Abstract
Data analysis in MRI usually entails a series of processing procedures. One of these procedures is noise assessment, which in the context of this work, includes both the identification of noise-only pixels and the estimation of noise variance (standard deviation). Although noise assessment is critical to many MRI processing techniques, the identification of noise-only pixels has received less attention than has the estimation of noise variance. The main objectives of this paper are therefore to demonstrate (a) that the identification of noise-only pixels has an important role to play in the analysis of MRI data, (b) that the identification of noise-only pixels and the estimation of noise variance can be combined into a coherent framework, and (c) that this framework can be made self-consistent. To this end, we propose a novel iterative approach to simultaneously identify noise-only pixels and estimate the noise standard deviation from these identified pixels in a commonly used data structure in MRI. Experimental and simulated data were used to investigate the feasibility, the accuracy and the stability of the proposed technique.
Keywords: noise identification, noise variance estimation, Rayleigh distribution, Gamma distribution, MR noise
I. INTRODUCTION
Magnetic resonance imaging (MRI) (1) is a rapidly expanding field and a widely used medical imaging modality possessing many noninvasive and quantitative techniques capable of probing functional activity (2) as well as tissue morphology in the brain (3,4). Data analysis in MRI is sophisticated and can be thought of as a “pipeline” of closely connected processing and modeling steps.
Because noise in MRI data affects all subsequent steps in this pipeline, e.g., from noise reduction (5) and image registration (6) to techniques for breaking the noise floor (7), parametric tensor estimation (8-11) and error propagation (12-17), accurate noise assessment has an important role in MRI studies.
Noise assessment in MRI usually means the estimation of Gaussian noise variance (or standard deviation (SD)) alone (18-23). Previously proposed methods for the estimation of Gaussian noise SD can be separated into two groups. In the first group, the Gaussian noise SD is estimated from a manually selected region-of-interest (ROI), while in the second group, it is estimated from an entire image or a volumetric data set automatically without human intervention.
A major problem faced by the first group of manual methods is lack of reproducibility of the results. The second group of automatic methods overcame this problem by bringing objectivity into the estimation process so that the Gaussian noise SD can be estimated without human input and that the results obtained can be reproduced. However, the most critical problem facing current automatic methods is the separation of pure noise from noisy signals and other artifacts because the way in which the automatic methods of Sijbers et al.(22) and of Chang et al. (21) work is by lumping the values of all the pixels from an entire image or from an entire volumetric data set into a one-dimensional array and then estimating the Gaussian noise SD from the histogram of this one-dimensional array. Complicated criteria and techniques have been developed by Sijbers et al.(22) and Chang et al. (21) to separate pure noise from noisy signals and other artifacts from the histogram alone. In this work, we introduce a simpler paradigm for performing noise assessment in MRI. The proposed method shows improved performance compared to previous methods, and may have application in other scientific and technological areas as well.
One of the major aims of this paradigm is to help us get out of the “one-dimensional” predicament faced by the automatic methods of Sijbers et al. and Chang et al. so that the separation of pure noise from noisy signals and other artifacts can be done more cleanly and simply. A moment of reflection will indicate that the identification of noise-only pixels should be a part of the paradigm in order to enhance the performance and accuracy of the estimation process, but the identification of noise-only pixels entails some a priori knowledge of the Gaussian noise SD. Therefore, any paradigm that attempts to make the identification of noise-only pixels a part of the overall noise assessment protocol will necessarily be iterative. Such a paradigm, if feasible, not only can improve the accuracy of the estimate of the Gaussian noise SD but also can provide spatial distributions of noise for further analysis or for quality control and calibration.
In this work, we will present one such paradigm. We will demonstrate that (a) the identification of noise-only pixels, which has not received much attention in MRI literature, is as important as—if not more important than—the estimation of Gaussian noise SD, (b) the identification of noise-only pixels and the estimation of Gaussian noise SD via the sample median (or the sample mean or other optimal sample quantiles, see Appendix B) can be combined into a single coherent framework of noise assessment, and (c) this framework can be made self-consistent, that is, it can be turned into a fixed-point iterative procedure.
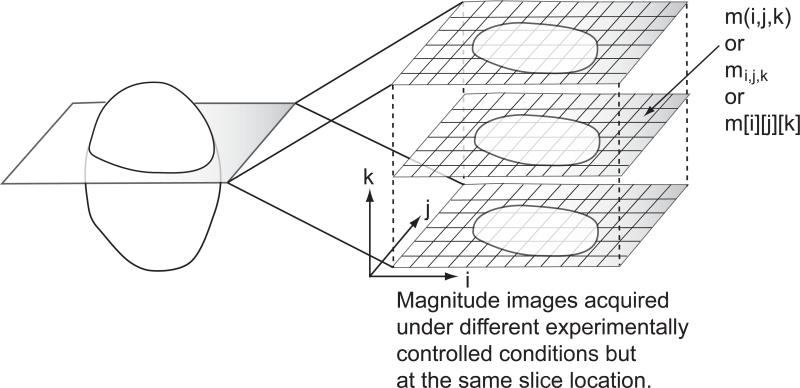
Briefly, we propose a novel approach to simultaneously identify noise-only pixels and estimate the Gaussian noise SD from a commonly used data structure (see Figure 1) in MRI. The data structure as shown in Figure 1 is ubiquitous in functional MRI and diffusion MRI. It is composed of a series of images acquired at the same physical (slice) location but not necessarily acquired under the same experimentally controlled conditions. Hereafter, we shall refer to the proposed technique as PIESNO, which stands for Probabilistic Identification and Estimation of Noise. PIESNO consists of two distinct parts that are connected dynamically in an iterative manner. The first part of PIESNO is the proposed probabilistic technique for identifying noise-only pixels, which is specifically formulated to deal with the data structure mentioned above. Here, it is assumed that the noise variance is uniform both within and across images in this data structure. The second part of PIESNO is the estimation of Gaussian noise SD via the sample median (or the sample mean or other optimal sample quantiles, see Appendix B), which will be outlined below.
Fig. 1.
The proposed data structure includes volumetric data composed of magnitude MR images that are acquired at the same slice location but not necessarily under identical experimentally controlled conditions.
The proposed probabilistic identification of noise-only pixels is designed to take advantage of this data structure to increase the discriminative power to identify noise-only pixels. Specifically, it identifies noise-only pixels through the distribution of the mean of a collection of measurements, shown as a vertical column of data along the k axis in Figure 1. Consequently, the discriminative power of the identification, which is related to the sharpness of the distribution of the mean, increases as the number of images within the proposed data structure increases. For completeness, we will show that the distribution of the mean used in this work is a Gamma distribution, which is well-known in MRI, e.g., see (24).
