using 28–30-mer primers spanning the site of the desired mutation, as described (31). Mutations were confirmed by sequencing.
The fusion protein was overexpressed in Escherichia coli strain BL21 (DE3) cells using 2 × YT medium supplemented with 50 mg/l ampicillin. The harvested culture was lysed by sonication in 20 mM Tris–HCl buffer (pH 7.0), containing 300 mM NaCl, 20 mM imidazole, 1 mM DTT, 1 mM phenylmethanesulfonylfluoride (PMSF) and protease inhibitor cocktail for general use (Nacalai Tesque). The lysate was applied to a Ni2+–NTA SuperFlow column (Qiagen), which was eluted with a 20–250-mM imidazole gradient, and the tag was removed by an overnight incubation with TEV protease at room temperature. Each of the tag-free wild-type and mutant CUG-PB1 RRM3 proteins was further purified by RESOURCE S column chromatography (GE Healthcare).
Nuclear magnetic resonance (NMR) spectroscopy
For NMR measurements, the samples were concentrated to 1.0–1.4 mM in 20 mM d-Tris–HCl buffer (pH 7.0), containing 100 mM NaCl, 1 mM 1,4-DL-dithiothreitol-d10 (d-DTT) and 0.02% NaN3 (in 90% H2O/10% D2O), using an Amicon Ultra-15 filter (5000 MWCO, Millipore). NMR experiments were performed at 25°C for the RNA-free form and at 15°C for the RNA-bound form on Bruker 700 and 800-MHz spectrometers (Bruker AV700 and Bruker AV800). The 1H, 15N, and 13C chemical shifts were referenced relative to the frequency of the 2H lock resonance of water. Backbone and side-chain assignments of CUG-BP1 RRM3 were obtained by using a combination of standard triple resonance experiments (32,33). 2D [1H, 15N]-HSQC and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra were used for the 1H, 15N and 13C assignments of the protein backbone. The 1H and 13C assignments of the nonaromatic side chains, including all prolines, were obtained using 2D [1H, 13C]-HSQC, and 3D HBHA(CO)NH, H(CCCO)NH, (H)CC(CO)NH, HCCH-COSY, HCCH-TOCSY and (H)CCH-TOCSY spectra. Assignments were checked for consistency with 3D 15N-edited [1H, 1H]-NOESY and 13C-edited [1H, 1H]-NOESY spectra. The 1H and 13C spin systems of the aromatic rings of Phe, Trp, His and Tyr were identified using 3D HCCH-COSY and HCCH-TOCSY experiments, and 3D 13C-edited [1H, 1H]-NOESY was used for the sequence-specific resonance assignment of the aromatic side chains. 3D HNHA, HN(CO)HB and HNHB spectra were used for the dihedral angle restraints for ϕ and χ1, respectively. NOESY spectra were recorded with mixing times of 80 and 150 ms. For the assignments of the RNA molecules, 2D filtered-NOESY (mixing times of 80 and 150 ms) and 2D filtered-TOCSY (mixing time of 30 ms) spectra were used. The sugar ring conformation was identified by the intensity of the cross peaks between H1′ and H2′ in the 2D TOCSY spectra. U1, G2, U3 and U5 were in the C2′–endo conformation. The NMR data were processed using NMRPipe (34). Analyses of the processed data were performed with the programs NMRView (35) and KUJIRA (36).
For the amide chemical-shift titration experiments, the RNA oligonucleotides [5′-UGUGUG-3′], [5′-UAUAUA-3′] and [5′-CUGCUG-3′] (Dharmacon) were dissolved in 20 mM d-Tris–HCl buffer (pH 7.0), containing 100 mM NaCl and 1 mM d-DTT, to make a 6-mM solution. 2D [1H, 15N]-HSQC spectra were recorded while increasing the concentration of the RNA relative to the CUG-BP1 RRM3 solutions (200 µM) to a final 1:2 ratio of CUG-BP1 RRM3:RNA.
The measurements of the nitrogen relaxation times, T1 and T2, and the proton–nitrogen heteronuclear NOEs were performed on a Bruker 600 MHz spectrometer equipped with a cryo-probe (Bruker AV 600) at 25°C, using 15N, 13C-labeled CUG-BP1 RRM3 at a concentration of 200 µM (37). Eight different values for the relaxation delay were recorded for the 15N T1 (T1 delays 5, 65, 145, 246, 366, 527, 757 and 1148 ms) and 15N T2 (T2 delays 32, 48, 64, 80, 96, 112, 128 and 144 ms) relaxation experiments. The 15N T1 and 15N T2 values were extracted using a curve-fitting subroutine included in the Sparky program (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco, CA, USA). The proton–nitrogen heteronuclear NOE values were calculated as the ratio between the cross-peak intensities with and without 1H saturation. The errors were estimated from the root mean square of the baseline noise in the two spectra (37).
Structure calculations
The three-dimensional structures of the free and complex forms of CUG-BP1 RRM3 were determined by combined automated NOESY cross-peak assignment (32,33,38) and structure calculations with torsion angle dynamics (39) implemented in the program CYANA 2.1 (40). Dihedral angle restraints for ϕ and ψ were obtained from the main-chain and 13Cβ chemical-shift values using the program TALOS (41), and by analyzing the NOESY and HNHA spectra. The χ1 angles of the protein side chains were estimated by inspecting the pattern of the inter- and intra-NOE intensities in conjunction with the 3D HNHB and HN(CO)HB spectra (42). In the RNA-free form, the results obtained from the 3D HNHB and HN(CO)HB spectra generally agreed with the pattern of the inter- and intra-NOE intensities. In the RNA-bound form, however, the qualities of the 3D HNHB and HN(CO)HB spectra were not sufficient. Therefore, the information for the χ1 angles was mainly obtained by the estimation of the pattern of the inter- and intra-NOE intensities, according to the method described by Powers et al. (42). For the determination of the three-dimensional structures of the RNA molecules, the intermolecular protein–RNA NOEs were manually assigned using 2D NOESY spectra with mixing times of 80 and 150 ms. The distance bounds for the protein–RNA NOEs were set as follows: the NOEs derived from the RNA molecule in the 2D NOESY spectra with a mixing time of 80 ms were divided into two groups with upper distance bounds of 3.5 and 5.0 Å, according to their intensity. Upper distance bounds of 6.0 Å were applied for the intermolecular NOEs that could only be identified from 2D NOESY spectra with a mixing time of 150 ms. In total, 90 intermolecular NOEs between CUG-BP1 RRM3 and RNA were used for the structure calculations.
