Abstract
AIM: To identify the distribution of hepatitis B virus (HBV) subgenotype and basal core promoter (BCP) mutations among patients with HBV-associated liver disease in Indonesia.
METHODS: Patients with chronic hepatitis (CH, n = 61), liver cirrhosis (LC, n = 62), and hepatocellular carcinoma (HCC, n = 48) were included in this study. HBV subgenotype was identified based on S or preS gene sequence, and mutations in the HBx gene including the overlapping BCP region were examined by direct sequencing.
RESULTS: HBV genotype B (subgenotypes B2, B3, B4, B5 and B7) the major genotype in the samples, accounted for 75.4%, 71.0% and 75.0% of CH, LC and HCC patients, respectively, while the genotype C (subgenotypes C1, C2 and C3) was detected in 24.6%, 29.0%, and 25.0% of CH, LC, and HCC patients, respectively. Subgenotypes B3 (84.9%) and C1 (82.2%) were the main subgenotype in HBV genotype B and C, respectively. Serotype adw2 (84.9%) and adrq+ (89.4%) were the most prevalent in HBV genotype B and C, respectively. Double mutation (A1762T/G1764A) in the BCP was significantly higher in LC (59.7%) and HCC (54.2%) than in CH (19.7%), suggesting that this mutation was associated with severity of liver disease. The T1753V was also higher in LC (46.8%), but lower in HCC (22.9%) and CH (18.0%), suggesting that this mutation may be an indicator of cirrhosis.
CONCLUSION: HBV genotype B/B3 and C/C1 are the major genotypes in Indonesia. Mutations in BCP, such as A1762T/G1764A and T1753V, might have an association with manifestations of liver disease.
Keywords: Basal core promoter mutation, Hepatitis B virus, Indonesia, Liver disease, Subgenotype
INTRODUCTION
Hepatitis B virus (HBV) infection is associated with a diverse clinical spectrum of liver damage ranging from asymptomatic carriers, chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (HCC)[1]. HBV, a member of the hepadnaviridae, is a relaxed circular double-stranded DNA virus, and is currently classified into 8 genotypes (A to H), which reflect its geographical distribution[2,3]. For instance, HBV genotype A is prevalent in Europe, Africa, and India[4]. HBV genotypes B and C are predominant in most parts of Asia, including China, Japan, and Indonesia[4–10]. Genotype D is common in the Mediterranean area, the Middle East and India, whereas genotype E is localized in sub-Saharan Africa[4,11–13]. Genotype F and H are only identified in Central and South America[4,14,15]. Genotype G has been found in France, Germany, and the United States[4,16–18].
Besides the differences in geographical distribution, there is growing evidence that the HBV genotype may also influence the clinical outcomes of liver disease. Among Asian patients who constitute approximately 75% of HBV carriers worldwide, it has been shown that HBV genotype C is more commonly associated with severe liver disease and the development of cirrhosis and HCC than HBV genotype B[19–23]. However, most of these studies were carried out in Taiwan and Japan, thus can not be generalized even for Asian countries.
In addition to HBV genotype, mutations in the core promoter, precore or HBx gene have been shown to have an association with severe liver disease. For instance, many studies have revealed that the double mutation in BCP (A1762T/G1764A) is associated with an increased risk of severe liver disease including HCC, and can be used as a prediagnostic biomarker of HCC[24–28]. The predominant mutation in the precore region of HBV which involved a G-to-A change at nucleotide 1896, and resulted in a premature stop codon at codon 28, was proved to be associated with increased HCC risk[23,28–30]. In addition, among HBV carriers, the A1762T/G1764A mutation is more frequently found in genotype C than genotype B[19,31]. However, an independent study on a comparison of HBV genotype C from Vietnam and Japan showed mutations at different positions in the core promoter/precore region of HBV[32], indicating that the effect of mutation on liver carcinogenesis may not be universal. In addition, some mutations in HBx protein, in particular for HBV genotype C, have been shown to be significantly associated with HCC. A Serine-to-Alanine mutation at codon 31 (S31A) in HBx protein[33], a Proline-to-Serine mutation at codon 38 (P38S) in HBx protein of HBV genotype C[34], and some other particular mutations in HBx protein were found to be associated with increased risk of HCC[35]. Those studies, however, were independently carried out in different countries (China Taiwan, Japan, and Korea), and resulted in three different results.
Despite various reports about the effect of HBV genotype and/or mutations on liver disease progression, the virological significance on liver carcinogenesis is not yet fully elucidated. In particular for Indonesia, some reports had been published regarding the distribution of HBV genotype[7–10,36], and only one study reported samples from CH, LC, and HCC[8]. Moreover, to the best of our knowledge there is no report on the distribution of BCP mutations and their possible association with clinical manifestations of liver disease. Thus, the aims of the present study were to identify the distribution of HBV genotype/subgenotype and BCP mutations in patients with different clinical status, and to investigate the association of HBV genotype/subgenotype or BCP mutations and liver disease progression in Indonesia.
MATERIALS AND METHODS
Samples
Serum samples were obtained from 171 patients with HBV-associated liver disease, comprising 61 CH patients (mean age 37.8 ± 13.0 years; male/female: 40/21), 62 LC patients (mean age 50.2 ± 11.6 years; male/female: 44/18), and 48 HCC patients (mean age 49.6 ± 10.4 years; male/female: 43/5). CH was defined as persistent seropositivity for HBsAg for at least 6 months. LC was diagnosed by liver function tests and ultrasonography. The diagnosis of HCC was on the basis of ultrasonography as well as an elevated serum α-fetoprotein (AFP) level (≥ 200 ng/mL), or liver biopsy samples by needle aspiration for samples in which the AFP level was low. Sera of CH, LC, and HCC patients were collected from Cipto Mangunkusumo Hospital, Gatot Soebroto Hospital, Klinik Hati, Jakarta, Siloam Hospital Lippo Karawaci, Tangerang, Mataram General Hospital, Mataram, and Wahidin Sudirohusodo Hospital, Makassar, from May 2006 until November 2008. All sera were hepatitis B surface antigen (HBsAg)-positive as determined by a commercially available enzyme-linked immunosorbent assay kit (Abbott Laboratories, Chicago, IL, USA). Blood samples were collected from each patient at the time of their clinical evaluation, separated into sera and stored at -70°C until viral DNA extraction. The study was approved by the Institutional Ethic Committee and informed consent was obtained from each patient.