Our technique for estimating Gaussian noise SD is based on the median method but we also provide other methods of estimation based on the sample mean and optimal sample quantiles. The median method is a simple formula of the Gaussian noise SD expressed in terms of the sample median of a collection of noise-only measurements. We chose the sample median for its ease of use. The theoretical reason behind our choice of the sample median over other slightly more optimal methods based on the sample quantile of a specific order is explicated in Appendix B.
Both the identification of noise-only pixels and the estimation of noise SD are integral parts of the proposed framework on noise assessment because they are connected dynamically and iteratively to identify noise and estimate noise SD in a self-consistent manner.
Both experimental and simulated data were used to investigate the feasibility, accuracy, stability and global property of the proposed framework. Our approach managed to tease apart two noise distributions through a simple global analysis based on a well-know graphical technique in nonlinear dynamics known as Cobweb (25).
A comparison between our technique and Sijbers’ (22) (hereafter referred to as the Sijbers Method) was performed. Our technique demonstrated a lower mean squared error in estimating the Gaussian noise SD and a combined method based on both our technique and the Sijbers Method was found to be the most optimal when the number of images within the data structure was above five.
II. METHODS
A. Theoretical background
In this section, we will first provide the necessary details about the distribution of the arithmetic mean of K independent Gamma random variables, which is also a Gamma distribution, and then establish the connection between the proposed data structure and this well-known distribution by a few simple changes of variables. Throughout this section, we will use the same notation as employed in (26).
It is known that magnitude MR signals, m (or mi,j,k in Figure 1), reconstructed from the sum-of-squares algorithm through an N -receiver-coil MRI system (27) follow a nonCentral Chi, , distribution of 2N degrees of freedom with the non-centrality parameter given by . The probability density function (PDF) of is given by (23,26):
| [1] |
where the PDF is zero for m < 0, η is the underlying (combined) signal intensity, σg is the Gaussian noise SD, and Ik is the kth-order modified Bessel function of the first kind. We should note that magnitude MR signals reconstructed from other parallel image reconstruction techniques may not follow nonCentral Chi distribution, see example the work of Dietrich et al. (28). Note that when N = 1, Eq.(1) reduces to the Rician PDF (29). In the context of MRI, the Rician PDF was first used by Henkelman (19) and Bernstein et al. (20). Later, it was popularized by Gudbjartsson and Patz (30). In what follows and for lack of a better name, we shall use the PDF of magnitude noise to refer to the PDF of magnitude MR signals in the absence of the underlying signal, i.e., η = 0.
In this work, the PDF of magnitude noise (i.e., η = 0) is more relevant than the PDF of magnitude signal (i.e., η ≠ 0). Therefore, we will derive the PDF of magnitude noise from Eq.[1] by setting the underlying signal to zero, i.e., η = 0. It can be shown that the PDF of magnitude noise is given by:
| [2] |
after IN–1 in Eq.[1] is replaced by its first term Taylor expansion about η = 0, which is . Note that when N = 1, Eq.[2] reduces to the Rayleigh PDF.
By a change of variables from m to (or ), it can be shown that t follows a particular form of the Gamma PDF:
| [3] |
where the Gamma PDF, fγ, is defined as (31):
| [4] |
and Γ is the Gamma function.
B. Sampling distribution of the mean of several observed m-values
The measurements mi,j,k's are drawn from the distribution defined in Eq.[2] and any statistic computed from a set of mi,j,k's will have to be drawn from a sampling distribution for that statistic. For simplicity, we have chosen the arithmetic mean as our statistic for the identification of noise-only pixels. Therefore, if one knows the distribution for the mean, which necessarily depends on σg, one can decide for any observed mean if that particular sample came from the noise-only distribution. In what follows we will provide the expression for the arithmetic of mean of the squares of mi,j,k's through the method of Characteristic function. The complete derivation is shown in Appendix A.
In short, the new random variable si,j representing the arithmetic mean of K independent Gamma random variables, {ti,j,1,ti,j,2,ħ,ti,j,K }, is given by
| [5] |
Further, the new random variable si,j related to K independent magnitude MR measurements, {mi,j,1, mi,j,2,ħ, mi,j,K}, drawn from the distribution of magnitude noise is given by:
| [6] |
The derivation in Appendix A uses only a few simple substitutions of variables, which takes advantage of the reproductive property of the Gamma distribution (32). Interested readers may find it interesting to compare the derivation presented in (24).
C. Probabilistic Identification of Noise-only Pixels
In order to probabilistically identify the noise-only pixels we need to define an upper and a lower threshold on s so that a given percentage of all noise-only voxels fall between these values. In order to do so we will need to derive the cumulative distribution function (CDF), and its inverse, of the distribution of s.
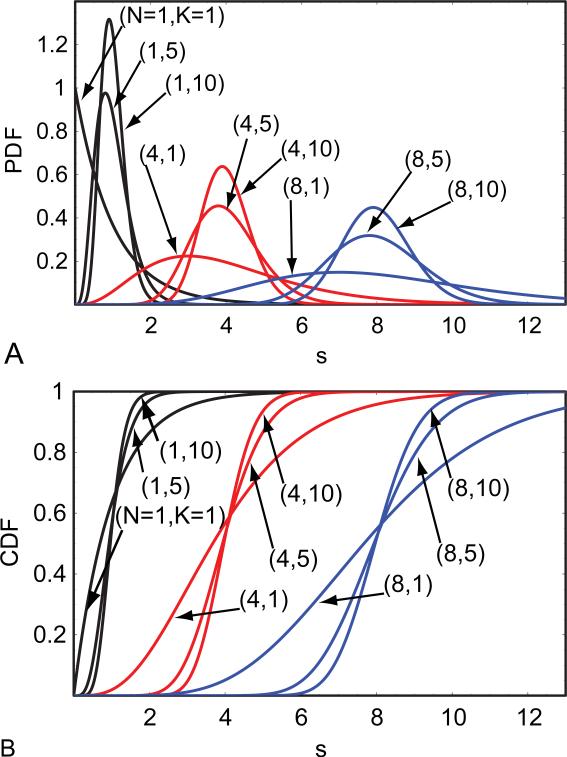
We will first define the CDF of s and its inverse to specify the lower and upper threshold values of s. A few CDF's of s are shown in Figure 2. We also assume that N and K are known. Additionally, the initial estimate of σg is required, which can be obtained automatically by the simple search method outlined below.
Fig. 2.
(A) PDF's and (B) CDF's of the mean of Gamma random variables with respect to different values of K and N.
Let the CDF of s with probability α be denoted by and its inverse, the inverse CDF of s, be denoted symbolically by . Note that the CDF of s and its inverse are related to each other by and . Note also that is given by Inverse Gamma Regularized [NK,1-α]/K in Mathematica (33).
The lower and upper threshold values of s, λ− and λ+, with probabilities respectively, α/2 and 1– (α/2), can be expressed in terms of the inverse CDF of s. Symbolically, they are given by and .
