Abstract
The efficient incorporation of influenza virus genome segments into virions is mediated by cis-acting regions at both ends of the viral RNAs. It was shown previously that nt 16–26 at the 3′ end of the non-structural (NS) viral RNA of influenza A virus are important for efficient virion incorporation and that nt 27–56 also contribute to this process. To understand further the signalling requirements for genome packaging, this study performed linker-scanning mutagenesis in the latter region and found that nt 27–35 made an appreciable contribution to the efficient incorporation of the NS segment. An NS vRNA library was then generated composed of an RNA population with randomized nucleotides at positions 16–35 such that the virus could select the sequences it required for virion incorporation. The sequences selected differed from the wild-type sequence and no conserved nucleotides were selected. The ability of non-wild-type sequences to function in this manner indicates that the incorporation of influenza A virus genome segments does not absolutely require specific sequences.
INTRODUCTION
Efficient packaging of the viral genome is a critical step in the viral life cycle. Viruses with segmented genomes face the added difficulty of incorporating at least one copy of each viral gene segment into each virion. The genome of influenza A virus comprises eight negative-sense, single-stranded viral RNA (vRNA) segments, and two models – random versus selective vRNA virion incorporation – have been proposed for the efficient incorporation of these segments into virions. Recently, we and others identified segment-specific virion incorporation signals in the polymerase subunits PA (de Wit et al., 2006; Liang et al., 2005, 2008; Marsh et al., 2008; Muramoto et al., 2006), PB1 (Liang et al., 2005, 2008; Marsh et al., 2008; Muramoto et al., 2006) and PB2 (Dos Santos Afonso et al., 2005; Gog et al., 2007; Liang et al., 2005, 2008; Marsh et al., 2008; Muramoto et al., 2006), and in the haemagglutinin (HA) (Marsh et al., 2007; Watanabe et al., 2003), nucleoprotein (NP) (Ozawa et al., 2007), neuraminidase (NA) (Fujii et al., 2003; Gog et al., 2007), M (Hutchinson et al., 2008) and non-structural (NS) (Fujii et al., 2005) segments. For every segment tested thus far, the viral incorporation signals are found at both ends of the vRNAs, although the relative contributions of the sequences at the 5′ and 3′ ends to efficient virion incorporation differ for each segment. The virion incorporation signals for each segment are thought to include sequences in the terminal non-coding and coding regions (Fujii et al., 2005; Liang et al., 2008). The mechanism(s) by which these sequences govern efficient virion incorporation remains unknown, but may include RNA–RNA interactions between the vRNA segments, or RNA–protein interactions between the vRNAs and viral or cellular proteins, or a combination of both.
We previously showed that, for the NS segment, nt 16–56 at the 3′ end of the vRNA contribute to efficient genome incorporation (Fujii et al., 2005). However, our study did not assess whether all of the nucleotides in this region are critical or whether specific nucleotides or particular structural features are essential to the process. Here, we address these points by further defining the segment region important for vRNA incorporation. By performing experiments in which virus selectively incorporated vRNA segments from a pool of recombinant segments containing randomized sequences in the region important for genome incorporation, we found that the incorporation of influenza A virus genome segments does not absolutely require specific sequences.
METHODS
Cells.
293T human embryonic kidney cells and COS-7 African green monkey kidney cells were maintained in Dulbecco's modified Eagle's medium with 10 % fetal calf serum (FCS) and antibiotics. Madin–Darby canine kidney (MDCK) cells were grown in minimal essential medium (MEM) containing 5 % newborn calf serum and antibiotics. All cells were maintained at 37 °C in 5 % CO2.
Plasmids.
The construction of plasmids for the transcription of influenza A/WSN/33 (H1N1) vRNAs under the control of the RNA polymerase I promoter (referred to as PolI plasmids) has been described previously (Neumann et al., 1999).
pPolINS(150)GFP(150) (Fig. 1a) was used to produce a negative-sense viral RNA containing the 3′ non-coding region and 150 nt of the 3′ coding region of the NS vRNA, the open reading frame of the enhanced green fluorescence protein (EGFP; Clontech), 150 nt of the 5′ coding region and the 5′ non-coding region of the NS vRNA (Fujii et al., 2005).