The structure calculations started from 200 randomized conformers and used the standard CYANA simulated annealing schedule (39), with 40 000 torsion angle dynamics steps per conformer. The 40 conformers with the lowest final CYANA target function values were subjected to restrained energy minimization in implicit solvent (generalized born solvation model) with the program AMBER9, using the AMBER 2003 force field (43). The restrained energy refinement consisted of three steps: an initial 500 steps of energy minimization, simulated annealing by 20 ps of Cartesian space molecular dynamics simulation (Supplementary Figure S1) and a final 2000 steps of energy minimization. Force constants of 32 kcal mol−1 Å−2 for distance restraints, 60/100 kcal mol–1 rad–2 for torsion angle restraints (protein/RNA) and 50 kcal mol–1 rad–2 for ω angles were used in the simulated annealing. The 20 conformers that were most consistent with the experimental restraints were then used for further analyses. PROCHECK-NMR (44) and MOLMOL (45) were used to validate and to visualize the final structures, respectively. The atomic coordinates for the ensemble of 20 energy-refined NMR conformers, representing the solution structures of CUG-BP1 RRM3 and the CUG–BP1 RRM3–RNA (UG)3 complex, have been deposited in the Protein Data Bank, with the accession codes 2RQ4 and 2RQC, respectively.
ITC measurements
ITC measurements were performed at 25°C by using a Microcal (Amherst, MA) VP-ITC calorimeter. Samples were buffered with 20 mM Tris (pH 7.0), containing 100 mM NaCl and 1 mM DTT, and were thoroughly degassed before use. At first, 2.0-ml solutions of the 20 and 50 μM wild-type CUG-BP1 RRM3 were prepared in the cell chamber. Then, a 20-fold higher concentration of five different hexameric RNAs, [5′-(UGUGUG)-3′], [5′-(UAUAUA)-3′], [5′-(CUGCUG)-3′], [5′-(CGUGUG)-3′] and [5′-(CGUAUG)-3′], were injected into the wild-type protein solution. In the same way, 2.0-ml solutions of the 20 and 50 μM CUG-BP1 RRM3 variants were prepared in the cell chamber, and the [5′-(UGUGUG)-3′] RNA was injected into the two variant protein solutions. Except for the wild-type protein plus the [5′-(CUGCUG)-3′] sequence, the heat generated due to dilution of the titrants was very small, and thus was ignored for the analysis. The data were analyzed with the Microcal ORIGIN software, using a binding model that assumes a single site of interaction.
RESULTS
Solution structure of the CUG-BP1 RRM3 domain
The 15N, 13C-labeled CUG-BP1 RRM3 (residues Leu382–Lys480) was prepared by a cell-free protein expression system (see ‘Materials and Methods’ section). In total, 99.4% of the main-chain and 92.0% of the side-chain atoms of residues 382–487 were assigned using multidimensional heteronuclear NMR spectroscopy (see ‘Materials and Methods’ section). We determined the solution structure of CUG-BP1 RRM3 on the basis of 1760 1H–1H distance restraints from nuclear Overhauser effect spectroscopy (NOESY) and 98 torsion angle restraints (Table 1). Among the 200 independently calculated structures, the 40 conformers with the lowest CYANA target function values were refined by restrained energy minimization (see ‘Materials and Methods’ section). The 20 conformers that were most consistent with the experimental restraints were used for further analyses.
Table 1.
Summary of determination and refinement statistics for the free and (UG)3-bound forms of CUG-BP1
| Free CUG-BP1 | CUG–BP1 RRM– (UG)3 complex |
||
|---|---|---|---|
| RRM | RNA | ||
| NMR constraints | |||
| Distance restraints | |||
| Total NOEs | 1760 | 1419 | 40 |
| Intra-residue | 473 | 408 | 31 |
| Inter-residue | |||
| Sequential (|i−j| = 1) | 394 | 320 | 9 |
| Medium-range (1 < |i−j| < 5) | 277 | 218 | |
| Long-range (|i−j| ≥ 5) | 652 | 473 | |
| Hydrogen bond restraintsa | 15 | 23 | |
| Protein–RNA intermolecular | 90 | ||
| Dihedral angle restraints | |||
| ϕ and ψ | 98b | 24c | |
| χ angle | 35 | 38 | |
| Sugar puckering | 4 | ||
| Structure statistics (40 structures) | |||
| CYANA target function (Å2) | 0.38 ± 0.02 | 0.44 ± 0.02 | |
| Residual NOE violations | |||
| Number >0.10 Å | 1 | 1 | |
| Maximum (Å) | 0.11 | 0.10 | |
| Residual dihedral angle violations | |||
| Number >5.0° | 0 | 0 | |
| Maximum (°) | 3.68 | 3.53 | |
| Energies of AMBER calculation (kcal/mol) | |||
| Mean AMBER energy | −3067.36 | −4285.54 | |
| Mean restraint violation energy | 3.851 | 5.055 | |
| Ramachandran plot statistics (%) | |||
| Residues in most favored regions | 86.6 | 85.4 | |
| Residues in additionally allowed regions | 12.6 | 13.2 | |
| Residues in generously allowed regions | 0.4 | 1.1 | |
| Residues in disallowed regions | 0.5 | 0.3 | |
| Average R.M.S.D. to mean structure (Å) | |||
| Protein backboned | 0.296 | 0.312 | |
| Protein heavy atomsd | 0.818 | 0.734 | |
| RNA heavy atomsd | 0.841 | ||
| Complex heavy atomsd | 0.814 | ||
aOnly used in CYANA calculation.
bFrom TALOS (41).
cFrom HNHA experiment.
dFor the calculated residues, the protein was Ala390–Phe433 and Phe444–Arg478, and the RNA was U1–G6.
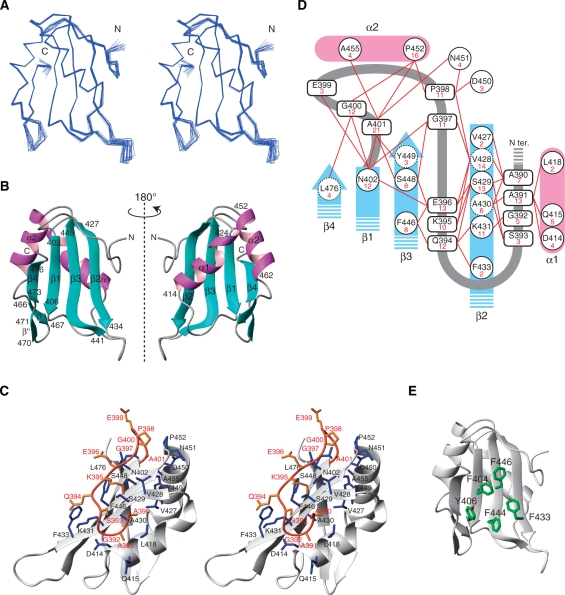
The segment spanning residues 402–476 adopts an RRM fold (βαββαβ) with a four-stranded anti-parallel β-sheet composed of residues 402–406 (β1), 427–434 (β2), 441–449 (β3) and 473–476 (β4) (Figure 2A and B). The β2 and β3 strands form an extra paired strand with a kink at Cys443 on β3 (Figure 2B and Supplementary Figure S2). Helix 1 (α1, 414–424) and helix 2 (α2, 452–462) connect β1–β2 and β3–β4, respectively (Figure 2B). In addition to the β1, β2, β3 and β4 strands, a short β-hairpin was identified between α2 and β4 (β′, Gln466-Ile467; β′′, Lys470–Arg471) (Figure 2B).