Thus, a collection of K independent magnitude MR signals {mi,j,1, mi,j,2,ħ, mi,j,K} is judged to contain only noise if satisfies the inequalities, λ− ≤ si,j ≤ λ+.
The test proposed here is a two-sided test. We should also mention that a one-sided test may be used instead of the two-sided test. But, the estimation step via the median or the quantile methods will no longer be as simple as the one presented below. In short, the median and the quantile methods will have be corrected for bias if these methods are to be used with a one-sided test.
D. Estimation of noise via the sample median, the sample mean or optimal quantiles
It is well known that the estimation of Gaussian noise SD can be obtained from the method of moments. For convenience, we provide here the method for estimating the Gaussian noise SD from the sample mean, denoted by <m>, of a collection of measurements m's (18,23,26):
| [7] |
where and the double factorial function is defined as n!!=n×(n- 2)×....
We should point out that the Gaussian noise SD, σg, can also be estimated from the sample quantile of a unique optimal order α* (Appendix B):
| [8] |
where qα* is the quantile of optimal order α*, see Table 1. Interestingly, these optimal quantiles approach the sample median as N increases, in the sense as described in Appendix B. A well known but slightly less optimal sample quantile is of course the sample median (i.e., α = 1/2), denoted by μ ≡ q1/2. Specifically, σg can be expressed as a function of both μ and N as follows:
| [9] |
Note that both the sample median and the median of the continuous PDF, Eq.[2], are denoted by the same symbol. Note also that the denominator of Eq.[9] can be computed in advance, see Table 2. Note also that when N = 1, Eq.[9] can be expressed analytically as .
Table 1.
The value of the optimal quantile order for different N and the value of the denominator of Eq.[8] for different N but at the optimal quantile order.
| N | Optimal quantile order α* | (2 Ps−1 ( α*| N,1))1/2 |
|---|---|---|
| 1 | 0.7968 | 1.7853 |
| 2 | 0.7306 | 2.2759 |
| 4 | 0.6722 | 3.0289 |
| 8 | 0.6254 | 4.1438 |
| 16 | 0.5900 | 5.7593 |
| 32 | 0.5642 | 8.0727 |
| 64 | 0.5456 | 11.3652 |
| 128 | 0.5323 | 16.0365 |
Table 2.
The value of the denominator of Eq.[9] for various N.
| N | (2 Ps−1(1/2| N,1))1/2 |
|---|---|
| 1 | 1.177410 |
| 2 | 1.832128 |
| 4 | 2.710003 |
| 8 | 3.916439 |
| 16 | 5.597844 |
| 32 | 7.958302 |
| 64 | 11.28423 |
E. The proposed framework of noise assessment (PIESNO)
PIESNO is best described through a step-by-step procedure as shown in Figure 3.
Fig. 3.
A step-by-step procedure of PIESNO.
Step 1 is on the identification of noise-only pixels. In practice, this step should be carried out on each of the pixels before moving on to Step 2 because dynamically adding new elements into the one dimensional array, Ω, of Step 2 is inefficient. Note also that the summation in Step 1 should be computed in advance.
In Step 2, the link between the identification of noise-only pixels and the estimation of Gaussian noise SD is established. Here, a one-dimensional array, Ω, is created. It is the union (in set-theoretic language) of all the arrays of K measurements that have been identified positively in Step 1. Note that the number of positive identifications is defined to be the number of the positively identified arrays. Finally, the sample median of Ω, μ, is selected. Note again that the sample mean of Ω or the sample quantile of Ω of a specific order may be used here.
Step 3 is the estimation of noise SD. The sample median, μ, of Ω computed in Step 2 is used to estimate σg via Eq.[9]. Note again that the sample mean and other optimal quantiles as defined in respectively Eq.[7] and Eq.[8] may be used instead of Eq.[9].
In Step 4, the iteration stops if Ω is empty, otherwise it will move through Steps 1 to 4 sequentially until convergence or the maximum number of iterations is reached. Here, a sequence of estimates of σg produced by the iterative procedure is considered convergent if the absolute difference between any successive estimates is less than some small number, say 10−10.
Note that the accuracy of the estimation of noise is dependent upon how the elements of Ω are distributed, which, in turn, is determined by the lower and upper threshold values of how the identification of noise is performed. In short, the two areas excluded by the threshold values in the PDF of s should be equal in order to ensure accurate estimate of the Gaussian noise SD.
F. An automatic method for determining a good initial estimate
PIESNO can be made automatic by a systematic search for a good initial estimate of σg. This systematic search begins by finding an upper bound, M, of σg. Here, M is estimated from the whole volumetric data through Eq.[9] (or Eq.[7] or Eq.[8]) where μ is taken to be the sample median (or the sample mean or the sample quantitle of a specific order) of the whole volumetric data.
Next, the interval from 0 to M is subdivided to generate a set of points, Φ = {M/l, 2M/l,ħ,(l−1)M/l, M} where l is some positive integer, say 100. Each point in Φ then serves as an initial solution. The best initial solution is the one that produces the highest number of positive identifications.
G. Graphical analysis of global behavior of the iterative procedure
In the previous section, we adopted the strategy of using the total number of positive identifications as a means to find a good initial estimate. This strategy is not foolproof but it works in many MR applications because it is generally true that there is only one distribution of magnitude noise. In this section, we will present a general strategy based on a well-known graphical technique in nonlinear dynamics known as Cobweb for analyzing the global behavior of the proposed iterative procedure (25). This general strategy is able to detect multiple distributions of magnitude noise based on the number of attracting fixed points.
First, let us define a function, Π, based on the first three steps of PIESNO:
| [10] |
In essence, Π takes in some value of the Gaussian noise SD, σn, and returns the next value of the Gaussian noise SD, σn+1, without going through the iterative process. Similarly, we can define T as a function of σn to tally the positive identifications at σn. It is important to note that Π(σn) is defined to be zero if T(σn) is zero. It should be clear that we can also generate the binary mask for a given σn. A plot of these two functions, and of a unit slope curve can provide an intuitive and informative snapshot of the global behavior of PIESNO for a given data set, see Section III.K for the results.