Viral DNA extraction and PCR amplification
HBV DNA was extracted from 200 μL serum using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, and 80 μL of eluted DNA was stored at -70°C until use. Full S gene was amplified by PCR with primers Fgp2 and Rgp2 (Table 1). The following cycling parameters were used for 40 cycles of PCR: denaturation at 95°C (30 s), annealing at 55°C (45 s) and elongation at 72°C (2 min). When the PCR amplification was negative, a nested PCR was carried out to amplify the preS region. Primers HBPr134 and HBPr135 (Table 1) were used as previously described for the first-round 35 cycles of PCR by the following cycling parameters[2]: denaturation at 95°C (1 min), annealing at 48°C (30 s) and elongation at 72°C (1 min). The second-round PCR was then performed using primers HBPr94 and HBPr134 (Table 1) with the same conditions as the first-round PCR except for annealing at 56°C (30 s). Similarly, HBx gene was amplified using primers Fgp3 and Rgp3 (Table 1). The cycling parameters were the same as that for S gene amplification, except with an elongation time of 1 min. A nested PCR was performed for PCR negative samples using primers HB1 and HB3 for the first round PCR [35 cycles: denaturation at 95°C (1 min), annealing at 48°C (30 s) and elongation at 72°C (1 min)] and using primers HB2 and HB3 for the second round PCR with the same parameters as the first-round PCR, but the annealing temperature was 46°C, as described previously[37]. Both sets of primers could amplify the full HBx gene. All PCR reactions were carried out by the PCR Core System (Promega, Madison, WI, USA). The PCR products were visualized on 1% agarose gel stained with ethidium bromide and purified using Wizard® SV Gel and the PCR Clean-Up System (Promega, Madison, WI, USA).
Table 1.
Primers used in this study
| Primer | Nucleotide sequence (5’→3’) | Position | Polarity | Reference |
| Full S | ||||
| Fgp2 | CGCCATGGGAGGTTGGTCTTCCAAACCTCG | 2848-2873 | Forward | This study |
| Rgp2 | GACAAGCTTAATGTATACCCAAAGACAAAAGAAAATTGG | 803-835 | Reverse | |
| PreS (nested PCR) | ||||
| HBPr94 | GGTAAAAAGGGACTCACGATG | 775-795 | Reverse | [2] |
| HBPr134 | TGCTGCTATGCCTCATCTTC | 414-433 | Forward | |
| HBPr135 | CAAAGACAAAAGAAAAATTGC | 803-822 | Reverse | |
| HBx | ||||
| Fgp3 | CGCCATGGCTGCTAGGCTGTGCTGCCAAC | 1374-1398 | Forward | This study |
| Rgp3 | CGCTCGAGGGCAGAGGGGAAAAAGTTGCATGGT | 1811-1838 | Reverse | |
| HBx (nested PCR) | ||||
| HB1 | GCCAAGTGTTTGCTGACGC | 1175-1193 | Forward | [37] |
| HB2 | CCATACTGCGGAACTCCTAG | 1266-1285 | Forward | |
| HB3 | AAAGTTGCATGGTGCTGGTG | 1804-1823 | Reverse |
Analysis of HBV genotype/subgenotype, serotype and HBx mutations
Nucleotide sequences of the PCR fragments were determined with the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA, USA) and the appropriate primers, and sequenced with 3130xl DNA sequencer (Applied Biosystems). All HBs and HBx gene sequences were edited manually and were aligned with reference sequences retrieved from GenBank, using the ClustalW program incorporated in Bioedit v7.0. HBV genotypes/subgenotypes were determined based on the homology in the S or preS gene. Phylogenetic trees were constructed by the neighbor-joining method. HBV serotypes were deduced on the basis of predicted amino acid sequences of HBsAg[3,38,39].
Statistical analysis
All statistical analyses were performed using SPSS 15.0 software for Windows (SPSS Inc., Chicago, IL, USA) and MedCalc® version 10.1.0.0 for Windows (MedCalc Software, Broekstraat, Mariakerke, Belgium). Significance differentiations for continuous variables were analyzed using t-test analysis. While the categorical variables were analyzed using the Fisher’s exact test and chi-square test. P < 0.05 were considered significant.
RESULTS
HBV genotypes/subgenotypes distribution and clinical diagnosis
Only HBV genotype B and C were detected in the samples, which were respectively distributed in 73.7% and 26.3% of the samples (Table 2). Among HBV genotype B, subgenotypes B2, B3, B4, B5 and B7 were identified, although subgenotype B3 was the major subgenotype identified (84.9% of all genotype B or 62.6% of total samples). HBV subgenotypes B2 and B4 were only found in CH and LC, respectively. HBV subgenotype B5 was found in LC and HCC, while subgenotype B7 was detected in all different clinical diagnoses of the samples. On the other hand, among HBV genotype C, subgenotypes C1, C2, and C3 were found, but subgenotype C1 was dominant (82.62% of all genotype C or 21.6% of total samples). HBV subgenotype C1 was distributed in all samples, but subgenotype C2 and C3 were not detected in HCC samples. Based on statistical analysis, there was no significant association between HBV genotype/subgenotype and a clinical diagnosis of liver disease (Table 2). Serotype distribution demonstrated that adw2 was the major serotype (62.6%) in the samples, followed by adrq+ (24.6%) (Table 2). Other serotypes such as adw, adw3, ayw, ayw1, and ayr were also found in a small number of the samples. Similar to genotype results, no association between serotype and clinical status of the liver disease was observed (Table 2).