Figure 2.
Solution structure of the third RRM domain of CUG-BP1. (A) The superimposed 20 conformers of CUG-BP1 RRM3 (Gly389-Arg478). Blue lines represent Cα traces (stereo view). (B) Ribbon representation of CUG-BP1 RRM3. The secondary structure elements and the sequence numbering are indicated. (C) Stereo view of a ribbon diagram of the interactions between the N-terminal extension and the RRM core. The N-terminal extension and the RRM are colored orange and white, respectively. Residues involved in the interaction are colored orange in the N-terminal extension and blue in the RRM. (D) Summary of the NOEs between the N-terminal extension and the RRM. The numbers of NOEs are indicated in red. (E) Aromatic residues on the β-sheet surface are represented in green (carbon) and red (oxygen).
The N-terminal extension of the CUG-BP1 RRM3 (residues 390–401) intimately interacts with the β-sheet surface and traverses its central region (Figure 2C). The N-terminal two residues, Ala390 and Ala391, are accommodated between the α1 helix and the β2 strand (Figure 2C). The region spanning residues Gly392–Gly397 adopts an extended form associated with the amino acid residues on the β-sheet surface (Asn402 on β1; Val428, Ser429, Ala430, Lys431 and Phe433 on β2; Phe446 and Ser448 on β3). Successively, the Pro398–Ala401 residues form a kink connecting the N-terminal extension with the RRM core, where it is associated with the amino acid residues Asn402 on β1, Val428 on β2, Ser448 and Tyr449 on β3, Asp450 and Asn451 in the β3-α2 loop, Pro452 and Ala455 in α2 and Leu476 on β4. As shown in Figure 2D, all of these interactions have been validated by the detection of the corresponding NOEs. The NMR dynamics analysis also indicated that the structure of this N-terminal extension is as rigid as the RRM core (Figure 3). To the best of our knowledge, this is the first report showing that the N-terminal extension preceding the RRM core covers the β-sheet surface and is involved in the formation of the RRM structure.
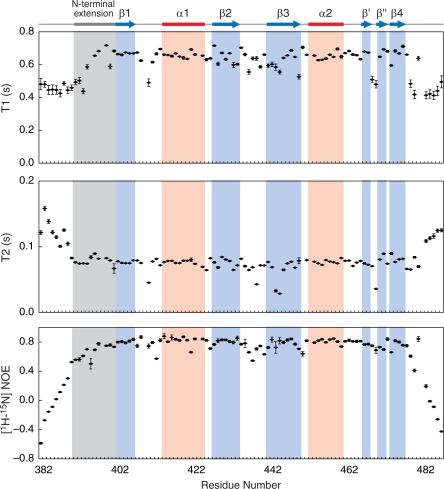
Figure 3.
Dynamics of the CUG-BP1 RRM3. Residues for which resonances disappeared are not shown (T1: K394, T2: D449). The T2 and heteronuclear NOE values of the N-terminal extension (averages of 0.08 s and 0.70 s, respectively) and the RRM core, indicated by the solid blue line (averages of 0.07 s and 0.78 s, respectively), were significantly (P-value of the t-test < 0.01) smaller (T2) or larger (heteronuclear NOE) than those from outside this region (averages of 0.10 s and 0.22 s for residues 383–389 and 477–486, respectively). In addition, the T1 values of the residues 394–401 in the N-terminal extension and in the RRM core (averages of 0.65 s and 0.64 s, respectively) were also significantly (P-value of the t-test < 0.01) larger than those of the rest of the molecule (average of 0.47 s for residues 383–393 and 477–486, respectively). Therefore, not only the RRM core but also the N-terminal extension is more rigid than the N- and C-terminal regions.
Two well-conserved amino-acid sequences that were identified in the RRMs are referred to as RNP1 and RNP2, which correspond to the β3 and β1 strands, respectively (Figure 1B). In the case of CUG-BP1 RRM3, five aromatic residues are located on the β-sheet surface: Phe404 and Tyr406 on β1, Phe433 on β2 as well as Phe444 and Phe446 on β3. Among these, Phe404, Phe444 and Phe446 are the well-conserved aromatic amino acid residues in RNP1 and RNP2 in the RRMs. However, the χ1 angle of Phe404 is –60° and that of Phe446 is 180° in the RNA-free form, which are different from those of the corresponding aromatic amino acid residues in the canonical RRMs (180° and –60°, respectively). On the other hand, Tyr406 and Phe433 are characteristic of the CUG-BP1 RRM3. They form a hydrophobic patch on the β-sheet surface (Figure 2E). Importantly, Phe446 interacts with Gln394 and Glu396 on the N-terminal extension (Figure 2D). In the canonical RRM fold, the aromatic amino acids corresponding to Phe404 and Phe446 are usually involved in stacking interactions with the base moieties of the RNA molecule.
Target sequence preference of the CUG-BP1 RRM3
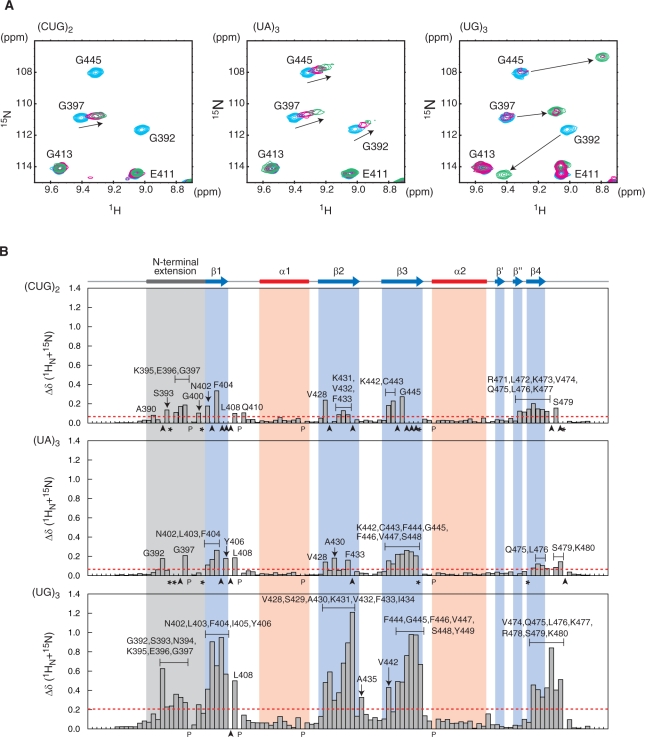
To clarify the RNA sequence preference of the CUG-BP1 RRM3, on the basis of the previous reports we selected three hexanucleotide RNAs, (UG)3, (UA)3 and (CUG)2, and examined their effects on the chemical-shift values for the main-chain 1H-15N resonances of the CUG-BP1 RRM3. In the cases of (UA)3 and (CUG)2, according to the increase in the RNA molar ratios (range from 1:0 to 1:2), some of these resonances shifted in a continuous manner (Figure 4A), indicating that for these RNA sequences, the exchange between the RNA-bound and RNA-free forms is fast on the NMR timescale. As shown in Figure 4B, some of the resonances originating from the residues located on the β-sheet surface and the N-terminal extension were significantly affected by the presence of the (CUG)2 and (UA)3 RNAs. Eleven residues (Gly397, Asn402, Phe404, Leu408, Val428, Phe433, Lys442, Cys443, Gly445, Gln475 and Leu476) were commonly affected by both of these RNAs. In addition, 14 residues (Ala390, Ser393, Lys395, Glu396, Gly400, Gln410, Lys431, Val432, Arg471, Leu472, Lys473, Val474, Lys477 and Ser479) were specifically affected by (CUG)2 and 10 residues (Gly392, Leu403, Tyr406, Ala430, Phe444, Phe446-Ser448, Ser479 and Lys480) were specifically affected by (UA)3. These results suggest that there may be a common key recognition mode for (CUG)2 and (UA)3, as well as a mechanism to distinguish between these two sequences.