III. RESULTS
H. Experimental data
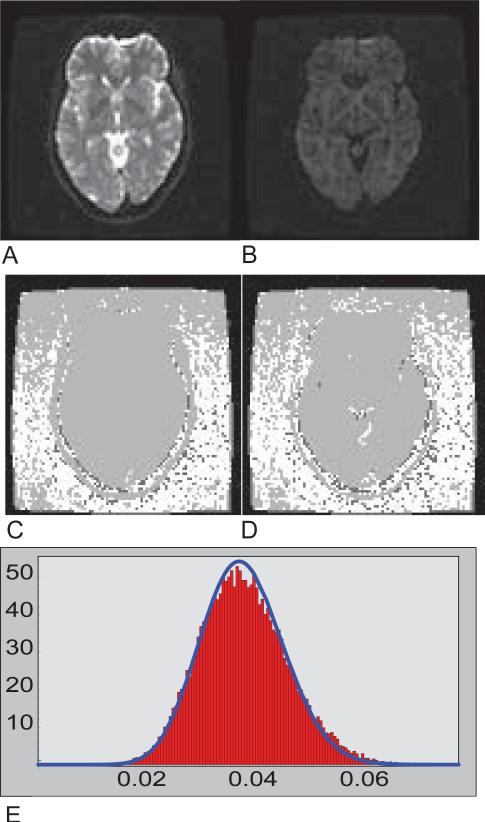
PIESNO was tested with a set of human brain data acquired on a 1.5 Tesla scanner (GE Medical Systems, Milwaukee, WI) based on the sum-of-squares reconstruction (27) with an 8-channel phased array coil, i.e., N = 8, using a single-shot spin-echo EPI sequence. Below are the specific experimental parameters: Field-Of-View (FOV) of 24cm × 24cm, 60 slices without gaps and with a slice thickness of 2.5mm, an image matrix of 96×96. Each diffusion weighted image (DWI) dataset consisted of 2 images without diffusion weighting (b=0 s/mm2) and 12 images with different gradient directions but with the same diffusion weighting of 1100 s/mm2, i.e., K = 14 at each slice location, see Figures 4A and 4B. If we set α to be 0.10, we have λ− = λ− = 6.798 and λ− = 9.282 for the case when K = 14. For this particular slice location, the initial estimate of σg was found to be 0.0106 through our automatic search method with l = 50. The final estimate of 0.0104 was obtained in 13 iterations in less than a second. Those regions that are classified as containing noise-only measurements are shown in white in Figure 4C.
Fig. 4.
(A) A T2-weighted image. (B) A diffusion-weighted image. (C) A quaternary image on the identification of noise. Positively identified pixels are white. In this case, the proposed method was applied to all 14 images (2 non diffusion-weighted + 12 diffusion-weighted images). Pixels in which the sij's are greater than the upper threshold are colored light gray. Pixels in which the sij's are nonzero but less than the lower threshold are colored dark gray. The black pixels belong to those sij's that are zero. (D) Another quaternary image on the identification of noise where the proposed method was applied to only the diffusion-weighted images. (E) The histogram of the noise array associated with (D) and the probability density function with N=8 and the Gaussian noise SD estimated from PIESNO.
Note that Figures 4C and 4D are quaternary images. Pixels that are classified as containing noise-only measurements are shown in white. Pixels whose sij's are greater than the upper threshold are shown in light gray. Pixels whose sij's are nonzero but less than the lower threshold are shown in dark gray, and finally, pixels whose sij's are zero are shown in black. The results of another simulation test where only the diffusion weighted images were analyzed are shown in Figure 4D and the histogram of the corresponding array of noise, Ω, is shown in Figure 4E. The probability density function of noise based the final estimate of the Gaussian noise SD produced by PIESNO is shown in Figure 4E. Further investigation shows that some regions excluded by PIESNO in the top of the field of view in both Figures 4C and 4D are due to low-level artifacts.
I. Numerical test using simulated data
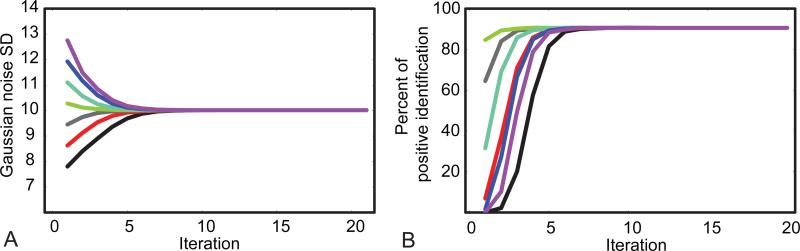
Simulated data were also used to gain qualitative insight into the numerical behavior of PIESNO. The automatic search method was not used in this test because the stability of PIESNO with respect to different initial estimates of σg was under investigation. Here, the same set of parameters was used, i.e., N = 8, K = 14, α = 0.10, λ− = 6.798 and λ− = 9.282, but the ground truth of σg, for simplicity, was set to 10 arbitrary units. We generated 5000 arrays of random samples with each array containing K = 14 random samples. Each random sample was generated according to the following expression, , where εi's and δi's are Gaussian distributed with mean zero and standard deviation of 10 arbitrary units. The results of this study are shown in Figure 5. Note that the number of positively identified regions was one out of 5000, when 7.805 and 12.75 were used as the initial estimates of σg. It is interesting to note that the proposed procedure converged to the result, which was close to the ground truth, even when it began with these estimates. Additionally, the final percent of positively identified samples was about 90.56% and the final estimate of σg was about 10.015. These results were independent of the initial solutions used.
Fig. 5.
(A) The estimated Gaussian noise SD as a function of the number of iterations with different initial solutions, i.e., 7.80, 8.62, 9.45, 10.27, 11.10, 11.92, 12.75, coded in color. (B) The percent of positive identification as a function of the number of iterations with the same set of initial solutions used in (A).
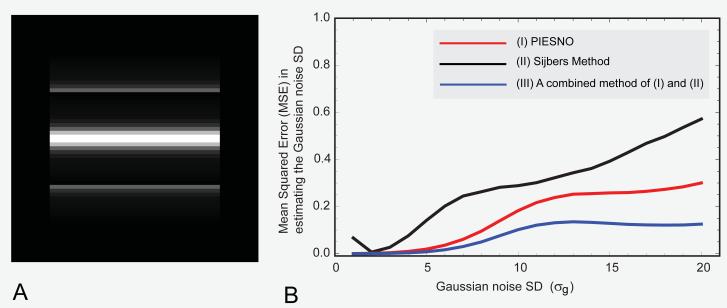
J. A comparison with Sijbers’ recently proposed technique using simulated data
A synthetic image matrix of 64 64 was constructed (Figure 6A) and used to compare the performance of three different methods, (I) PIESNO, (II) Sijbers’ recently proposed method (22) and (III) a method that combines both (I) and (II) by using one of the outputs of PIESNO, i.e., Ω, as an input to (II). The performance measure used was the mean squared error in estimating the Gaussian noise SD. Further, the performance of these methods in terms of average time needed to carry out a single estimation of noise variance was also evaluated.
Fig. 6.
(A) The ground truth image, a synthetic image with different signal intensities ranging from 0 to 260 (arbitrary units). (B) The mean squared error (in estimating the Gaussian noise SD) as a function of the Gaussian noise SD. Note that the mean squared error in estimating the Gaussian noise SD is computed from K=14 and 5000 repeated measurements (images) for each Gaussian noise SD on the horizontal axis.