Table 2.
HBV genotype and serotype distribution in samples with different clinical diagnosis
| Characteristics |
n(%) in each clinical diagnosis |
||||||||||
| CH (n = 61) | LC (n = 62) |
HCC |
Total (n = 171) |
P |
|||||||
| With LC (n = 12) | Without LC (n = 36) | All HCC (n = 48) | CH vs LC | CH vs All HCC | LC vs All HCC | CH vs LC vs All HCC | |||||
| Genotype and subgenotype | |||||||||||
| B | B2 | 5 (8.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (2.9) | NS | NS | NA | 0.007 |
| B3 | 38 (62.3) | 41 (66.1) | 8 (66.7) | 20 (55.6) | 28 (58.3) | 107 (62.6) | NS | NS | NS | NS | |
| B4 | 0 (0.0) | 1 (1.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.6) | NS | NA | NS | NS | |
| B5 | 0 (0.0) | 0 (0.0) | 2 (16.7) | 3 (8.3) | 5 (10.4) | 5 (2.9) | NA | 0.034 | 0.032 | 0.002 | |
| B7 | 3 (4.9) | 2 (3.2) | 0 (0.0) | 3 (8.3) | 3 (6.3) | 8 (4.7) | NS | NS | NS | NS | |
| Total genotype B | 46 (75.4) | 44 (71.0) | 10 (83.3) | 26 (72.2) | 36 (75.0) | 126 (73.7) | NS | NS | NS | NS | |
| C | C1 | 12 (19.7) | 13 (21.0) | 2 (16.7) | 10 (27.8) | 12 (25.0) | 37 (21.6) | NS | NS | NS | NS |
| C2 | 1 (1.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.6) | NS | NS | NA | NS | |
| C3 | 2 (3.3) | 5 (8.1) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (4.1) | NS | NS | NS | NS | |
| Total genotype C | 15 (24.6) | 18 (29.0) | 2 (16.7) | 10 (27.8) | 12 (25.0) | 45 (26.3) | NS | NS | NS | NS | |
| No. HBV genotype B/C (%-B) | 46/15 (75.4) | 44/18 (71.0) | 10/2 (83.3) | 26/10 (72.2) | 36/12 (75.0) | 126/45 (73.7) | NS | NS | NS | NS | |
| Genotype and serotype | |||||||||||
| B | adw | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (2.8) | 1 (2.1) | 1 (0.6) | NA | NS | NS | NS |
| adw2 | 38 (62.3) | 39 (62.9) | 9 (75.0) | 21 (58.3) | 30 (62.5) | 107 (62.6) | NS | NS | NS | NS | |
| adw3 | 3 (4.9) | 1 (1.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (2.3) | NS | NS | NS | NS | |
| ayw | 0 (0.0) | 1 (1.6) | 0 (0.0) | 1 (2.8) | 1 (2.1) | 2 (1.2) | NS | NS | NS | NS | |
| ayw1 | 5 (8.2) | 3 (4.8) | 0 (0.0) | 4 (11.1) | 4 (8.3) | 12 (7.0) | NS | NS | NS | NS | |
| C | adrq+ | 14 (23.0) | 17 (27.4) | 2 (16.7) | 9 (25.0) | 11 (22.9) | 42 (24.6) | NS | NS | NS | NS |
| adw2 | 0 (0.0) | 1 (1.6) | 0 (0.0) | 1 (2.8) | 1 (2.1) | 2 (1.2) | NS | NS | NS | NS | |
| ayr | 1 (1.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.6) | NS | NS | NA | NS | |
CH: Chronic hepatitis; LC: Liver cirrhosis; HCC: Hepatocellular carcinoma; NS: Not significant; NA: Not applicable; HBV: Hepatitis B virus.
HBx and basal core promoter mutations
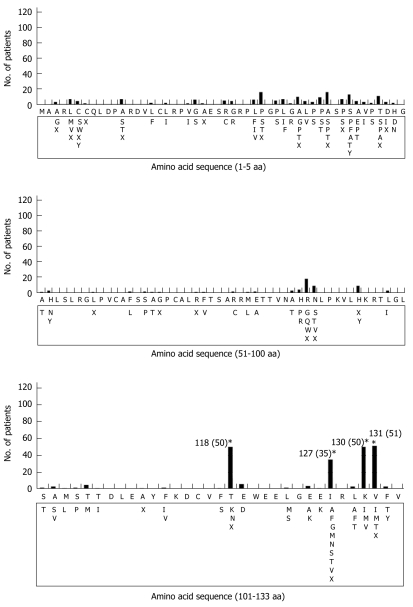
Initially, amino acid sequences of HBx from the samples were aligned and compared with reference sequences of amino acids retrieved from GenBank (accession no. BAA23459 and BAD86602 for HBV genotype B and C, respectively). Several amino acid changes were observed in both HBV genotype B and C. The prevalence of four amino acid substitutions (T118N, I127N/T/S, K130M and V131I) in HBV genotype B were significantly different between CH and LC, and three of them (T118N, K130M and V131I) showed a significant difference in prevalence between CH, LC and HCC, but none of them was significantly different between LC and HCC (Figure 1, Table 3). In contrast, none of the amino acid substitutions showed any significant difference in prevalence in the different clinical status in HBV genotype C (data not shown).
Figure 1.
Distribution and frequencies of the amino acid mutations in the 133 amino acids of HBx protein of HBV genotype B observed in the present study. Reference sequence of HBV genotype B (accession no. BAA23459) is shown at the top and mutations are shown below the reference sequence. Stars indicate the major substitutions observed and values in parentheses are number of patients with respective mutation.