Figure 4.
NMR chemical-shift perturbation of CUG-BP1 RRM3 upon RNA binding. (A) Close-up views of the 1H-15N HSQC spectrum of CUG-BP1 RRM3, showing selected amide shift changes in the absence (cyan) and presence (ratio of CUG-BP1 RRM3:RNA = 1:0.4, red, and 1:1, green) of the RNAs [5′-CUGCUG-3′] (left), [5′-UAUAUA-3′] (center) and [5′-UGUGUG-3′] (right). (B) Quantification of the chemical-shift perturbation values of CUG-BP1 RRM3 upon binding to RNAs (ratio protein:RNA = 1:2). The perturbation values were obtained from the [1H,15N] HSQC spectrum. The absolute values of the chemical-shift change Δδ(15N + 1HN) were calculated as follows: Δδ(15N + 1HN) = ((δ15N/6.5)2 + δ1H2)1/2. The baseline of the amide perturbation was defined as the average of the smallest 70% for (UA)3 and (CUG)2 and 65% for (UG)3. The perturbation values greater than the baseline (i.e. 0.02, 0.02 and 0.07 p.p.m., respectively) plus three times the standard deviation of the baseline (i.e. 0.04, 0.04 and 0.13 p.p.m., respectively) were considered as significant perturbations (i.e. from above, the significant levels are 0.06, 0.06 and 0.20 p.p.m., respectively, indicated by red dashed lines). Black letters indicate amino acid residues with significant chemical shift changes. The residues with resonances that disappeared after the addition of the RNA are indicated by arrowheads. The residues with resonances that could not be assigned after the addition of the RNA are indicated by asterisks.
On the other hand, the effect of (UG)3 on the 1H-15N HSQC spectrum was strikingly different from those of (CUG)2 and (UA)3. Some of the main-chain 1H-15N resonances of the free form gradually disappeared, and correspondingly, new resonances of the bound form appeared. This indicated that the exchange between the RNA-bound and RNA-free forms was slow on the NMR time scale (Figure 4A). Almost all of the crosspeaks from the residues of the β-sheet surface (β1: Asn402, Leu403, Phe404, Ile405 and Tyr406; β2: Val428, Ser429, ALA430, Lys431, Val432, Phe433 and Ile434, β3: Lys442, Phe444, Gly445, Phe446, Val447, Ser448 and Tyr449; β4: Val474, Gln475 and Leu476) were significantly affected upon RNA binding (Figure 4B). Moreover, the crosspeaks originating from the N-terminal extension (Gly392, Ser393, Gln394, Lys395, Glu396 and Gly397), the β1-α1 loop (Leu408), the β2-β3 loop (Ala435) and the C-terminal region (Lys477, Glu478, Leu479 and Lys480) were also affected (Figure 4B). Among the 11 residues commonly affected by the (CUG)2 and (UA)3 RNAs, nine residues (Asn402, Phe404, Leu408, Val428, Phe433, Lys442, Gly445, Gln475 and Leu476) were also affected by the (UG)3 RNA. The quantitative analysis of the perturbation values clearly indicated that the CUG-BP RRM3 binds much more tightly to the (UG)3 RNA than to the (CUG)2 and (UA)3 RNAs (Figure 4B). Taken together, these data suggest that the (UG)3 RNA is the most preferred target RNA for the CUG-BP1 RRM3, among the sequences examined.
Solution structure of the CUG-BP1 RRM3–(UG)3 RNA complex
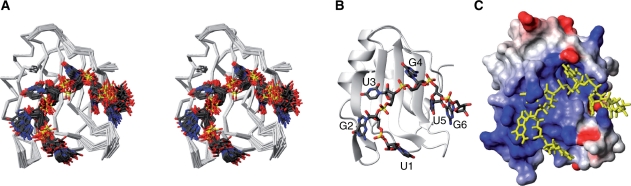
The NMR chemical-shift perturbation analyses demonstrated that the CUG-BP1 RRM3 prefers the (UG)3 RNA. Therefore, we determined the solution structure of CUG-BP1 RRM3 in complex with the (UG)3 RNA (Figure 5). Using multidimensional heteronuclear NMR spectroscopy, 99.4% of the main-chain and 92.0% of the side-chain resonances of residues 382–487 of CUG-BP1 were assigned, as well as 82.6% of the hydrogen atoms in the (UG)3 RNA molecule. The solution structure of the complex was determined using 1549 1H–1H distance restraints from the NOESY spectra, including 90 intermolecular and 40 intra-RNA distance restraints (Table 1 and Supplementary Figure S3). Among the 200 independently calculated structures, the 40 conformers with the lowest CYANA target functions were refined by restrained energy minimization (see ‘Materials and Methods’ section). The 20 conformers that were most consistent with the experimental restraints were used for further analyses (Figure 5A).
Figure 5.
Structure of the CUG–BP1–(UG)3 complex. (A) Backbone traces of the 20 conformers of the CUG–BP1–(UG)3 complex (stereo view). The CUG-BP1 RRM3 backbone is colored white. The RNA is shown in dark gray (carbon), red (oxygen), blue (nitrogen) and yellow (phosphorus). (B) Ribbon representation of the CUG–BP1–(UG)3 complex. The color scheme is the same as in (A). (C) Electrostatic potential surface of CUG-BP1 RRM3 in the complex with (UG)3. The RNA is colored yellow.
The RNA molecule traverses the positively charged surface of the β-sheet, surrounding the β2–β3 loop protruding from the β-sheet surface (Figure 5B and C). U1 is located in the groove formed by the β2–β3 and α2–β4 loops. G2 and U3 are held on the β-sheet surface. G4 intrudes into the pocket formed by the N-terminal extension and the β-sheet surface. U5 and G6 are wedged between the N-terminal extension and β2. These structural features were supported by NOE information (Table 1 and Supplementary Figure S3).