Note that the following data structure, {{0, 2160}, {2, 176}, {4, 176}, {6, 176}, {8, 176}, {10, 176}, {12, 176}, {14, 176}, {28, 176}, {48, 176}, {96, 176}, {192, 88}, {260, 88}}, shows all distinct elements of our synthetic image together with their multiplicities, e.g., there are 2160 elements with null signal intensity, 176 elements with signal intensity of 2 (arbitrary unit), and so on.
For convenience, the same set of parameters was used, i.e., K = 14, α = 0.10, except for N, which in this case was 1 because the Sijbers method was specifically designed for dealing with the Rician/Rayleigh data. Therefore, we had λ− = 0.604 and λ+ = 1.476. For simplicity, the 14 images (K = 14) were drawn from repeated measurements.
In this comparison, 5000 volumetric data sets each of which contains 64 64 14 random samples were generated for each value of σg based on the sum-of-squares algorithm. The values of σg range from 1 to 20 in a uniform step of 1. Each volumetric data set served as an input to both PIESNO and the Sijbers method. The initial number of bins for the Sijbers method was 1200 but it is well known that one of the advantages of the Sijbers method is the ability to find an optimal number of bins. The results are shown in Figures 6B. PIESNO achieved lower mean squared error in estimating the Gaussian noise SD than did the Sijbers method. In addition, it is clear that PIESNO can improve the performance of the Sijbers method (or other methods of noise variance estimation) when the noise ensemble, Ω, is used as an input to the Sijbers method.
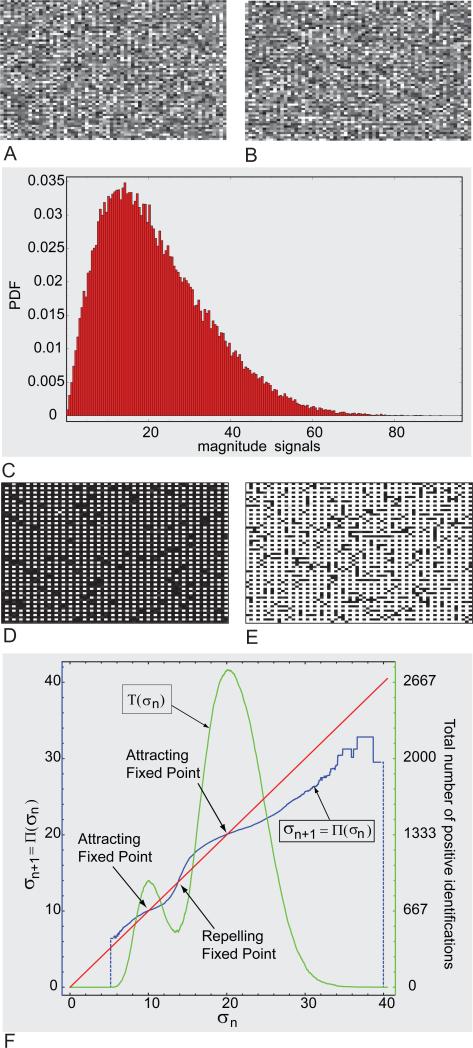
K. Graphical analysis of global behavior of PIESNO using a simulated data set
A synthetic volumetric data of 64 64 16 was constructed. Each of the 16 images was created with random numbers drawn from two Rayleigh distributions, σg = 10 and σg = 20, based on the following rule. If both indices of a pixel location, (i,j), are even number, then a random number drawn from Rayleigh distribution of σg = 10 will be placed on this pixel location; otherwise, a random number drawn from the other Rayleigh distribution will be placed on this pixel location. Figures 7A and 7B are representative of the noisy images constructed using the rule mentioned above. Figure 7C is a histogram of the whole volumetric data. Note that it is impossible to infer from this histogram that there are two (Rayleigh) distributions.
Figure 7.
(A-B)Two representative noisy images. (C) Histogram of the noisy volumetric data. (D-E) Binary masks indicating two spatial distributions of noise drawn from two different Rayleigh distribution. (F) Graphical analysis of PIESNO.
Based on the technique outlined in Section II.G, it is possible to detect multiple Rayleigh distributions through PIESNO. Figures 7D and 7E are binary masks computed at two different attracting fixed points shown in Figure 7F. In Figure 7F, it is clear that the function, T, which is the total number of positive identifications, peaks near the attracting fixed points. The nonzero trough of the function, T, seems to coincide with the repelling fixed points. The results showed that the number of attracting fixed points coincides with the number of clusters of noise with different probability distributions.
IV. DISCUSSION
Earlier approaches such as those of Sijbers et al. (22) and Chang et al. (21) are general and can be applied to any 3D volumetric data set. These approaches can also be applied to the particular 3D volumetric data considered in this paper.
In a typical functional or diffusion MRI data set, one usually has many images for a particular slice location and these images are usually acquired under slightly different experimentally-controlled conditions. The data structure considered in this work requires nothing more than a simple rearrangement of data and so is the data structure considered by Sijbers et al. and Chang et al. The data structure considered by them is a 3D volumetric data set consisted of distinct images across multiple slice locations. There is nothing special about the latter data structure or the data structure used in this paper because the basic assumption about the property of the Gaussian noise SD is the same. Namely, the Gaussian noise SD is assumed to be uniform either across multiple slice locations or across multiple images of the same location, e.g., if the readout bandwidth is maintained at the same level for all the images. Consequently, it is superfluous to regard the data structure used by Sijbers et al. and Chang et al. to be better or more appropriate than the data structure used in this work for estimating the Gaussian noise SD.
Our approach is designed for the particular 3D volumetric data mentioned above (multiple images acquired with different experimental conditions but at the same slice location) and cannot be applied to any 3D volumetric data. Our method takes advantage of this particular 3D volumetric data structure to increase the discriminative power of the identification of noise-only pixel. We should point out that our approach may not be optimal when K is small (less than 5). But, in high angular resolution diffusion MRI or functional MRI, K is usually greater than 5.
The initial motivation behind this work was simply to take advantage of the multiplicity of the images within the proposed data structure to increase the discriminative power of the identification of noise-only pixels, and to develop a simple and self-consistent framework of noise assessment that incorporates both the identification of noise-only pixels and the estimation of Gaussian noise SD.
This paper can be thought of as a sequel to but independent of our recent paper on a signal-transformational framework for breaking the noise floor in MRI and other imaging sciences (7). The present method for estimating the Gaussian noise SD and the technique proposed in (7) represent our major attempt to decouple the fixed point formula of SNR (26) into two self-consistent approaches for estimating the underlying signal and the Gaussian noise SD, respectively. The advantage of this decoupling is substantial because the estimation of the Gaussian noise SD can be obtained from a much larger collection of samples. As a consequence, the precision of the Gaussian noise SD estimate will be significantly increased, and in turn, the precision of the underlying signal intensity estimate will also be increased (7).