Table 3.
Frequencies of some HBx mutations in HBV genotype B according to different clinical diagnosis
| Amino acid substitutions |
n(%) in each clinical diagnosis |
|||||||||
| CH (n = 46) | LC (n = 44) |
HCC |
Total (n = 126) |
P |
||||||
| With LC (n = 10) | Without LC (n = 26) | All HCC (n = 36) | CH vs LC | CH vs All HCC | LC vs All HCC | CH vs LC vs All HCC | ||||
| T118N | 23 (50.0) | 6 (13.6) | 4 (40.0) | 7 (26.9) | 11 (30.6) | 40 (31.7) | < 0.001 | NS | NS | 0.028 |
| I127N/T/S | 5 (10.9) | 16 (36.4) | 2 (20.0) | 2 (7.7) | 4 (11.1) | 25 (19.8) | 0.009 | NS | 0.019 | NS |
| K130M | 8 (17.4) | 23 (52.3) | 5 (50.0) | 11 (42.3) | 16 (44.4) | 47 (37.3) | 0.001 | 0.015 | NS | 0.006 |
| V131I | 8 (17.4) | 22 (50.0) | 4 (40.0) | 11 (42.3) | 15 (41.6) | 45 (35.7) | 0.002 | 0.030 | NS | 0.012 |
The four substituted amino acids located in BCP region, the corresponding nucleotides (C1726A/T1727(C/T) corresponding to T118N, T1753V corresponding to I127N/T/S, and A1762T/G1764A corresponding to K130M and V131I) were analyzed (Table 4). Mutations at positions 1762 and 1764 (corresponding to K130M and V131I amino acid substitutions), either as a double mutation or an independent mutation, were significantly higher in LC and HCC than CH. Particularly, the double mutation (A1762T/G1764A) which was found in 19.7%, 59.7% and 54.2% of CH, LC and HCC, respectively (P < 0.001). There was no significant difference in the prevalence of the double mutation between HCC with and without cirrhosis (41.7% and 58.3%). Analysis of the nucleotide at position 1753 showed that a T-to-V (A/G/C) mutation (corresponding to I127N/T/S amino acid substitutions) was significantly higher in LC (46.8%) compared with CH (18.0%) and HCC (22.9%) (P = 0.004), suggesting that this mutation could be an indicator of liver cirrhosis. Moreover, the prevalence of T1753V mutation was also not significantly different between HCC with cirrhosis (16.7%) and that without cirrhosis (25.0%) (data not shown).
Table 4.
Prevalence of HBx and core promoter mutations in samples with different clinical diagnosis
| Characteristics |
n(%) in each clinical diagnosis |
|||||||||
| CH (n = 61) | LC (n = 62) |
HCC |
Total (n = 171) |
P |
||||||
| With LC (n = 12) | Without LC (n = 36) | All HCC (n = 48) | CH vs LC | CH vs All HCC | LC vs All HCC | CH vs LC vs All HCC | ||||
| Genotype B/C (%B) | 46/15 (75.4) | 44/18 (71.0) | 10/2 (83.3) | 26/10 (72.2) | 36/12 (75.0) | 126/45 (73.7) | NS | NS | NS | NS |
| BCP mutations | ||||||||||
| C1726A/T1727(C/T) | 24 (39.3) | 8 (12.9) | 4 (33.3) | 7 (19.4) | 11 (22.9) | 43 (34.1) | 0.002 | NS | 0.015 | 0.003 |
| T1753V | 11 (18.0) | 29 (46.8) | 2 (16.7) | 9 (25.0) | 11 (22.9) | 51 (40.5) | 0.015 | NS | 0.018 | 0.004 |
| A1762T | 12 (19.7) | 38 (61.3) | 6 (50.0) | 21 (58.3) | 27 (56.3) | 77 (61.1) | < 0.001 | < 0.001 | NS | < 0.001 |
| G1764A | 13 (21.3) | 38 (61.3) | 5 (41.7) | 21 (58.3) | 26 (54.2) | 77 (61.1) | < 0.001 | 0.0002 | NS | < 0.001 |
| C1766T | 1 (1.6) | 3 (4.8) | 1 (8.3) | 1 (2.8) | 2 (4.2) | 6 (4.8) | NS | NS | NS | NS |
| T1768A | 1 (1.6) | 2 (3.2) | 1 (8.3) | 2 (5.6) | 3 (6.3) | 6 (4.8) | NS | NS | NS | NS |
| A1762T/G1764A | 12 (19.7) | 37 (59.7) | 5 (41.7) | 21 (58.3) | 26 (54.2) | 75 (59.5) | < 0.001 | 0.0004 | NS | < 0.001 |
In addition, C1726A/T1727 (C/T) mutations (corresponding to T118N substitution) were significantly higher in CH (39.3%) than in LC (12.9%) and HCC (22.9%). These results suggested that these mutations were reversely associated with severity of liver disease. In another words, single nucleotide polymorphisms (SNPs) in C1726/T1727 have an association with liver disease manifestations. The distribution of SNPs in 1726/1727 is shown in Table 5. In HBV genotype B, most of nucleotides in 1726 were A or C. The percentage of 1726A was significantly higher in CH (52.2%) than in LC (20.5%) and HCC (33.3%) (P = 0.009), while 1726C was more prevalent in LC (79.5%) and HCC (66.7%) than in CH (41.3%) (P = 0.001). On the other hand, most of the nucleotides in 1727 were T, however there was no significant difference in the percentage of 1727T in CH (95.7%), LC (88.6%), and HCC (97.2%) (P = 0.279). These results suggested that SNP in 1726, but not in 1727, of HBV genotype B was associated with the development of liver disease. In contrast, no association between SNP in the same positions in HBV genotype C and progression of liver disease was observed (Table 5).