The mechanism of sequence-specific RNA recognition by the CUG-BP1 RRM3
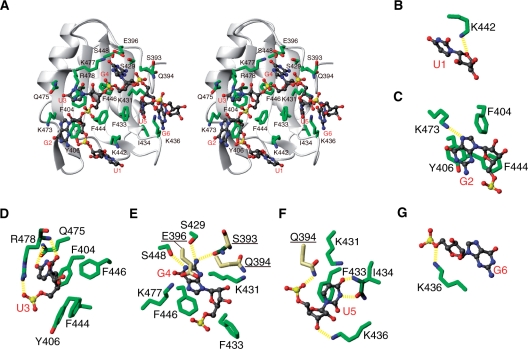
As shown in Figure 6, the RNA bases of G2–U5 are recognized by the CUG-BP1 RRM3 through extensive stacking interactions and hydrogen bonds (see also Supplementary Table S1). The G2 base is stacked with Tyr406 and forms a hydrogen bond between the N7 nitrogen atom and the Hζ atom of Lys473 (Figure 6C). The U3 base is stacked with the well-conserved Phe404 of the RNP2 motif, and its functional moieties are recognized by hydrogen bonds between the O2 atom of U3 and the HN proton of Arg478, as well as the O4 atom of U3 and the Hϵ atom of Gln475 in the pocket formed by Phe404, Tyr406, Phe444, Phe446, Gln475 and Arg478 (Figure 6D). In addition, the solution structure of the complex indicated a possible hydrogen bond between the OP2 atom of the U3 phosphate group and the guanidyl proton Hη of Arg478 (Figure 6D). The G4 base was stacked with the conserved Phe446 of the RNP1 motif, and its functional moieties were recognized by hydrogen bonds between the H1 atom of G4 and the Oγ atom of Ser448, the H21 proton of G4 and the Oγ atom of Ser429, and the H22 proton of G4 and the carbonyl O atom of Ser393, in a pocket formed by Ser393, Gln394, Glu396, Ser429, Lys431, Phe433, Phe446, Ser448 and Lys477 (Figure 6E). Upon binding to the (UG)3 RNA, the χ1 angles of Phe404 and Phe446 changed to 180° and –60°, respectively. The N-terminal extension (Ser393, Gln394 and Glu396) plays an important role in forming this pocket. The Lys477 ζ protons are located in the proximity of the G4 nucleotide and interact with the O6, N7 and OP2 atoms. At the same time, the Lys477 ζ protons could interact with the Oϵ atom of Glu396 in the N-terminal extension (Figure 6E). However, the chemical shifts of the ζ protons of Lys477 were indistinguishable from each other, and the side chain conformation of Lys477 was not defined well enough to unambiguously identify the partner directly interacting with G4. The U5 base was stacked with Phe433 on the β2 strand, and the H3 and O4 atoms of the base were recognized by hydrogen bonds with the main-chain carbonyl and amide groups of Ile434, in a concave region formed by Gln394, Lys431, Phe433, Ile434 and Lys436 (Figure 6F). Furthermore, two hydrogen bonds were formed between the OP2 atom of U5 and the Hϵ atom of Gln394, and between the O2′ atom of U5 and the Hζ atom of Lys436 (Figure 6F). On the other hand, U1 and G6 are peripheral residues, and they do not form stacking interactions or base-mediated hydrogen bonds with the CUG-BP1 RRM3. The complex structure suggests that the sugar moieties of U1 and G6 may form hydrogen bonds to the protein between the O4′ atom of U1 and the Hζ atom of Lys442, and between the O5′ atom of G6 and the Hζ atom of Lys436 (Figure 6B and G).
Figure 6.
Complex formation between CUG-BP1 RRM3 and the (UG)3 RNA. (A) Ribbon representation of the complex forming interactions (stereo view). The side chains for the RNA-recognition in CUG-BP1 RRM3 and the RNA molecule are represented as follows: green, carbon in the protein; dark gray, carbon in RNA; red, oxygen; blue, nitrogen; yellow, phosphorus. (B–G Close-up views of the RNA-recognition by CUG-BP1 RRM3. The color scheme is the same as in (A), except for the carbon atoms in the main chain (dark green) and in the N-terminal extension (yellow) of the protein. The hydrogen bonds were calculated by MOLMOL, and are represented by yellow dashed lines (see also Supplementary Table S1).
Collectively, these findings indicate that the nucleotides G2–U5 are primarily recognized by the CUG-BP1 RRM3. Among them, U3, G4 and U5 form the core sequence that confers high specificity through extensive hydrogen bonding, while G2 provides moderate specificity with a single intermolecular hydrogen bond (see also ‘Discussion’ section and Figure 7 for details).
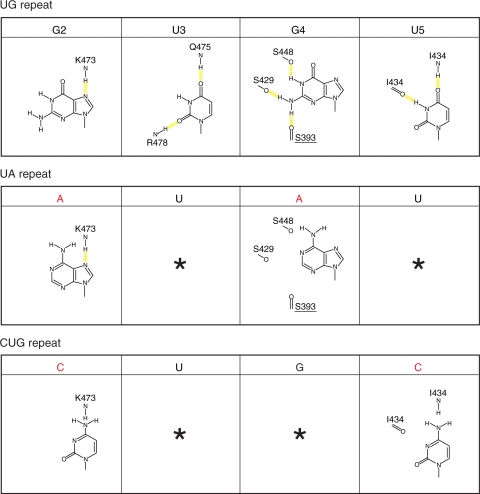
Figure 7.
RNA recognition mechanism by the CUG-BP1 RRM3 domain. CUG-BP1 RRM3 prefers the UG repeat sequence, rather than the UA and the CUG repeat sequences. The hydrogen bonds are represented by yellow dashed lines. Asterisks indicate the same type of nucleotide as that in the UG repeats.
ITC experiment for the recognition of the CUG-BP1 RRM3
In order to investigate the discrimination between the purine nucleotides A and G by the CUG-BP1 RRM3, we compared the binding activities of the CUG-BP1 RRM3 for two hexamers, [5′-(UGUGUG)-3′] and [5′-(UAUAUA)-3′], by ITC measurements. Consistent with the results from the NMR titration experiment, the CUG-BP1 RRM3 bound to the [5′-(UGUGUG)-3′] sequence with a Kd value of 1.9 μM. Meanwhile, the Kd value for the [5′-(UAUAUA)-3′] sequence was 1.1 mM (Table 2 and Supplementary Figure S4A and B). In agreement with the NMR titration experiments, the binding activity for [5′-(CUGCUG)-3′] was much weaker than for [5′-(UGUGUG)-3′]. In the case of the [5′-(CUGCUG)-3′] sequence, however, a large amount of heat was produced by hydration during the ITC measurement, and we could not fit the raw data to a theoretical curve to obtain an accurate Kd value (Table 2 and Supplementary Figure S4C). Furthermore, the complex structure revealed that the UGU trinucleotide is the key sequence for the binding. Especially, the fourth guanine nucleotide in the [5′-(UGUGUG)-3′] sequence plays a crucial role in binding to the CUG-BP1 RRM3. Thus, we examined the effect of a single alteration at the position of the fourth guanine nucleotide in the target sequence. Assessing the effect of the single alteration of the guanine nucleotide at the fourth position in the [5′-(UGUGUG)-3′] sequence is difficult, because there are two UGU units in the [5′-(UGUGUG)-3′] sequence (U1–G2–U3 and U3–G4–U5, respectively). Thus, we exchanged the uridine nucleotide at the first position for a cytosine nucleotide, so only one UGU unit was present in the hexamer sequence. We selected the two hexamers, [5′-(CGUGUG)-3′] and [5′-(CGUAUG)-3′], and compared their dissociation constants (Table 2 and Supplementary Figure S4D and E). As a result, the CUG-BP1 RRM3 could bind to the [5′-(CGUGUG)-3′] sequence with a Kd value of 4.0 μM. In contrast, the Kd value for the [5′-(CGUAUG)-3′] sequence was 0.9 mM. The single alteration of the fourth guanine nucleotide dramatically abolished the binding efficiency between the CUG-BP1 RRM3 and the target RNA (Table 2 and Supplementary Figure S4D and E).