The proposed identification of noise-only pixels is a simple and highly discriminative method for identifying noise because its identification criterion entails a few simple arithmetic operations and its discriminative power is proportional to the number of images (i.e., K) within the proposed data structure. PIESNO can be viewed as a clustering algorithm for finding different noise-only regions. Note that the input parameters of PIESNO can be obtained readily, e.g., see (34) for a routine on the CDF of the Gamma distribution. Note also that its discriminative power decreases as the number of combined channels (i.e., N) increases, see Figure 2.
In this work, we use a quaternary image rather than a binary image to distinguish different types of pixel. In essence, a quaternary image can provide useful visual information about the four distinct decision-making regions (from the PDF) to which a particular sij belongs. For example, PIESNO seems to be able to exclude low-level artifacts in the top of the field of view in both Figures 4C and 4D. Interestingly, the values of these low-level artifacts are still higher than the upper threshold. Note that some pixels in dark gray are persistently distributed at the interface between the brain and the skull. The values of these pixels are nonzero but less than the lower threshold. The source of this peculiar phenomenon may be due to the use of the Fermi filter in the image reconstruction. Finally, regions excluded by PIESNO that are outside of the tissue region may provide useful information about spatial distribution of low-level artifacts.
The proposed estimation of Gaussian noise SD via the median method is an efficient and robust method for estimating the Gaussian noise SD because sample median can be found very efficiently, e.g., see (35); the sample median is a statistically robust measure, that is, robust against outliers. Furthermore, the sample mean or the other slightly more optimal quantiles developed in Appendix B may be used instead of the sample median.
The proposed framework of noise assessment, PIESNO, shows how both the identification of noise-only pixels and the estimation of Gaussian noise SD can be connected in a dynamic and iterative manner. PIESNO was shown to be beneficial to methods of estimation such as that of Sijbers and coworkers. That is, we showed that PIESNO can be combined with other noise variance estimation methods, e.g., (21,22,36), to achieve better performance in terms of the mean squared error in estimating the Gaussian noise SD. We remark that the proposed method is slightly more general than the maximum likelihood based approach proposed by Sijbers and coworkers (22) because our method is applicable to larger class of magnitude data than just Rayleigh-distributed data (i.e., N = 1).
PIESNO is general and can be adapted to other imaging sciences by using a different PDF and CDF of interest. An important application of PIESNO is the assessment of noise in the brain region. Specifically, PIESNO can be used to evaluate the quality of images acquired at high diffusion weighting through one of the high angular resolution diffusion imaging (HARDI) techniques (37-42). HARDI images (acquired at the same slice location and the same diffusion weighting, but with either different or the same gradient directions) can be arranged into the proposed data structure. These HARDI images can then be analyzed via PIESNO to determine whether a region of interest (ROI) has any pixels that contain only noise. It should be clear that this type of noise assessment is also applicable to fMRI images.
Here, we discuss some limitations of our approach. PIESNO cannot be expected to perform well for very small K because the identification of noise may be less stable and may not produce sufficient number of elements in Ω for the estimation of noise via the median method to be useful as an estimator. Nevertheless, one can still estimate the Gaussian noise SD from a region-of-interest (ROI) in the image background via the median method alone or the optimal quantile method developed in Appendix B or other classical formulae based on the mean or the standard deviation of the measurements in that ROI (19). Based on our experience with numerous simulation tests on the same simulated data used in Figure 6, PIESNO was found to perform well when K was greater than 5. At K=5, the mean squared error of PIESNO in estimating the Gaussian noise SD is slightly higher than the Sijbers method but the combined method remains the best method in terms of mean squared error in estimating the Gaussian noise SD. At present, we do not have a rigorous statement about the convergence of PIESNO but it has never failed to converge in all cases we tested. For some qualitative features the convergence of PIESNO, see Appendix C.
In conclusion, we have demonstrated that it is useful and logical to combine both the identification of noise-only pixels and the estimation of noise variance into a single coherent framework of noise assessment. This is an important paradigm for noise assessment in MRI and in other fields. We have also demonstrated the convergence of the proposed method and presented a simple graphical analysis for understanding global behavior of our iterative schemes.
ACKNOWLEDGMENTS
C.G. Koay dedicates this work to Felix Koay. He is very grateful to Drs. J. Sijbers and D. Poot for providing the Matlab software that implements (22), and to Dr. D. Poot for help with the software. The authors are grateful to the reviewers for very thoughtful and helpful comments, to Liz Salak for editing the paper, to Dr. Peter J. Basser for critically reading the paper, to Dr. Alan Barnett for his help in the pulse sequence, to Dr. Stefano Marenco for sharing the experimental data, and to Ms. Lindsay Walker and Dr. Lin-Ching Chang for help in image formatting. C.G. Koay was supported in part by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institute on Drug Abuse, the National Institute of Mental Health, and the National Institute of Neurological Disorders and Stroke as part of the NIH MRI Study of Normal Pediatric Brain Development with supplemental funding from the NIH Neuroscience Blueprint. PIESNO is freely available upon request (http://sites.google.com/site/piesnoformri/).
Appendix A
In a few steps, we will use the Characteristic functions (43) of fγ and pγ to derive the PDF of the arithmetic mean of K independent Gamma random variables with PDF fγ(t|N,1), Eq.[4]. First, the Characteristic function of fγ can be shown to be:
| [A1] |
where . Therefore, the Characteristic function of pγ ≡ fγ(t|N,1) is given by:
| [A2] |
Second, the Characteristic function of the PDF of the arithmetic mean of K independent Gamma random variables can be expressed as:
| [A3] |
| [A4] |
Thus, the PDF of the arithmetic mean of K independent Gamma random variables is a Gamma distribution given by:
| [A5] |
where s (or ) is a new random variable denoting the arithmetic mean of K independent Gamma random variables, (or ), with PDF fγ(t|N,K = 1). A few PDF's of s are shown in Figure 2.