Table 5.
Single nucleotide polymorphisms in 1726/1727 of HBV Genotype B and C
| SNP |
n(%) in each HBV genotype and clinical diagnosis |
|||||||||||
|
Genotype B |
Genotype C |
|||||||||||
| CH (n = 46) | LC (n = 44) |
HCC |
P | CH (n = 15) | LC (n = 18) |
HCC |
P | |||||
| With LC (n = 10) | Without LC (n = 26) | All HCC (n = 36) | With LC (n = 2) | Without LC (n = 10) | All HCC (n = 12) | |||||||
| 1726A | 24 (52.2) | 9 (20.5) | 4 (40.0) | 8 (30.8) | 11 (33.3) | 0.009 | 13 (86.7) | 13 (72.2) | 2 (100.0) | 7 (70.0) | 9 (75.0) | NS |
| 1726C | 19 (41.3) | 35 (79.5) | 6 (60.0) | 18 (69.2) | 24 (66.7) | 0.001 | 2 (13.3) | 5 (27.8) | 0 (0.0) | 3 (30.0) | 4 (33.3) | NS |
| 1726T | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| 1727A | 1 (2.2) | 2 (4.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 8 (53.3) | 11 (61.1) | 2 (100.0) | 2 (20.0) | 4 (33.3) | NS |
| 1727C | 0 (0.0) | 1 (2.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| 1727G | 1 (2.2) | 2 (4.5) | 0 (0.0) | 1 (3.8) | 1 (2.8) | NS | 4 (26.7) | 2 (11.1) | 0 (0.0) | 5 (50.0) | 5 (41.7) | NS |
| 1727T | 44 (95.7) | 39 (88.6) | 10 (100.0) | 25 (96.2) | 35 (97.2) | NS | 3 (20.0) | 5 (27.8) | 0 (0.0) | 3 (30.0) | 3 (25.0) | NS |
| 1726A/1727A | 1 (2.2) | 1 (2.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 7 (46.7) | 10 (55.6) | 2 (100.0) | 2 (20.0) | 4 (33.3) | NS |
| 1726A/1727G | 1 (2.2) | 1 (2.3) | 0 (0.0) | 1 (3.8) | 1 (2.8) | NS | 4 (26.7) | 2 (11.1) | 0 (0.0) | 5 (50.0) | 5 (41.7) | NS |
| 1726A/1727T | 22 (47.8) | 7 (15.9) | 4 (40.0) | 7 (26.9) | 11 (30.6) | 0.007 | 2 (13.3) | 1 (5.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS |
| 1726C/1727A | 0 (0.0) | 1 (2.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 1 (6.7) | 1 (5.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS |
| 1726C/1727G | 0 (0.0) | 1 (2.3) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NS | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | NA |
| 1726C/1727T | 19 (41.3) | 32 (72.7) | 6 (60.0) | 18 (69.2) | 24 (66.7) | 0.006 | 1 (6.7) | 4 (22.2) | 0 (0.0) | 3 (30.0) | 3 (25.0) | NS |
Comparison of BCP and HBx mutations between genotype B and C
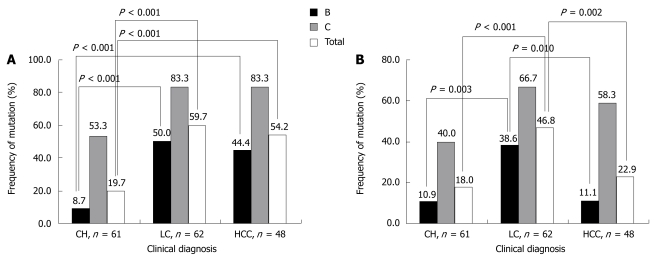
The percentage of cases with A1762T/G1764A mutation was significantly higher in genotype C than genotype B, regardless of clinical status: 53.3% vs 8.7% in CH, 83.3% vs 50.0% in LC, and 83.3% vs 45.5% in HCC (Figure 2A). From an analysis of total samples of these two genotypes, it was shown that the percentage of A1762T/G1764A mutation in genotype C was higher than that of genotype B (73.3% vs 33.3%, P < 0.001). Similar to the results of A1762T/G1764A mutation, T1753V mutation also showed a significantly different distribution between genotypes C (55.6%) and B (20.6%) with P < 0.001 (Table 6 and Figure 2B). When T1753V mutation was observed in each clinical status, its prevalence in HBV genotype C and B were 40.0% vs 10.9% in CH, 66.7% vs 38.6% in LC, and 58.3% vs 11.1% in HCC. In contrast, C1726A/T1727(C/T) mutation was more frequent in HBV genotype B (31.7%) than genotype C (6.7%) (P = 0.002).
Figure 2.
The prevalence of A1762T/G1764A (A) and T1753V (B) mutations in the samples with different clinical diagnoses. The number on each histogram represents the percentage of each mutation in each group. P values between the groups are shown respectively.
Table 6.