Table 2.
Isothermal titration calorimetry data for the interactions between CUG-BP1 RRM3 and various RNAs
| Protein | RNA | Kd | ΔH | ΔS | Stoichiometry |
|---|---|---|---|---|---|
| (μM) | (kcal mol–1) | (cal K–1 mol–1) | (n) | ||
| CUG-BP1 RRM3 Wild-type | [5′-(UG)3-3′] | 1.9 | −19.1 | −37.8 | 1.1 |
| [5′-(UA)3-3′] | 1100.0 | −916.2 | −3060.0 | 1.0 | |
| [5′-(CUG)2-3′] | NDa | NDa | NDa | NDa | |
| [5′-CGUGUG-3′] | 4.0 | −14.6 | −24.2 | 1.3 | |
| [5′-CGUAUG-3′] | 900.0 | −62.6 | −196.0 | 1.0 | |
| CUG-BP1 RRM3 variant (AGS/QQQ) | [5′-(UG)3-3′] | 2.5 | −19.3 | −39.3 | 0.9 |
| CUG-BP1 RRM3 variant (AGS/PQQ) | [5′-(UG)3-3′] | 2.9 | −20.5 | −43.2 | 1.0 |
aNot detected.
RNA-binding activities of the RRM3s among the CELF/Bruno-like family members
A comparison of the RRM3s among the CELF/Bruno-like family members revealed that the QKEGPEG sequence in the N-terminal extension (corresponding to Gln394–Gly400 in CUG-BP1 RRM3) and the amino acid residues that are involved in the RNA recognition in the RRM body are very well conserved. On the other hand, the amino acid residues (Ala390-–Ser393) in the CUG-BP1 RRM3 connected to the conserved QKEGPEG sequence are slightly more diverse among the family members (Figure 1B), although Ala391 provides many NOEs to the amino acid residues on the β2 strand and the α1 helix (Figure 2D), and it seems to play an important role in anchoring the N-terminal extension on the β-sheet. To examine the effect of these differences in the N-terminal extension on the RNA-binding activities of the family members, we produced two variants of the CUG-BP1 RRM3 (384–480), in which the residues Ala391–Gly392–Ser393 were replaced by Gln–Gln–Gln or Pro–Gln–Gln, respectively. The dissociation constants Kd for binding the (UG)3 hexanucleotide were 2.5 μM for Gln–Gln–Gln and 2.9 μM for Pro–Gln–Gln. These values are almost the same as that for the wild-type CUG-BP1 RRM3 (Table 2 and Supplementary Figure S4F and G). This suggests that in spite of the slight sequence diversity in the N-terminal extension among the family members, except for the Bruno like 4 protein, the RRM3s exhibit similar RNA-binding specificities.
DISCUSSION
Several previous reports have described the RNA-binding mode and the specificity for the members of the CELF/Bruno-like family. In this study, we solved the solution structure of the third RRM of CUG-BP1 (CUG-BP1 RRM3), an important member of the CELF/Bruno-like family, by NMR. The chemical-shift perturbation analysis revealed that CUG-BP1 RRM3 specifically recognizes the UG repeat sequence, rather than the UA and CUG repeat sequences. Further structural analyses identified a unique molecular mechanism for the recognition of UG repeats by the CUG-BP1 RRM3. The characteristic features of the RNA recognition of the CUG-BP1 RRM3 are the involvement of its N-terminal extension for the formation of the specific nucleotide binding pocket and the unique placement of the four aromatic amino acids (Phe404, Tyr406, Phe433 and Phe446) involved in the stacking interaction of the nucleotide bases on the β-sheet surface.
The N-terminal extension forms a unique binding pocket in CUG-BP1 RRM3
The C-terminal extensions of the RRM core reportedly adopt a specific secondary structure and play important roles in the formation of the individual RRM structures, as well as in the specific recognition of RNA molecules (15,46). For example, the C-terminal RRM of La protein and the first RRM of U1A protein both have a C-terminal extension that forms an α-helix covering the hydrophobic β-sheet surface of the RRM in the absence of RNA molecules. Especially for the U1A protein, the C-terminal α-helix also plays an important role in the stacking of the bases of the RNA molecules (15,46). On the other hand, in the case of PTBP RRM2 and RRM3, the C-terminal extensions lack distinct secondary structures, and they traverse the β-sheet surface of the RRM body and continue to pair with the β2 strand to form a fifth β-strand (21).
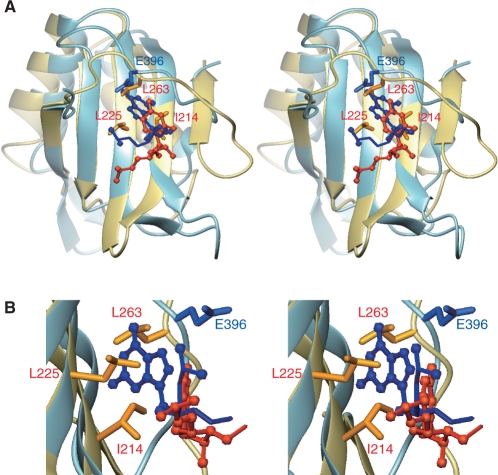
The CUG-BP1 RRM3 and the PTBP RRM2 and RRM3 share the common feature that the extension peptide segments (N-terminal for CUG-BP1 and C-terminal for PTBP, respectively) traverse the center of the β-sheet surface of the RRM core and continuously snuggle up to the second β-strand (Figure 8). Although the locations of the extended peptide segments on the β-sheet are almost the same in CUG-BP1 RRM3 and PTBP RRM2 and RRM3 (Figure 8), the roles of these segments in the recognition of the RNA molecules differ from each other. Namely, in CUG-BP1 RRM3, the characteristic segment Gln394-Gly400 of the N-terminal extension forms a deep pocket that binds the fourth guanine nucleotide with the canonical aromatic amino acid residues on RNP1 (Phe446) (Figure 9). This was confirmed by our ITC experiments. The directions of the side chains of Phe404 and Phe446 are important for the formation of the deep pocket on the β-sheet surface with the overlaid peptide. In the free form of CUG-BP1 RRM3, the χ1 angles of Phe404 and Phe446 are –60° and 180°, respectively. In these configurations, the pocket for the G4 nucleotide is not apparent on the surface of RRM3 (Figure 9A and C). However, binding to the (UG)3 hexanucleotide changes the χ1 angles for these residues, to 180° for Phe404 and –60° for Phe446, which is necessary for the formation of the deep binding pocket for the G4 nucleotides (Figure 9B and D) [The change in the χ1 angles mentioned above was confirmed by the change in the pattern of the intra-residue NOEs. For example, in the RNA-free form, the ζ proton of Phe446 exhibits several NOEs for Hγ1 of Glu396 and for Hβ1 of Ser448, reflecting the fact that the χ1 angle of Phe446 is 180°. In contrast, in the complex form, these NOEs are missing and the ζ proton of Phe446 exhibits NOEs for the ϵ protons of Phe444, indicating that the χ1 angle of Phe446 turned to –60° (Supplementary Figure S5)]. Conceivably, the strict discrimination of the target RNA is thus achieved, as many interactions with the target are necessary to compensate for the energy that is associated with the conformational changes of these aromatic amino acids for the formation of the binding pocket (Figure 9).