Appendix B
The connection between the quantile of order α, denoted by qα, and σg of Eq.[2] is given by the definition of the quantile in a continuous PDF:
| [B1] |
Equation [B1] can also be expressed in terms of the CDF of s, Ps(λ|N,K) = α, by making these substitutions, K = 1, and :
| [B2] |
Equivalently, Eq.[B2] can be expressed in terms of the inverse CDF of s:
| [B3] |
Hence,
| [B4] |
Although a self-contained discussion on the theoretical justifications for using the sample median as an estimator of the Gaussian noise SD is beyond the scope of the paper, it is still useful to point out that the sample quantile of any order α (0<α<1) is a consistent asymptotically normal estimator with several desired properties, e.g., page 215 of Dudewicz (44). Further, the Gaussian noise SD can be estimated from quantiles of different orders as given in Eq.[A4]. It can be shown that the sample median, which is the sample quantile of order 0.5, has higher relative efficiency than most quantiles of other orders as N increases. We note that the expression for the expected variance of the quantile of order α is given by (44):
| [B5] |
Based on Eqs[B4,B5], it can be shown that the expected SD for estimating the Gaussian noise SD through the quantile of order α can be expressed as:
| [B6] |
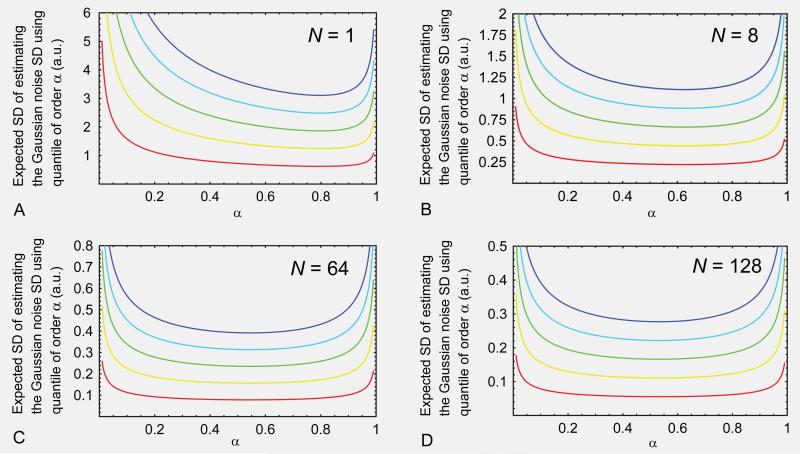
Figures 8A, 8B, 8C and 8D show the expected SD for estimating the Gaussian noise SD through the quantile of order α with N equal to 1, 8, 64 and 128, respectively, and with different Gaussian noise SDs, σg = 1, 2, 3, 4 and 5, coded in different hues ranging respectively from red to blue.
Fig. 8.
Expected SD for estimating the Gaussian noise SD using the quantile of order α with (A) N=1, (B) N=8, (C) N=64, and (D) N=128, and with different Gaussian noise SDs, i.e, σg = 1, 2, 3, 4, and 5 (arbitrary units), which were indicated with hues ranging respectively from red to blue.
The optimal quantile order, α*, can be found by finding the minimum point of Eq.[B6]. It can be shown that the optimal quantile order is independent of σg. The numerical values of the optimal quantile order for different N are listed in Table A. For completeness, the values of the denominator of Eq.[B4] evaluated at the optimal quantile order are also listed in Table A. It is clear that the use of sample median is expedient and reasonable because we expect the optimal quantile order to approach ½ as N increases. It is also clear that the sample median used in estimating the Gaussian noise SD may be replaced by the sample mean.
Appendix C
Although we do not have a rigorous and general statement about the convergence of the proposed framework, the proposed framework has never failed to converge in all the cases we tested. In this appendix, we discuss some qualitative features of the convergence of PIESNO.
Let us assume that there is only one attracting fixed point, denoted by σFP, which is unbeknown to us. In what follows, we will show that if σn is some estimate, which results in nonempty noise array Ω, then we have the following conditions for the next estimate, σn+1 : (A) if σn>σFP then σn>σn+1>σFP, and (B) if σn<σFP then σn<σn+1<σFP. Based on these two cases we can conclude that σn converges to σFP as n increases.
Let us assume that σn>σFP, then we can write σn = βσFP with β>1. In many simulations we carried out with K>5, we found that σn+1 seems to be close the value . The same is true when σn<σFP and σn = βσFP with β<1. Note that the data structure used in these tests was 128×128×K with α = 0.1 and N = 1 to N = 64. For cases with K<5, (A) and (B) remain true but we did not find to be true.
For example, let the dimensions of the volumetric data be 128×128×K with K = 8, 12, 16 and 20. Let us take σFP = 50 as the ground truth, α = 0.1, N = 1 and σn = βσFP with β = 1.3. Then, we have and the following results for K = 8, 12, 16 and 20, respectively: , 56.55, 56.95, and 57.16. As K increases, the estimate will be more and more variable because fewer and fewer of the si,j are within the acceptance interval, [λ−, λ+]. Finally, the results for K = 8, 12, 16 and 20, with N = 8 and are respectively as follows: , 52.31, 52.54, and 52.66.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lauterbur PC. Image formation by induced local interactions: Examples employing nuclear magnetic resonance. Nature. 1973;242(5394):190–191. [PubMed] [Google Scholar]
- 2.Ogawa S, Lee TM, Kay AR, Tank DW. Brain Magnetic Resonance Imaging with Contrast Dependent on Blood Oxygenation. Proceedings of the National Academy of Sciences. 1990;87(24):9868–9872. doi: 10.1073/pnas.87.24.9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basser PJ, Mattiello J, Le Bihan D. MR diffusion tensor spectroscopy and imaging. Biophys J. 1994;66(1):259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuch DS, Reese TG, Wiegell MR, Van JW. Diffusion MRI of complex neural architecture. Neuron. 2003;40(5):885–895. doi: 10.1016/s0896-6273(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 5.Nowak RD. Wavelet-based Rician noise removal for magnetic resonance imaging. IEEE Transactions on Image Processing. 1999;8(10):1408–1419. doi: 10.1109/83.791966. [DOI] [PubMed] [Google Scholar]
- 6.Rohde GK, Barnett AS, Basser PJ, Pierpaoli C. Estimating intensity variance due to noise in registered images: Applications to diffusion tensor MRI. NeuroImage. 2005;26(3):673–684. doi: 10.1016/j.neuroimage.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 7.Koay CG, Özarslan E, Basser PJ. A signal transformational framework for breaking the noise floor and its applications in MRI. Journal of Magnetic Resonance. 2009 doi: 10.1016/j.jmr.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koay CG, Chang L-C, Carew JD, Pierpaoli C, Basser PJ. A unifying theoretical and algorithmic framework for least squares methods of estimation in diffusion tensor imaging. Journal of Magnetic Resonance. 2006;182(1):115–125. doi: 10.1016/j.jmr.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 9.Chang L-C, Jones DK, Pierpaoli C. RESTORE: Robust estimation of tensors by outlier rejection. Magnetic Resonance in Medicine. 2005;53(5):1088–1095. doi: 10.1002/mrm.20426. [DOI] [PubMed] [Google Scholar]
- 10.Basser PJ, Mattiello J, Le Bihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. (Series B).Journal of Magnetic Resonance. 1994;103(3):247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- 11.Jones DK, Basser PJ. Squashing peanuts and smashing pumpkins: How noise distorts diffusion-weighted MR data. Magnetic Resonance in Medicine. 2004;52(5):979–993. doi: 10.1002/mrm.20283. [DOI] [PubMed] [Google Scholar]