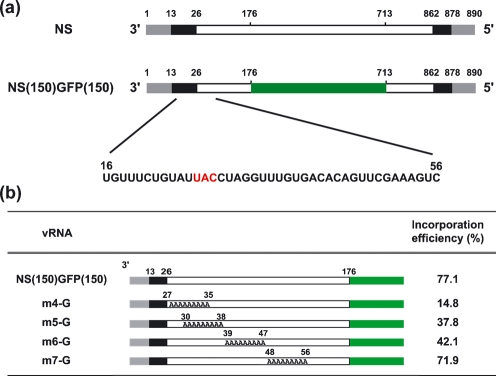
Fig. 1.
Schematic diagram of NS(150)GFP(150) vRNAs. (a) Schematic diagrams of the NS and NS(150)GFP(150) segments. The grey bars represent the conserved influenza virus promoter regions. The black and white bars indicate the segment-specific non-coding and coding regions, respectively. In NS(150)GFP(150), the centre portion of the NS coding region is replaced with the coding region for GFP (green). The numbers indicate the nucleotide positions from the 3′ end of the NS vRNA. The initiation codon is indicated in red. (b) Schematic diagrams of NS(150)GFP(150) vRNAs containing A substitutions in the 3′ end. The viral promoter, non-coding and coding regions are represented by grey, black and white bars, respectively, whilst the green bar represents the GFP coding region. The nucleotides replaced with A residues are shown. The numbers indicate the nucleotide position from the 3′ end of the vRNA. The incorporation efficiencies were determined as described in Methods. Representative data from three independent experiments are shown.
To generate plasmid pPolIm4-G, which encodes a recombinant NS(150)GFP(150) segment in which nt 27–35 at the 3′ end of the viral RNA are replaced with A residues (Fig. 1b), inverse PCR (Ochman et al., 1988) was performed with the back-to-back primers NSDG-UP2 (5′-GAATTCGAAGACAAAACACTGTGTCAAGCTTTCAGGTAGATTG-3′) and NSDG-DW2-2 (5′-GAATTCGAAGACAATGTTAAAAAAAAATATGTCTTTGTCACCCTGC-3′). Additional mutants containing A residues at nt 30–38, 39–47 or 48–56 at the 3′ end of the viral RNA were generated similarly. All plasmids were sequenced to ensure that unwanted mutations were not introduced.
pPolIHA//NS(HAΨ) (Fig. 2a) produced a negative-sense, bicistronic HA/NS segment comprising the 3′ non-coding region of the HA vRNA, the HA coding region, a second 3′ non-coding region of the HA vRNA (including the viral promoter), the overlapping open reading frames of both the NS1 and NS2 proteins, 80 nt derived from the carboxyl-terminal HA coding region that harbours the HA vRNA packaging signal (indicated as ‘Ψ’) and the 5′ HA non-coding sequence. To generate this plasmid, pPolIHA, which allows the transcription of the HA segment of A/WSN/33 (H1N1) virus, was amplified by inverse PCR (Ochman et al., 1988) using the back-to-back primers ANST-F (5′-CACACACGTCTCTGATTAGGATTTCAGAAATATAAGG-3′) and HADW-1 (5′-CACACACGTCTCTTGCTTCAGATGCATATTCTGCACTG-3′). The PCR product was ligated with an NS gene fragment that also contained the 3′ non-coding region of the HA segment, in addition to 80 nt of the HA coding region and the HA 5′ non-coding region (see Fig. 2a).
Fig. 2.
Selection of efficient incorporation signals from a pool of randomized sequences (‘sequence trapping’). (a) Representation of HA//NS(HAΨ) bicistronic vRNA. The grey, red and orange bars indicate the HA non-coding sequence, the HA coding sequence and the NS coding sequence, respectively; Ψ depicts the 5′ region of the HA segment that is required for efficient incorporation. (b) Generation of recombinant influenza virus containing a bicistronic HA/NS segment. 293T cells were transfected with a plasmid for the expression of HA//NS(HAΨ) or HA vRNA, together with other plasmids for the production of influenza virus; the plasmid for the production of NS vRNA was omitted. Forty-eight hours later, the supernatant was tested in plaque assays. (c) Schematic diagram of the system for the selection of sequences important for efficient NS vRNA incorporation. 293T cells were transfected with a mixture of plasmids for the expression of an NS packaging library and other plasmids for the production of influenza virus. At 48 h post-transfection, the supernatant from plasmid-transfected cells was titrated in plaque assays on MDCK cells. GFP-positive plaques were picked and the sequence of the NS segment was determined.