Figure 8.
Superposition of the 3D structures of the CUG-BP1 RRM3 (light blue) with the fourth guanine nucleotide (blue), and the PTBP RRM2 (gold) with the fourth uridine nucleotide (orange) (PDB ID 2ADB) (stereo view). (A) The structures are superposed on the main-chain atoms of the β-sheet. The side chains of I214, L225 and L263 for the PTBP RRM2 and that of Q396 for the CUG-BP1 RRM3 are also shown. (B) Close-up view.
Figure 9.
Comparison of the transparent surface representations and the ribbon representations of the CUG-BP1 RRM3. (A and C) RNA-free form. (B and D) (UG)3 RNA-bound form. The side chains of the aromatic amino acid residues (F404, Y406, F433, F444 and F446) on the β-sheet are colored orange. In addition, the N-terminal extension (Ala390–Ala401) is colored red.
In contrast, in the case of PTBP RRM2, no RNA recognition pocket is formed (Figure 8). The location of Glu396 in CUG-BP1 RRM3 is occupied by hydrophobic amino acids on the C-terminal segment (Leu263 for PTBP RRM2) that form a rigid hydrophobic core, with the aliphatic amino acid residues on the β-sheet surface (I214 and L225 in PTBP RRM2) located at the positions corresponding to Phe404 and Phe446 in CUG-BP1 RRM3. Thus, there is no pocket between the C-terminal segment and the hydrophobic amino acid residues on the β-sheet. Consequently, in the case of the PTBP RRM2, the target RNA mainly contacts the bottom area of the β-sheet, which has a shallow interaction surface.
As described above, although the characteristic segment (Gln394–Gly400) of CUG-BP1 RRM3 is well conserved among the CELF/Bruno-like family members (Figure 1B), slight variations exist among them in the region of Ala391–Ser393, just before the conserved segment. However, ITC experiments for the variants of CUG-BP1 RRM3, with the sequence Pro–Gln–Gln or Gln–Gln–Gln instead of the Ala–Gly–Ser sequence corresponding to other members of CELF/Bruno-like family, revealed that the difference in the region has little effect on the RNA-binding activities. This suggests that all of the CELF/Bruno-like family member RRM3s exhibit the same RNA preference and have common functional features.
A BLAST search for the CUG-BP1 RRM3 sequence, including the N-terminal segment (Ala390–Tyr486), indicated that the first RRM of RBMS3 (RNA-binding motif, single-stranded-interacting protein 3) is similar to the CUG-BP1 RRM3 (E-value 67). Comparisons of the amino acid sequences of their N-terminal extensions revealed similar amino acid sequences within these RRMs (Figure 1B). Especially, Glu396 in CUG-BP1 RRM3 is also conserved in RBMS3. As mentioned below, Glu396 in CUG-BP1 RRM3 plays an important role in the formation of the hydrogen-bond network for RNA recognition (Figure 6E). Thus, it is likely that these proteins have an RNA-recognition mode similar to that of CUG-BP1 RRM3.
Specific RNA recognition mechanism by the CUG-BP1 RRM3 domain
The extensive stacking interactions provided by the four aromatic rings of Phe404, Tyr406, Phe433 and Phe446 enable the strong binding of single-stranded RNA by CUG-BP1 RRM3. Three of these aromatic rings, Phe404, Phe446 and Phe433, form the pockets that recognize U3, G4 and U5, respectively (Figure 6). This explains why the UG repeat is much more preferable than the UA or CUG repeat.
The base of U3 fits in the pocket by stacking with Phe404, and the O2 and O4 atoms of the U3 base are recognized by hydrogen bonds. Unlike a uridine base, a cytosine base at this position cannot form a hydrogen bonding network. In addition, purine bases may sterically clash within the pocket, and they would lack hydrogen acceptors around this position (Figure 6). Therefore, other bases besides uridine may not be recognized at this position.
Many of the functional moieties of G4 are well recognized through the hydrogen bonds within the pocket of Phe446, as described in the ‘Results’ section. As mentioned above, we excluded the interacting partner for Lys477 in Figure 7. However, it is likely that Glu396 and Lys477 cooperatively form a network of hydrogen bonds that recognize the G4 base. Interestingly, the Glu396 and Lys477 residue pair is well conserved among the CELF/Bruno-like family members. The substitution of G4 by adenine is predicted to significantly reduce the interaction with the protein (Figure 7, middle panel). Actually, ITC experiments for the RNA sequences [5′-(CGUGUG)-3′] and [5′-(CGUAUG)-3′] revealed the importance of the fourth guanine nucleotide in the RNA sequence. Interestingly, among the known RRM–RNA complex structures, when a guanine nucleotide resides in the position corresponding to G4 in the CUG-BP1 RRM3, all of the base conformations of these guanine nucleotides adopted a syn conformation (25). However, the base conformation of G4 in the CUG-BP1 RRM3 adopted an anti conformation, suggesting a novel aspect of the RRM–RNA interaction.
Similarly, as the O4 and H3 of the U5 base are well recognized by the main chain amide and carbonyl groups, its substitution by a cytosine base is predicted to result in the loss of all of these hydrogen bonds, and to cause steric hindrance, due to the amino group attached to C4 (Figure 7, lower panel). Therefore, we concluded that this position should be occupied by a uridine residue. Taken together, the UGU triplet is the core for the sequence-specific RNA recognition by the CUG-BP1 RRM3.
In the case of the G2 nucleotide, no obvious pocket is formed for the base recognition. However, the base of G2 is stacked with Tyr406, which is characteristic of the CUG-BP1 RRM3, and only one hydrogen bond is formed with the side chain of Lys473 (Figure 6). This hydrogen bond would be preserved by a substitution with an adenine base, without a conformational change of the protein (Figure 7, middle panel). We concluded that the key RNA sequence for the CUGBP1 RRM3 is the tetraplet (G/A)UGU. In this way, the present structural study revealed the anomalous recognition mechanism of the tetraplet (G/A)UGU sequence by the CUG-BP1 RRM3. Among the several specific RNA elements reported as targets of CELF/Bruno-like family members, the GU-rich sequences [otherwise known as a GRE (9)], the embryo deadenylation element [EDEN; U(A/G) repeat (6)], the Bruno responsive element [also referred to as repeat elements of uridine and purine, UREs (3)] and the class III AU-rich element (8) of (pre-) mRNAs contain UGU units. Importantly, in many cases, the UGU sequences are scattered within repeats of similar UAU units. The strict discrimination of short UGU units from repeats of UAU units by CUG-BP1 RRM3 may thus be necessary for the function of CELF/Bruno-like family members.