Comparison of core promoter mutations in HBV genotype B and C
| Characteristics |
HBV genotype |
P | ||
| B | C | All | ||
| No. (%) of patients | 126 (73.7) | 45 (26.3) | 171 (100.0) | < 0.001 |
| Age (mean ± SD) | 44.3 ± 12.8 | 49.4 ± 13.3 | 45.6 ± 13.1 | 0.024 |
| Male/Female (%Male) | 95/31 (75.4) | 32/13 (71.1) | 127/44 (74.3) | NS |
| No. (%) of A1762T/G1764A | 42 (33.3) | 33 (73.3) | 75 (43.9) | < 0.001 |
| No. (%) of T1753V | 26 (20.6) | 25 (55.6) | 51 (29.8) | < 0.001 |
| No. (%) of C1726A/T1727 (C/T) | 40 (31.7) | 3 (6.7) | 43 (25.1) | 0.002 |
DISCUSSION
Identification of viral as well as host factors associated with the development of severe liver disease including HCC may have important clinical implications in the management of patients with HBV infection. Many studies have suggested that HBV genotype might play an important role in the development of severe liver diseases. However, it is also widely accepted that HBV genotypes appear to show varying geographic patterns in their distribution which means the association between HBV genotype and the severity of liver disease may differ from one region to another. For instance, studies from Taiwan and Japan demonstrated that HBV genotype C is associated more with severe liver disease that HBV genotype B[19–23]. Since HBV genotype B and C are the main genotypes transmitted in these areas, the investigators compared between genotype B and C. However, another study from Alaska showed that there was a significant association between HBV genotype F and the development of HCC among native Alaskan people[40]. This means that the association between HBV genotype and severity of liver disease could be different, depending on the area and the genotype of HBV transmitted in that particular area. From this study, a cross-sectional analysis of subjects from several different centers in Indonesia, it was found that HBV genotype B was the major genotype and no association between HBV genotype as well as serotype and clinical status of liver disease was observed. Analysis of data from a prospective cohort study, however, is needed to further elucidate the association between HBV genotype and manifestations of liver disease in Indonesian HBV carriers.
Many studies have demonstrated that virus mutations, including mutations of HBx protein, BCP, and precore are linked to the severity and outcome of HBV infection. A study from Taiwan reported that the amino acid substitution at codon 31 of HBx protein (S31A) was frequently found in HCC patients and was predicted to have an association with HCC development[33]. A Japanese group also reported that a mutation at codon 38 (P38S) of HBV genotype C was associated with HCC development[34]. Recently, it was reported that mutations in HBx protein (V5M/L, P38S, H94Y, I127T/N, K130M and V131I) from Korean patients are linked with severity of liver disease[35]. HBx protein analysis of samples in the present study showed that I127N/T/S, K130M and V131I amino acid substitutions are associated with severe liver disease, especially with liver cirrhosis (Table 3). However, no association between S31A as well as P38S mutations and liver disease progression was found, which is different from previous studies in Taiwan, Japan, and Korea[33–35].
It is well known that the double mutation (A1762T/G1764A) in BCP is associated with an increased risk of liver disease. For instance, the frequency of double mutation (A1762T/G1764A) increased with advancing clinical status in Taiwanese patients [3%, 11%, 32% and 64% in asymptomatic carriers (AC), LC, CH, and HCC groups, respectively][24]. A recent report from China has also demonstrated that the incidence of double mutation increased along with the progression of liver disease; the percentage of the double mutation was 33%, 56% and 85% in CH, LC, and HCC groups, respectively[31]. In Indonesian patients, however, the A1762T/G1764A double mutation was increased in CH from 19.7% to 59.7% in LC and was slightly decreased in HCC (54.2%) (Table 4). These results suggest that the double mutation is associated with severe liver disease. In addition, analysis of the nucleotide at position 1753 showed that a T-to-V (A/G/C) mutation increased to 46.8% in LC from 18.0% in CH, but dramatically decreased in HCC (22.9%) (Table 4), suggesting that this mutation is associated with liver cirrhosis rather than HCC. In contrast, analysis of sera or plasma from Japanese subjects with AC, CH, LC and HCC infected with HBV genotype C showed that the percentage of T1753V mutation increased with progression of liver disease[41]. It is also reported that T1753V mutation was higher in HCC (53.2%) compared with LC (18.8%) and CH (9.8%)[31]. These results were inconsistent with the present study, particularly in LC and HCC. These discrepancies might be associated with HCC status; most of HCC cases in the present study were without cirrhosis. Another possibility is that most of the samples analyzed in the previous reports were HBV genotype C, whereas most of samples in the present study were HBV genotype B.
The most interesting finding of the present study is the association of SNP at position 1726 of HBV genotype B, but not genotype C, and severity of liver disease. Since HBV genotype B is the major genotype in Indonesia, this finding is important for the management and prevention of HBV carriers from developing more advanced disease such as liver cirrhosis and HCC in Indonesia. This association, however, has to be confirmed by analyzing more samples. A comparison of mutation prevalence between HBV genotype B and C showed that the percentage of T1753V and A1762T/G1764A mutations were higher in genotype C than in genotype B (Table 6). These results are in accordance with previous findings from Taiwan and China[20,31].
In summary, the present study demonstrated that HBV genotype B and C were detected among HBV-associated liver disease patients in Indonesia, and genotype B was predominant. It was found that HBV genotype, as well as the serotype, might not be associated with an increased risk of HCC. The A1762T/G1764A and T1753V mutations in BCP can be used as an indicator for progression of liver disease in Indonesian patients.
COMMENTS
Background
Hepatitis B virus (HBV) genotype, mutations in the core promoter, precore or HBx gene have been shown to have an association with severe liver disease. The aims of the study were to identify the distribution of HBV subgenotype and basal core promoter (BCP) mutations among patients with HBV-associated liver disease in Indonesia, and analyze the possible association between HBV genotype and/or BCP mutations and severity of liver disease among Indonesian patients.
Research frontiers
Although there were some reports on the distribution of HBV genotype in Indonesia, the association between HBV genotype and/or BCP mutations and liver disease progression has not been investigated. Therefore it is important to have information not only related to the distribution of HBV genotype/subgenotype and BCP mutations in patients with different clinical status, but also the association of HBV genotype/subgenotype and/or BCP mutations and liver disease progression in Indonesia.
Innovations and breakthroughs
The present study demonstrated that only HBV genotype B and C were detected among HBV-associated liver disease patients in Indonesia, and genotype B was predominant. It was found that HBV genotype, as well as the serotype, might not be associated with an increased risk of hepatocellular carcinoma (HCC). The double mutation (A1762T/G1764A) was associated with progression of liver disease, while T1753V mutation could be used as an indicator of liver cirrhosis rather than HCC. In addition, SNP in 1726 has an association with manifestations of liver disease.