Generation of recombinant influenza virus or virus-like particles (VLPs).
293T cells (1×106) were transfected with plasmids designed to express the influenza vRNAs and all of the viral proteins, as described by us previously (Neumann et al., 1999). Briefly, DNA and transfection reagent were mixed [2 μl TransIT-293 (μg DNA)−1], incubated at room temperature for 10 min and added to the cells. Six hours later, the DNA/transfection reagent mixture was replaced with Opti-MEM (Invitrogen) containing 1 % FCS. At 48 h post-transfection, the virus- or VLP-containing supernatant derived from the transfected 293T cells was harvested and used in the experiments described below.
Determination of the incorporation efficiency of recombinant NS(150)GFP(150) vRNA.
Twenty-four hours after being infected with influenza VLPs or virions, GFP-positive cells [representing the number of VLPs or virions containing an NS(150)GFP(150) vRNA] were counted. Cells were then fixed and processed for indirect immunostaining with an anti-NP antibody (described below). The number of NP-positive cells represented the total number of infectious VLPs or virions. Hence, the ratio of GFP-positive cells to NP-positive cells represented the incorporation efficiency of a recombinant NS(150)GFP(150) vRNA.
Construction of a library containing randomized NS packaging sequences.
To generate a library that contained randomized packaging sequences at positions 16–35 at the 3′ end of the NS vRNA, we performed degenerative PCR mutagenesis using plasmid pPolINS(150)GFP(150) as template. The region was divided into two halves (NSRG1, nt 16–26; and NSRG2, nt 27–35), and nucleotides in each half were randomized by performing inverse PCR with primers containing degenerate nucleotides (see Table 1). The PCR products were self-ligated and propagated in Escherichia coli strain DH5α, which resulted in 104 independent clones per library.
Table 1.
Primers used for degenerative PCR
The nucleotides where degeneracy was introduced are shown in bold. The BbsI site is underlined.
| Library | Forward primer | Reverse primer | ||
|---|---|---|---|---|
| Name | Sequence (5′→3′) | Name | Sequence (5′→3′) | |
| pPolI-NSRG1 | NSRG-UP1 | GAATTCGAAGACAAGGTGNNNNNNNNNNNATGGATCCAAACACTGTGTC | NSRG-DW1 | GAATTCGAAGACAACACCCTGCTTTTGC |
| pPolI-NSRG2 | NSRG-UP2 | GAATTCGAAGACAAAACACTGTGTCAAGCTTTCAGGTAGATTG | NSRG-DW2 | GAATTCGAAGACAATGTTNNNNNNNNNTATGTCTTTGTCACCCTGC |
Selection of packaging sequences from a pool of randomized sequences.
293T cells (1×106) were transfected with one of the NS packaging signal libraries, with pPolIHA//NS(HAΨ) and with plasmids expressing the remaining six viral RNAs (i.e. PB2, PB1, PA, NP, NA and M) and all of the viral proteins. At 48 h post-transfection, the virus-containing supernatant of the transfected 293T cells was harvested and incubated with MDCK cells for 1 h at 37 °C in 5 % CO2. Cells were then washed with MEM containing 5 % BSA and overlaid with MEM containing 1 % agarose. At 48 h post-infection, GFP-expressing plaques were picked and amplified in liquid culture.
To identify the packaging sequences selected by this method, viral RNAs were extracted by using an Isogen-LS RNA extraction kit (Nippon Gene) and reverse transcribed with SuperScript III (Invitrogen) and a primer corresponding to the 3′ end of the viral RNAs (U12-A: 5′-AGCAAAAGCAGG-3′). The NS(150)GFP(150) gene fragments containing selected packaging sequences were amplified with PfuUltra (Stratagene) and the gene-specific primers NSNTR-UP3 (5′-GAATTCGAAGACTAACCCAGCAAAAGCAGGGTG-3′) and GFDW-2 (5′-GAATTCGAAGACTATTATTACTTGTACAGCTCGTCCATGCCG-3′). The NS–GFP gene was then sequenced with an Applied Biosystems 3100 Auto Sequencer by using cycle sequencing dye terminator chemistry (Perkin Elmer) with the GFP-specific primer GFDW-3 (5′-AACAGCTCCTCGCCCTTGCTCACC-3′).