The CUG triplet has been considered as the target sequence for the CUG-BP1 protein (1). However, the UGU triplet or UG-repeat, rather than the CUG triplet, is also reportedly important for the binding of the CUG-BP1 family proteins (3,7,26,47). This notion is quite consistent with the present study on the functional importance of CUG-BP1 RRM3.
The roles of the two RNA-binding sites in the CUG-BP1 protein
Previously, it was reported that RRM1-RRM2 of CUG-BP1 could also cooperatively bind to the UG-repeats (26). However, the N-terminal extensions of the CUG-BP1 RRM1 and RRM2 lack sequences homologous to that in RRM3. In addition, the characteristic aromatic residues located on the β-sheet in the CUG-BP1 RRM3 (Tyr406, Phe433, Phe444 and Phe446) are not conserved in the CUG-BP1 RRM1 and RRM2. Furthermore, a previous study suggested that RRM1 and RRM2 mainly bind to the CUG-repeat sequence, in contrast to RRM3 (27). This suggests that the RRM1–RRM2 di-domains in CUG-BP1 could bind to the target RNA in a different manner than the RRM3 mono-domain, and that CUG-BP1 targets various RNA sequences by utilizing the distinct RNA-binding affinities of the two independent RNA-binding sites: RRM1–RRM2 and RRM3.
The present structural study revealed the substantial role of CUG-BP1 RRM3 in the recognition and discrimination of the target RNA. The precise structural analysis of the two N-terminal RRMs will lead to a comprehensive understanding of the functions of this protein family.
ACCESSION NUMBERS
Protein Data Bank, with the accession codes 2RQ4 and 2RQC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The RIKEN Structural Genomics/Proteomics Initiative (RSGI); the National Project on Protein Structural and Functional Analyses of the Ministry of Education, Culture, Sports, Science and Technology of Japan; the Human Frontier Science Program (HFSP) (to the Muto research groups); and by a Grant-in-Aid for Scientific Research of the Japan Society for the Promotion of Science (JSPS) and by the Volkswagen Foundation (to P.G.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr T. Nagata and Ms. S. Suzuki for help with the NMR data analysis, structure calculations, and structure refinement. We also thank Dr T. Matsuda, Dr Y. Tomo, Dr M. Aoki, Dr S. Watanabe, Dr T. Harada, Dr T. Nagira, Ms. E. Seki, Mr K. Hanada, Mr M. Ikari, Ms. Y. Fujikura and Ms. Y. Kamewari-Hayami for sample preparation. We are grateful to Dr K. Kurimoto for helpful discussions about the manuscript preparation. We would like to thank Ms. A. Ishii and Ms. T. Nakayama for help with the manuscript preparation.
Footnotes
Present address: Tatsuhiko Someya, Graduate School of Life and Environmental Sciences, University of Tsukuba, 1-1-1 Tennodai, Tsukuba-shi, Ibaraki 305-8572, Japan.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 3.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7:133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 4.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27:4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreau C, Paillard L, Mereau A, Osborne HB. Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie. 2006;88:515–525. doi: 10.1016/j.biochi.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17:278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J. Biol. Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- 9.Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, et al. Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell. 2008;29:263–270. doi: 10.1016/j.molcel.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Nagai K, Oubridge C, Ito N, Avis J, Evans P. The RNP domain: a sequence-specific RNA-binding domain involved in processing and transport of RNA. Trends Biochem. Sci. 1995;20:235–240. doi: 10.1016/s0968-0004(00)89024-6. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Canadillas JM, Varani G. Recent advances in RNA-protein recognition. Curr. Opin. Struct. Biol. 2001;11:53–58. doi: 10.1016/s0959-440x(00)00164-0. [DOI] [PubMed] [Google Scholar]
- 15.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 16.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B′′-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 17.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J. 2006;25:5126–5137. doi: 10.1038/sj.emboj.7601385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature. 1999;398:579–585. doi: 10.1038/19242. [DOI] [PubMed] [Google Scholar]
- 20.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 21.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 22.Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL. Structural basis for polypyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol. Cell. 2006;23:49–59. doi: 10.1016/j.molcel.2006.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, Tanaka Hall TM. Structural basis for recognition of AU-rich element RNA by the HuD protein. Nat. Struct. Biol. 2001;8:141–145. doi: 10.1038/84131. [DOI] [PubMed] [Google Scholar]
- 24.Abe R, Sakashita E, Yamamoto K, Sakamoto H. Two different RNA binding activities for the AU-rich element and the poly(A) sequence of the mouse neuronal protein mHuC. Nucleic Acids Res. 1996;24:4895–4901. doi: 10.1093/nar/24.24.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auweter SD, Oberstrass FC, Allain FH. Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res. 2006;34:4943–4959. doi: 10.1093/nar/gkl620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori D, Sasagawa N, Kino Y, Ishiura S. Quantitative analysis of CUG-BP1 binding to RNA repeats. J. Biochem. 2008;143:377–383. doi: 10.1093/jb/mvm230. [DOI] [PubMed] [Google Scholar]
- 27.Timchenko LT. Myotonic dystrophy: the role of RNA CUG triplet repeats. Am. J. Hum. Genet. 1999;64:360–364. doi: 10.1086/302268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 29.Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J. Biomol. NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- 31.Ito W, Ishiguro H, Kurosawa Y. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene. 1991;102:67–70. doi: 10.1016/0378-1119(91)90539-n. [DOI] [PubMed] [Google Scholar]
- 32.Bax A. Multidimensional nuclear magnetic resonance methods for protein studies. Curr. Opin. Struct. Biol. 1994;4:738–744. [Google Scholar]
- 33.Kay LE. NMR methods for the study of protein structure and dynamics. Biochem. Cell Biol. 1997;75:1–15. doi: 10.1139/o97-023. [DOI] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 37.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 38.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 39.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 40.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 41.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 42.Powers R, Garrett DS, March CJ, Frieden EA, Gronenborn AM, Clore GM. The high-resolution, three-dimensional solution structure of human interleukin-4 determined by multidimensional heteronuclear magnetic resonance spectroscopy. Biochemistry. 1993;32:6744–6762. doi: 10.1021/bi00077a030. [DOI] [PubMed] [Google Scholar]
- 43.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 44.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 45.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 46.Jacks A, Babon J, Kelly G, Manolaridis I, Cary PD, Curry S, Conte MR. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure. 2003;11:833–843. doi: 10.1016/s0969-2126(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 47.Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem. J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.