Applications
The double mutation (A1762T/G1764A) can be used for the prediction of severe liver disease including cirrhosis and HCC, whereas the T1753V mutation is a predictor of liver cirrhosis in Indonesian patients. In addition, SNP in 1726 can also be used for the prediction of liver disease severity.
Terminology
HBs; HBs gene encode the surface protein of HBV that consist of preS1, preS2, and S. HBx; HBx gene encode functional X protein. BCP; BCP can be considered a part of HBx gene that regulate the core gene expression.
Peer review
The study provides a identify HBV subgenotype and basal core promoter (BCP) mutations distribution among HBV-associated liver disease patients in Indonesia. The work is of theoretical and practical importance.
Acknowledgments
The authors appreciate the assistance of Ivan Stevanus Chandra, Griskalia Christine, and Shinta Soraya in sample collection, and Theresia Imelda Octavia in technical assistance. The authors also appreciate Dr. David Vaux (La Trobe University, Australia) for critical reading.
Supported by MRIN Funding, Budget No. cc041/2007 and cc041/2008
Peer reviewer: Vasiliy I Reshetnyak, MD, PhD, Professor, Scientist Secretary of the Scientific Research Institute of General Reanimatology, 25-2, Petrovka Str., 107031, Moscow, Russia
S- Editor Tian L L- Editor Webster JR E- Editor Ma WH
References
- 1.Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 2.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, Rossau R. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 3.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 4.Liu WC, Phiet PH, Chiang TY, Sun KT, Hung KH, Young KC, Wu IC, Cheng PN, Chang TT. Five subgenotypes of hepatitis B virus genotype B with distinct geographic and virological characteristics. Virus Res. 2007;129:212–223. doi: 10.1016/j.virusres.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 5.Du H, Li T, Zhang HY, He ZP, Dong QM, Duan XZ, Zhuang H. Correlation of hepatitis B virus (HBV) genotypes and mutations in basal core promoter/precore with clinical features of chronic HBV infection. Liver Int. 2007;27:240–246. doi: 10.1111/j.1478-3231.2006.01400.x. [DOI] [PubMed] [Google Scholar]
- 6.Orito E, Ichida T, Sakugawa H, Sata M, Horiike N, Hino K, Okita K, Okanoue T, Iino S, Tanaka E, et al. Geographic distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology. 2001;34:590–594. doi: 10.1053/jhep.2001.27221. [DOI] [PubMed] [Google Scholar]
- 7.Sastrosoewignjo RI, Sandjaja B, Okamoto H. Molecular epidemiology of hepatitis B virus in Indonesia. J Gastroenterol Hepatol. 1991;6:491–498. doi: 10.1111/j.1440-1746.1991.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 8.Lusida MI, Surayah , Sakugawa H, Nagano-Fujii M, Soetjipto , Mulyanto , Handajani R, Boediwarsono , Setiawan PB, Nidom CA, et al. Genotype and subtype analyses of hepatitis B virus (HBV) and possible co-infection of HBV and hepatitis C virus (HCV) or hepatitis D virus (HDV) in blood donors, patients with chronic liver disease and patients on hemodialysis in Surabaya, Indonesia. Microbiol Immunol. 2003;47:969–975. doi: 10.1111/j.1348-0421.2003.tb03457.x. [DOI] [PubMed] [Google Scholar]
- 9.Nurainy N, Muljono DH, Sudoyo H, Marzuki S. Genetic study of hepatitis B virus in Indonesia reveals a new subgenotype of genotype B in east Nusa Tenggara. Arch Virol. 2008;153:1057–1065. doi: 10.1007/s00705-008-0092-z. [DOI] [PubMed] [Google Scholar]
- 10.Mulyanto , Depamede SN, Surayah K, Tsuda F, Ichiyama K, Takahashi M, Okamoto H. A nationwide molecular epidemiological study on hepatitis B virus in Indonesia: identification of two novel subgenotypes, B8 and C7. Arch Virol. 2009;154:1047–1059. doi: 10.1007/s00705-009-0406-9. [DOI] [PubMed] [Google Scholar]
- 11.Zekri AR, Hafez MM, Mohamed NI, Hassan ZK, El-Sayed MH, Khaled MM, Mansour T. Hepatitis B virus (HBV) genotypes in Egyptian pediatric cancer patients with acute and chronic active HBV infection. Virol J. 2007;4:74. doi: 10.1186/1743-422X-4-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara K, Tanaka Y, Orito E, Ohno T, Kato T, Sugihara K, Hasegawa I, Sakurai M, Ito K, Ozasa A, et al. Distribution of HBV genotypes among HBV carriers in Benin:phylogenetic analysis and virological characteristics of HBV genotype E. World J Gastroenterol. 2005;11:6410–6415. doi: 10.3748/wjg.v11.i41.6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 14.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059–2073. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 15.Nakano T, Lu L, Hu X, Mizokami M, Orito E, Shapiro C, Hadler S, Robertson B. Characterization of hepatitis B virus genotypes among Yucpa Indians in Venezuela. J Gen Virol. 2001;82:359–365. doi: 10.1099/0022-1317-82-2-359. [DOI] [PubMed] [Google Scholar]
- 16.Deterding K, Constantinescu I, Nedelcu FD, Gervain J, Nemecek V, Srtunecky O, Vince A, Grgurevic I, Bielawski KP, Zalewska M, et al. Prevalence of HBV genotypes in Central and Eastern Europe. J Med Virol. 2008;80:1707–1711. doi: 10.1002/jmv.21294. [DOI] [PubMed] [Google Scholar]
- 17.Vieth S, Manegold C, Drosten C, Nippraschk T, Gunther S. Sequence and phylogenetic analysis of hepatitis B virus genotype G isolated in Germany. Virus Genes. 2002;24:153–156. doi: 10.1023/a:1014572600432. [DOI] [PubMed] [Google Scholar]