Immunostaining.
All staining procedures were conducted at room temperature. VLP-infected cells were fixed for 30 min with 4 % paraformaldehyde in PBS and permeabilized with 0.1 % Triton X-100 for 30 min. The samples were then incubated with an anti-NP antibody at a 1 : 1000 dilution in PBS for 30 min. After washing three times with PBS, the samples were reacted with biotinylated antibody (Vector Laboratories) at a 1 : 100 dilution in PBS for 30 min and then treated with VECTASTAIN Elite ABC Reagent (Vector Laboratories) according to the manufacturer's instructions. Samples were visualized by their reaction with diaminobenzidine tetrahydrochloride (Sigma).
Predicting vRNA secondary structure.
RNA-folding calculations were performed by using the computer programs mfold (version 3.2) (Walter et al., 1994; Zuker, 1989) on the web server (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) and vsfold (version 5.23) (Dawson et al., 2006, 2007) on the web server (http://www.rna.it-chiba.ac.jp/∼vsfold/vsfold5/).
RESULTS
Nucleotides 27–35 at the 3′ end of the NS vRNA are important for efficient NS vRNA virion incorporation
Our previous study suggested an important role for the nucleotides at positions 16–56 at the 3′ end of the NS vRNA of A/WSN/33 (H1N1) virus for efficient incorporation of this segment into the virion (Fujii et al., 2005). A recombinant NS vRNA possessing the 3′ non-coding region, 150 nt of the 3′ NS coding region, the GFP coding region, 150 nt of the 5′ NS coding region and the 5′ non-coding region [NS(150)GFP(150); Fig. 1a] was incorporated efficiently into virions (Fujii et al., 2005), unlike a recombinant NS(150)GFP(150) vRNA that lacked the nucleotides at positions 16–26, which are part of the NS non-coding region (Fig. 1a) (Fujii et al., 2005). Nucleotides 27–56 are part of the NS coding region (Fig. 1a) and the substitution of silent nucleotides into this region (i.e. nucleotides that did not affect the amino acid sequence) also interfered with the efficient incorporation of the NS vRNA (Fujii et al., 2005).
To define further the region critical for NS vRNA packaging, we performed linker-scanning analysis by producing four mutant pPolINS(150)GFP(150) plasmids that contained A residue substitutions at nt 27–35, 30–38, 39–47 or 48–56 (Fig. 1b). We also introduced A residue substitutions at nt 16–26 of the pPolINS(150)GFP(150) plasmid; however, this plasmid was not subjected to further analyses, as the efficiency of GFP expression from this plasmid was much lower than that of the other plasmids (data not shown). The resulting plasmids were transfected into 293T cells together with plasmids for the production of the remaining seven viral RNAs and all ten viral proteins. Forty-eight hours later, VLPs in the supernatants of the transfected cells were harvested and used to infect MDCK cells. At 24 h post-infection, the number of GFP-positive cells [representing the number of VLPs that contained the mutant NS(150)GFP(150) vRNA] and the number of NP-positive cells (representing the total number of VLPs) were determined. The incorporation efficiency was defined as the percentage of VLPs that contained a mutant NS(150)GFP(150) vRNA. The substitution of nucleotides at positions 27–35 substantially reduced vRNA incorporation efficiencies (Fig. 1b, m4-G), identifying this region as critical for efficient packaging. By contrast, the substitution of nucleotides at positions 48–56 did not appreciably affect virion incorporation efficiency (Fig. 1b, m7-G), whilst A residue substitutions at positions 30–38 or 39–47 of the 3′ region had moderate effects (Fig. 1b, m5-G and m6-G).
Does efficient virion incorporation require specific nucleotides?