- 12.Koay CG, Chang LC, Pierpaoli C, Basser PJ. Error propagation framework for diffusion tensor imaging via diffusion tensor representations. IEEE Transactions on Medical Imaging. 2007;26(8):1017–1034. doi: 10.1109/TMI.2007.897415. [DOI] [PubMed] [Google Scholar]
- 13.Anderson AW. Theoretical analysis of the effects of noise on diffusion tensor imaging. Magnetic Resonance in Medicine. 2001;46(6):1174–1188. doi: 10.1002/mrm.1315. [DOI] [PubMed] [Google Scholar]
- 14.Koay CG, Nevo U, Chang L-C, Pierpaoli C, Basser PJ. The elliptical cone of uncertainty and its normalized measures in diffusion tensor imaging. IEEE Transactions on Medical Imaging. 2008;27(6):834–846. doi: 10.1109/TMI.2008.915663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones DK. Determining and visualizing uncertainty in estimates of fiber orientation from diffusion tensor MRI. Magnetic Resonance in Medicine. 2003;49(1):7–12. doi: 10.1002/mrm.10331. [DOI] [PubMed] [Google Scholar]
- 16.Chang L-C, Koay CG, Pierpaoli C, Basser PJ. Variance of estimated DTI-derived parameters via first-order perturbation methods. Magnetic Resonance in Medicine. 2007;57(1):141–149. doi: 10.1002/mrm.21111. [DOI] [PubMed] [Google Scholar]
- 17.Jeong H-K, Anderson AW. Characterizing fiber directional uncertainty in diffusion tensor MRI. Magnetic Resonance in Medicine. 2008;60(6):1408–1421. doi: 10.1002/mrm.21734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edelstein WA, Bottomley PA, Pfeifer LM. A signal-to-noise calibration procedure for NMR imaging systems. Med Phys. 1984;11(2):180–185. doi: 10.1118/1.595484. [DOI] [PubMed] [Google Scholar]
- 19.Henkelman RM. Measurement of signal intensities in the presence of noise in MR images. Med Phys. 1985;12(2):232–233. doi: 10.1118/1.595711. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein MA, Thomasson DM, Perman WH. Improved detectability in low signal-to-noise ratio magnetic resonance images by means of a phase-corrected real reconstruction. Med Phys. 1989;16(5):813–817. doi: 10.1118/1.596304. [DOI] [PubMed] [Google Scholar]
- 21.Chang L-C, Rohde GK, Pierpaoli C. An automatic method for estimating noise-induced signal variance in magnitude-reconstructed magnetic resonance images. SPIE Medical Imaging: Image processing. 2005;5747:1136–1142. [Google Scholar]
- 22.Sijbers J, Poot D, den Dekker AJ, Pintjens W. Automatic estimation of the noise variance from the histogram of a magnetic resonance image. Physics in Medicine and Biology. 2007;52(5):1335–1348. doi: 10.1088/0031-9155/52/5/009. [DOI] [PubMed] [Google Scholar]
- 23.Constantinides CD, Atalar E, McVeigh ER. Signal-to-noise measurements in magnitude images from NMR phased arrays. Magnetic Resonance in Medicine. 1997;38(5):852–857. doi: 10.1002/mrm.1910380524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen AH, Kirsch JE. Analysis of noise in phase contrast MR imaging. Med Phys. 1996;23(6):857–869. doi: 10.1118/1.597729. [DOI] [PubMed] [Google Scholar]
- 25.Strogatz SH. Nonlinear dynamics and chaos. Perseus Publishing; Cambridge: 1994. [Google Scholar]
- 26.Koay CG, Basser PJ. Analytically exact correction scheme for signal extraction from noisy magnitude MR signals. Journal of Magnetic Resonance. 2006;179(2):317–322. doi: 10.1016/j.jmr.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magnetic Resonance in Medicine. 1990;16:192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich O, Raya JG, Reeder SB, Ingrisch M, Reiser MF, Schoenberg SO. Influence of multichannel combination, parallel imaging and other reconstruction techniques on MRI noise characteristics. Magnetic Resonance Imaging. 2008;26(6):754–762. doi: 10.1016/j.mri.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Rice SO. Mathematical analysis of random noise. Bell System Technical Journal. 1944;23 and 24 [Google Scholar]
- 30.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magnetic Resonance in Medicine. 1995;34(6):910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casella G, Berger RL. Statistical Inference. Duxbury; Pacific Grove: 2002. [Google Scholar]
- 32.Rao CR. Linear statistical inference and its applications. Wiley; New York: 1973. [Google Scholar]
- 33.Wolfram S. The Mathematica book. Wolfram Media; Champaign: 2003. [Google Scholar]
- 34.Press WH, Flannery BP, Teukolsky SA, Vetterling WT. Numerical recipes in C. Cambridge University Press; New York: 1992. [Google Scholar]
- 35.Sedgewick R. Algorithms in Java. Addison Wesley; Boston: 2003. [Google Scholar]
- 36.Brummer ME, Mersereau RM, Eisner RL, Lewine RRJ. Automatic detection of brain contours in MRI data sets. IEEE Transactions on Medical Imaging. 1993;12(2):153–166. doi: 10.1109/42.232244. [DOI] [PubMed] [Google Scholar]
- 37.Özarslan E, Shepherd TM, Vemuri BC, Blackband SJ, Mareci TH. Resolution of complex tissue microarchitecture using the diffusion orientation transform (DOT). NeuroImage. 2006;31(3):1086–1103. doi: 10.1016/j.neuroimage.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 38.Tuch DS, Reese TG, Wiegell MR, Makris N, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging reveals intravoxel white matter fiber heterogeneity. Magnetic Resonance in Medicine. 2002;48(4):577–582. doi: 10.1002/mrm.10268. [DOI] [PubMed] [Google Scholar]
- 39.Tuch DS. Q-ball imaging. Magnetic Resonance in Medicine. 2004;52(6):1358–1372. doi: 10.1002/mrm.20279. [DOI] [PubMed] [Google Scholar]
- 40.Tuch DS, Weisskoff RM, Belliveau JW, Wedeen VJ. High angular resolution diffusion imaging of the human brain. Philadelphia: 1999. p. 321. [Google Scholar]
- 41.Anderson AW. Measurement of fiber orientation distributions using high angular resolution diffusion imaging. Magnetic Resonance in Medicine. 2005;54(5):1194–1206. doi: 10.1002/mrm.20667. [DOI] [PubMed] [Google Scholar]
- 42.Frank LR. Characterization of anisotropy in high angular resolution diffusion-weighted MRI. Magnetic Resonance in Medicine. 2002;47(6):1083–1099. doi: 10.1002/mrm.10156. [DOI] [PubMed] [Google Scholar]
- 43.Papoulis A. Probability, random variables, and stochastic processes. McGraw-Hill; New York: 1965. [Google Scholar]
- 44.Dudewicz EJ. Introduction to statistics and probability. Holt, Rinehart & Winston; New York: 1976. [Google Scholar]