- 18.Chu CJ, Keeffe EB, Han SH, Perrillo RP, Min AD, Soldevila-Pico C, Carey W, Brown RS Jr, Luketic VA, Terrault N, et al. Hepatitis B virus genotypes in the United States: results of a nationwide study. Gastroenterology. 2003;125:444–451. doi: 10.1016/s0016-5085(03)00895-3. [DOI] [PubMed] [Google Scholar]
- 19.Orito E, Mizokami M, Sakugawa H, Michitaka K, Ishikawa K, Ichida T, Okanoue T, Yotsuyanagi H, Iino S. A case-control study for clinical and molecular biological differences between hepatitis B viruses of genotypes B and C. Japan HBV Genotype Research Group. Hepatology. 2001;33:218–223. doi: 10.1053/jhep.2001.20532. [DOI] [PubMed] [Google Scholar]
- 20.Kao JH. Hepatitis B virus genotypes and hepatocellular carcinoma in Taiwan. Intervirology. 2003;46:400–407. doi: 10.1159/000074999. [DOI] [PubMed] [Google Scholar]
- 21.Chan HL, Wong ML, Hui AY, Hung LC, Chan FK, Sung JJ. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J Clin Microbiol. 2003;41:1277–1279. doi: 10.1128/JCM.41.3.1277-1279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan HL, Hui AY, Wong ML, Tse AM, Hung LC, Wong VW, Sung JJ. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53:1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuen MF, Tanaka Y, Mizokami M, Yuen JC, Wong DK, Yuan HJ, Sum SM, Chan AO, Wong BC, Lai CL. Role of hepatitis B virus genotypes Ba and C, core promoter and precore mutations on hepatocellular carcinoma: a case control study. Carcinogenesis. 2004;25:1593–1598. doi: 10.1093/carcin/bgh172. [DOI] [PubMed] [Google Scholar]
- 24.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124:327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 25.Kuang SY, Jackson PE, Wang JB, Lu PX, Munoz A, Qian GS, Kensler TW, Groopman JD. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc Natl Acad Sci USA. 2004;101:3575–3580. doi: 10.1073/pnas.0308232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka Y, Mukaide M, Orito E, Yuen MF, Ito K, Kurbanov F, Sugauchi F, Asahina Y, Izumi N, Kato M, et al. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J Hepatol. 2006;45:646–653. doi: 10.1016/j.jhep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 27.Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258–1265. doi: 10.1086/502978. [DOI] [PubMed] [Google Scholar]
- 28.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 2007;27:1356–1363. doi: 10.1111/j.1478-3231.2007.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Precore/basal core promoter mutants and hepatitis B viral DNA levels as predictors for liver deaths and hepatocellular carcinoma. World J Gastroenterol. 2006;12:6620–6626. doi: 10.3748/wjg.v12.i41.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Mitri MS, Cassini R, Morsica G, Bagaglio S, Andreone P, Loggi E, Muratori P, Bernardi M. Virological analysis, genotypes and mutational patterns of the HBV precore/core gene in HBV/HCV-related hepatocellular carcinoma. J Viral Hepat. 2006;13:574–581. doi: 10.1111/j.1365-2893.2006.00726.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Tanaka Y, Huang Y, Kurbanov F, Chen J, Zeng G, Zhou B, Mizokami M, Hou J. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong BX, Yano Y, Seo Y, Phuong TM, Tanaka Y, Kato H, Miki A, Utsumi T, Azuma T, Trach NK, et al. Variations in the core promoter/pre-core region in HBV genotype C in Japanese and Northern Vietnamese patients. J Med Virol. 2007;79:1293–1304. doi: 10.1002/jmv.20934. [DOI] [PubMed] [Google Scholar]
- 33.Yeh CT, Shen CH, Tai DI, Chu CM, Liaw YF. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene. 2000;19:5213–5220. doi: 10.1038/sj.onc.1203903. [DOI] [PubMed] [Google Scholar]
- 34.Muroyama R, Kato N, Yoshida H, Otsuka M, Moriyama M, Wang Y, Shao RX, Dharel N, Tanaka Y, Ohta M, et al. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45:805–812. doi: 10.1016/j.jhep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 35.Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 36.Lusida MI, Nugrahaputra VE, Soetjipto , Handajani R, Nagano-Fujii M, Sasayama M, Utsumi T, Hotta H. Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. J Clin Microbiol. 2008;46:2160–2166. doi: 10.1128/JCM.01681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, Jin Y, Qian G, Tu H. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol. 2008;49:718–725. doi: 10.1016/j.jhep.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 38.Norder H, Courouce AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, Robertson BH, Locarnini S, Magnius LO. Genetic diversity of hepatitis B virus strains derived worldwide: genotypes, subgenotypes, and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 39.Purdy MA, Talekar G, Swenson P, Araujo A, Fields H. A new algorithm for deduction of hepatitis B surface antigen subtype determinants from the amino acid sequence. Intervirology. 2007;50:45–51. doi: 10.1159/000096312. [DOI] [PubMed] [Google Scholar]
- 40.Livingston SE, Simonetti JP, McMahon BJ, Bulkow LR, Hurlburt KJ, Homan CE, Snowball MM, Cagle HH, Williams JL, Chulanov VP. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195:5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi K, Ohta Y, Kanai K, Akahane Y, Iwasa Y, Hino K, Ohno N, Yoshizawa H, Mishiro S. Clinical implications of mutations C-to-T1653 and T-to-C/A/G1753 of hepatitis B virus genotype C genome in chronic liver disease. Arch Virol. 1999;144:1299–1308. doi: 10.1007/s007050050588. [DOI] [PubMed] [Google Scholar]