Our results suggested that the nucleotides at positions 16–26 (Fujii et al., 2005) and 27–35 (this study) at the 3′ end of the NS vRNA are important for efficient virion incorporation. These findings prompted the question: are there specific nucleotides critical for efficient packaging? To answer this question, we devised a strategy that allowed only sequences mediating efficient vRNA packaging to be selected from a pool of random packaging sequences (see below and Fig. 2c). First, we generated a bicistronic HA/NS segment, HA//NS(HAΨ), that expressed HA and the two proteins encoded by the NS segment (NS1 and NS2) (Fig. 2a). In this bicistronic segment, the HA and NS1/NS2 coding sequences were separated by an influenza virus promoter region derived from the 3′ end of the influenza viral RNA; the duplication of the viral promoter ensured the expression of more than one viral protein from one viral gene segment (Flick & Hobom, 1999; Machado et al., 2003). For efficient genome incorporation, HA//NS(HAΨ) also contained the HA packaging signal (Ψ) (Watanabe et al., 2003) (Fig. 2a). To test its functionality, we transfected cells with a plasmid encoding the HA//NS(HAΨ) vRNA, together with plasmids encoding the remaining vRNA segments with the exception of the NS vRNA, and plasmids encoding all of the viral proteins (thus generating a seven-segment virus). At 48 h post-transfection, we detected 104 p.f.u. virus ml−1 in the culture supernatant of plasmid-transfected 293T cells (Fig. 2b). As a negative control, we transfected cells with the plasmid expressing the wild-type HA vRNA segment instead of HA//NS(HAΨ) and, as expected, no plaques were detected (Fig. 2b).
The successful generation of a recombinant influenza virus containing a bicistronic HA/NS vRNA then allowed us to randomize portions of the NS(150)GFP(150) vRNA in an attempt to select for specific sequences required for efficient virion incorporation. Using pPolINS(150)GFP(150) vRNA as a template, we introduced random nucleotides at positions 16–26 or 27–35 at the 3′ end of the NS vRNA to create two libraries of randomized packaging sequences. Although the total number of possible sequences for NSRG1 and NSRG2 was much larger (411=4.2×106 and 49=2.6×105, respectively) than the pool size (104 independent clones per library; see Methods), these libraries allowed us to uncover any sequence trends towards the efficient incorporation of recombinant NS segment into the virion. Cells were transfected with one of the libraries, with the plasmid for the synthesis of the bicistronic HA/NS vRNA and with plasmids for the synthesis of the remaining vRNA and viral proteins (Fig. 2c). At 48 h post-transfection, we harvested the virus-containing supernatant of transfected 293T cells and performed plaque assays in MDCK cells (Fig. 2c). From a total of 5×103 plaques for both libraries tested, we obtained 29 GFP-positive plaques, representing viruses that had incorporated an NS(150)GFP(150) vRNA from the pool of randomized sequences.
We next determined the sequences of the selected packaging signals of 12 viruses recovered for positions 16–26 and 17 viruses for positions 27–35 (Table 2). The 12 sequences for positions 16–26 were different from the wild-type sequence and from each other. Although there were no conserved nucleotides, there was a nucleotide preference for certain positions: eight of the 12 selected sequences contained an A at position 16 or a U at position 19, nine of the 12 contained an A at position 20 and five of the 12 contained all three of these nucleotides. Thus, this sequence displayed some nucleotide preference clustered at positions 16–20, whilst the rest of the sequence (positions 21–26) was random.
Table 2.
Nucleotide sequences selected by sequence trapping
RT-PCR products from mutant viruses were sequenced directly. Nucleotides identical to the wild-type are shown in bold.
| Sequence no. | Nucleotides16–26 | Incorporationefficiency (%)* | Nucleotides27–35 | Incorporationefficiency (%)* |
|---|---|---|---|---|
| Wild-type | UGUUUCUGUAU | 79.0 | UACCUAGGU | 79.0 |
| 1 | AAGUGAACAAU | 78.5 | GUACCUGGU (3)† | 59.5 |
| 2 | AAGUACGCGAU | 74.2 | GGUGUUCUA | 56.3 |
| 3 | ACGUAGCUUUA | 67.4 | UUGUUUUAU (9)† | 55.2 |
| 4 | ACGUAGCUUUU | 66.0 | GUCUCGUGU | 54.8 |
| 5 | ACGAUACGACA | 65.5 | UUCUUAUUU | 48.5 |
| 6 | UAAACGUGUGG | 60.2 | GUGUUUUGA | 47.0 |
| 7 | ACGUAACAUUG | 59.1 | UGUGUUUUU | 37.4 |
| 8 | ACGUAAGCCAC | 56.5 | ||
| 9 | UUAUAGUCUAU | 46.3 | ||
| 10 | UCAUAUACAAU | 44.2 | ||
| 11 | AUACAGAGUCA | 43.5 | ||
| 12 | UUACAAGGCCA | 36.2 |
*Incorporation efficiencies were determined as described in Methods. Representative data from three independent experiments are shown.
†Number of clones possessing the indicated sequence.
For positions 27–35, the results were similar to those for positions 16–26; i.e. none of the recovered sequences was identical to that of the wild-type. One sequence motif was found in nine of the viral clones, while another was detected in three clones, suggesting that they may support more efficient virion incorporation than others. The most dominant motif was characterized by a high U content; these Us were in the same position as the U in the wild-type sequence, and also prevalent in other positions.
Do selected NS vRNA packaging signals mediate efficient virion incorporation?
To confirm that the selected sequences do indeed function as specific NS vRNA incorporation signals, we tested their virion incorporation efficiencies by infecting cells with the respective recombinant viruses and determining the ratios of GFP-positive cells to the total number of infected cells (i.e. the number of NP-positive cells) (Table 2). Most of the mutant NS(150)GFP(150) vRNAs were incorporated into more than 50 % of the virions. In fact, some of the sequences tested (e.g. sequences 1 and 2 for positions 16–26, see Table 2) were as efficient as the wild-type sequence in mediating virion incorporation of the respective vRNAs. For some sequences tested, however, the incorporation rates were below 50 %. Given that the vRNAs encoding these sequences were obtained from GFP-expressing plaques, the levels of incorporation must have been adequate to induce a detectable level of GFP expression in plaques.
As we did not find conserved nucleotides among the trapped sequences, we looked for secondary structures that may be common to these sequences. Using the programs mfold and vsfold, we did not find any conserved secondary structures at positions 1–56 (data not shown). Together, our findings indicate that wild-type nucleotide sequences are not absolutely required for efficient NS vRNA virion incorporation. Moreover, no specific nucleotides are required, at least at positions 16–35, although some nucleotide preferences were noted. Overall, these findings suggest that structural features (albeit not identifiable by the current analysis), rather than specific nucleotides, are likely to play an important role in influenza NS vRNA packaging. Further studies are needed to reveal the precise role of these regions in genome packaging.
DISCUSSION
Viral genome packaging is mediated by the cis-acting region of the genome (Ball, 2007). Here, we investigated nucleotides in the NS segment that affect the incorporation efficiency of this vRNA segment. Mutational analyses described here and our previous paper (Fujii et al., 2005) revealed that the nucleotides at positions 16–35 of the 3′ end of the NS vRNA are critical for its efficient incorporation. Interestingly, we found that the nucleotide sequence was neither conserved among the trapped sequences nor identical to that of the wild-type. Two different lineages have been demonstrated in the NS segment (Treanor et al., 1989), suggesting that some variations in the region required for NS vRNA incorporation can be tolerated. The extent of sequence variation found in our study exceeds the variation found between the two lineages of NS segments. Thus, our results clearly demonstrated that sequences other than that of the wild-type allow incorporation of the influenza A virus NS segment into virions. Recently, by using a similar vRNA selection strategy, we found that the 27 nt sequence at the 5′ end of the coding region in the M segment can also accommodate highly diverse sequences (Ozawa et al., 2009), supporting our findings with the NS segment.
How are the packaging signals on the influenza virus RNA recognized? In infected host cells, the influenza viral genome segments form viral ribonucleoprotein (vRNP) complexes in which the vRNAs are associated with nucleoprotein and three polymerase proteins (Palese, 2007). Unlike the genomic RNAs of paramyxoviruses (e.g. simian virus 5, Sendai virus and Newcastle disease virus; Heggeness et al., 1980; Kingsbury & Darlington, 1968; Lynch & Kolakofsky, 1978) and rhabdoviruses (such as vesicular stomatitis virus; Blumberg et al., 1983; Heggeness et al., 1980), whose RNAs are covered by viral NPs, the genomic RNAs of influenza vRNPs are sensitive to RNase and chemical modification (Klumpp et al., 1997). This finding suggests that the influenza vRNAs wrap around the NP. It may be that the nucleotides at positions 16–35 of the NS segment are exposed and have enough flexibility to form a secondary or tertiary structure that can be recognized by other viral or host molecules.
The packaging of the multi-segmented influenza virus genome into virions is a biologically intriguing event. Two hypotheses have been proposed for the mechanism by which the influenza virus genome is packaged. The random-packaging hypothesis suggests that each viral RNA segment possesses a common packaging signal that allows random incorporation of the RNA segments into the virions (Bancroft & Parslow, 2002). The selective-packaging hypothesis is based on the concept that each viral RNA segment possesses a unique packaging signal that is required for its incorporation into the virion (McGeoch et al., 1976; Odagiri & Tashiro, 1997). In this study, we examined the NS vRNA sequence and revealed that nucleotides at positions 16–35 are important for efficient genome incorporation. Importantly, these nucleotides do not share homology with other viral segments (data not shown), suggesting that these nucleotides form a unique signalling sequence for the NS segment incorporation. Thus, this finding strongly supports the selective-packaging hypothesis.
Here, we explored a ‘sequence-trapping’ approach to identifying sequences that allow efficient influenza virus genome packaging. Using this method, we showed that no specific nucleotides are required for efficient genome incorporation. These results are biologically relevant, as they were obtained in the context of replicating virus. We previously used a similar strategy to identify specific amino acids in the NS2 (NEP) protein that are required for nuclear export (Iwatsuki-Horimoto et al., 2004). This approach can, therefore, be useful in determining the nucleotide or amino acid requirements for functional RNA and protein domains.
Acknowledgments
We thank members of our laboratories for helpful discussions. We also thank Krisna Wells for excellent technical assistance, Gabriele Neumann for critical review of the manuscript and Susan Watson for editing the manuscript. This work was supported by CREST and ERATO (Japan Science and Technology Agency) and by a grant-in-aid for Specially Promoted Research from the Ministries of Education, Culture, Sports, Science and Technology, and by grants-in-aid from the Ministry of Health, Labour and Welfare of Japan, and by National Institute of Allergy and Infectious Disease Public Health Service research grants. K. F. is the recipient of a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
References
- Ball, L. A. (2007). Virus replication strategies. In Fields Virology, 5th edn, pp. 119–139. Edited by D. M. Knipe & P. M. Howley. Philadelphia, PA: Lippincott Williams & Wilkins.
- Bancroft, C. T. & Parslow, T. G. (2002). Evidence for segment-nonspecific packaging of the influenza A virus genome. J Virol 76, 7133–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg, B. M., Giorgi, C. & Kolakofsky, D. (1983). N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32, 559–567. [DOI] [PubMed] [Google Scholar]
- Dawson, W., Fujiwara, K., Kawai, G., Futamura, Y. & Yamamoto, K. (2006). A method for finding optimal RNA secondary structures using a new entropy model (vsfold). Nucleosides Nucleotides Nucleic Acids 25, 171–189. [DOI] [PubMed] [Google Scholar]
- Dawson, W. K., Fujiwara, K. & Kawai, G. (2007). Prediction of RNA pseudoknots using heuristic modeling with mapping and sequential folding. PLoS One 2, e905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, E., Spronken, M. I., Rimmelzwaan, G. F., Osterhaus, A. D. & Fouchier, R. A. (2006). Evidence for specific packaging of the influenza A virus genome from conditionally defective virus particles lacking a polymerase gene. Vaccine 24, 6647–6650. [DOI] [PubMed] [Google Scholar]
- Dos Santos Afonso, E., Escriou, N., Leclercq, I., van der Werf, S. & Naffakh, N. (2005). The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5′ end of the PB2 segment. Virology 341, 34–46. [DOI] [PubMed] [Google Scholar]
- Flick, R. & Hobom, G. (1999). Transient bicistronic vRNA segments for indirect selection of recombinant influenza viruses. Virology 262, 93–103. [DOI] [PubMed] [Google Scholar]
- Fujii, Y., Goto, H., Watanabe, T., Yoshida, T. & Kawaoka, Y. (2003). Selective incorporation of influenza virus RNA segments into virions. Proc Natl Acad Sci U S A 100, 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, K., Fujii, Y., Noda, T., Muramoto, Y., Watanabe, T., Takada, A., Goto, H., Horimoto, T. & Kawaoka, Y. (2005). Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J Virol 79, 3766–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gog, J. R., Afonso Edos, S., Dalton, R. M., Leclercq, I., Tiley, L., Elton, D., von Kirchbach, J. C., Naffakh, N., Escriou, N. & Digard, P. (2007). Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res 35, 1897–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness, M. H., Scheid, A. & Choppin, P. W. (1980). Conformation of the helical nucleocapsids of paramyxoviruses and vesicular stomatitis virus: reversible coiling and uncoiling induced by changes in salt concentration. Proc Natl Acad Sci U S A 77, 2631–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson, E. C., Curran, M. D., Read, E. K., Gog, J. R. & Digard, P. (2008). Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J Virol 82, 11869–11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto, K., Horimoto, T., Fujii, Y. & Kawaoka, Y. (2004). Generation of influenza A virus NS2 (NEP) mutants with an altered nuclear export signal sequence. J Virol 78, 10149–10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury, D. W. & Darlington, R. W. (1968). Isolation and properties of Newcastle disease virus nucleocapsid. J Virol 2, 248–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp, K., Ruigrok, R. W. & Baudin, F. (1997). Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J 16, 1248–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Hong, Y. & Parslow, T. G. (2005). cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J Virol 79, 10348–10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y., Huang, T., Ly, H., Parslow, T. G. & Liang, Y. (2008). Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J Virol 82, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, S. & Kolakofsky, D. (1978). Ends of the RNA within Sendai virus defective interfering nucleocapsids are not free. J Virol 28, 584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado, A. V., Naffakh, N., van der Werf, S. & Escriou, N. (2003). Expression of a foreign gene by stable recombinant influenza viruses harboring a dicistronic genomic segment with an internal promoter. Virology 313, 235–249. [DOI] [PubMed] [Google Scholar]
- Marsh, G. A., Hatami, R. & Palese, P. (2007). Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J Virol 81, 9727–9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, G. A., Rabadan, R., Levine, A. J. & Palese, P. (2008). Highly conserved regions of influenza A virus polymerase gene segments are critical for efficient viral RNA packaging. J Virol 82, 2295–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch, D., Fellner, P. & Newton, C. (1976). Influenza virus genome consists of eight distinct RNA species. Proc Natl Acad Sci U S A 73, 3045–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto, Y., Takada, A., Fujii, K., Noda, T., Iwatsuki-Horimoto, K., Watanabe, S., Horimoto, T., Kida, H. & Kawaoka, Y. (2006). Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J Virol 80, 2318–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, G., Watanabe, T., Ito, H., Watanabe, S., Goto, H., Gao, P., Hughes, M., Perez, D. R., Donis, R. & other authors (1999). Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A 96, 9345–9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., Gerber, A. S. & Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri, T. & Tashiro, M. (1997). Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol 71, 2138–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, M., Fujii, K., Muramoto, Y., Yamada, S., Yamayoshi, S., Takada, A., Goto, H., Horimoto, T. & Kawaoka, Y. (2007). Contributions of two nuclear localization signals of influenza A virus nucleoprotein to viral replication. J Virol 81, 30–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa, M., Maeda, J., Iwatsuki-Horimoto, K., Watanabe, S., Goto, H., Horimoto, T. & Kawaoka, Y. (2009). Nucleotide sequence requirements at the 5′ end of the influenza A virus M RNA segment for efficient virus replication. J Virol 83, 3384–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese, P. & Shaw, M. L. (2007). Orthomyxoviridae: the viruses and their replication. In Fields Virology, 5th edn, pp. 1647–1689. Edited by D. M. Knipe & P. M. Howley. Philadelphia, PA: Lippincott Williams & Wilkins.
- Treanor, J. J., Snyder, M. H., London, W. T. & Murphy, B. R. (1989). The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology 171, 1–9. [DOI] [PubMed] [Google Scholar]
- Walter, A. E., Turner, D. H., Kim, J., Lyttle, M. H., Muller, P., Mathews, D. H. & Zuker, M. (1994). Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci U S A 91, 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Watanabe, S., Noda, T., Fujii, Y. & Kawaoka, Y. (2003). Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J Virol 77, 10575–10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (1989). On finding all suboptimal foldings of an RNA molecule. Science 244, 48–52. [DOI] [PubMed] [Google Scholar]