Abstract
The FTO gene was recently identified as a susceptibility locus for both obesity and type 2 diabetes by whole-genome association analyses of several European populations. We tested for an association between FTO risk alleles and obesity and diabetes in a well-characterized multiethnic cohort of postmenopausal women in the United States. We genotyped two most significantly associated single-nucleotide polymorphisms (SNPs) (rs9939609 and rs8050136) in intron 1 of FTO gene in a nested case–control study of 1,517 diabetes cases and 2,123 controls from the Women’s Health Initiative–Observational Study (WHI-OS). The allelic frequencies of either rs9939609 or rs8050136 differed widely across four ethnic groups. The frequency of the rare allele A of rs9939609 among controls was much lower in Asians/Pacific Islanders (17%) than in blacks (45%), whites (40%), and Hispanics (31%). We found significant associations of rs9939609 with BMI and waist circumference in white and Hispanic women, but not among black and Asian/Pacific Islander women. On average, each copy of the risk-allele A at rs9939609 was significantly associated with 0.45 kg/m2 increase in BMI (95% confidence interval (CI): 0.16–0.74; P = 0.004) and 0.97 cm increase in waist circumference (95% CI: 0.21–0.65; P = 0.0002). Similar results were observed for rs8050136. However, we found no significant genetic associations with diabetes risk, either within the full study sample or in any ethnic group. In conclusion, common genetic variants in the intron 1 of FTO gene may confer a modest susceptibility to obesity in an ethnicity-specific manner, but may be unlikely to contribute to a clinically significant diabetes risk.
INTRODUCTION
The human FTO gene on chromosome 16ql2.2 was recently identified as a susceptibility locus for both obesity and type 2 diabetes by whole-genome association analyses of several European populations (1–4). FTO protein was found to be ubiquitously expressed in various tissues with relatively high levels in the hypothalamus and pancreatic islets (1). FTO protein could, therefore, be a key link between central nervous system and energy balance. FTO protein has recently been identified as a member of the nonheme and 2-oxoglutarate-dependent oxygenase with nucleic acid demethylase activity (5). The mechanisms underlying the role of FTO demethylase activity in the control of energy homeostasis, however, remain to be defined.
Findings from whole-genome association studies have initially pointed to its modest genetic associations with both obesity and diabetes in individuals of European ancestry (1–3). In a genome-wide scan in 1,924 diabetes cases and 2,938 controls for type 2 diabetes susceptibility genes, Frayling et al. first reported on the association of a set of single-nucleotide polymorphisms (SNPs) within intron 1 of the FTO gene with BMI (1). In particular, the significant additive association of rs9939609 with BMI was reproduced in 13 cohorts comprised of 38,759 European whites (1). The minor allele A at rs9939609 has a relatively high frequency in European populations (45%), and the homozygous carriers of this allele (16%) had increased risks of being overweight, obese, or diabetic as compared with the noncarriers in European adults (1). Recent genome-wide genotype data from a UK sample of 1,924 cases and 2,938 controls (3) and a Finnish sample of 1,161 type 2 diabetes cases and 1,174 normal glucose-tolerant controls (2) replicated this FTO–obesity association and identified another SNP in intron 1, rs8050136, which seemed to provide the strongest statistical evidence (3). Of note, the diabetes associations with both rs9939609 and rs8050136 appeared to be largely mediated through their high associations with BMI (1–3). Furthermore, there is evidence to suggest ethnic differences for the associations of FTO variants and obesity and/or diabetes (6,7), but very few data are available from a well-characterized multiethnic cohort of US populations, especially American minority groups.
To provide such information, we examined the associations of these two common SNPs newly identified in the FTO gene with obesity and diabetes risk employing a large case–control study nested in the Women’s Health Initiative–Observational Study (WHI-OS), an ethnically diverse cohort of postmenopausal women aged 50–79 years including whites, blacks, Hispanics, and Asian/Pacific Islanders. We also investigated whether and to what extent the FTO variants may quantitatively affect obesity-associated intermediate metabolic traits, including insulin resistance, pancreatic beta-cell function, systemic inflammation, and endothelial dysfunction.
METHODS AND PROCEDURES
Study population
The WHI-OS is a longitudinal study designed to examine the association between clinical, socioeconomic, behavioral, and dietary risk factors and subsequent incidence of health outcomes, including cardiovascular disease and diabetes mellitus. Details of the scientific rationale, eligibility, and other design aspects have been described elsewhere (8,9). At baseline, all WHI-OS participants completed screening and enrollment questionnaires, underwent a physical examination, and provided fasting blood samples (after an overnight fast for at least 12 h). The study has been reviewed and approved by human subjects review committees at each participating institution, and signed informed consent was obtained from all women enrolled.
WHI-OS participants were followed by annual mailed self-administered questionnaires (medical history and exposure updates) and an additional clinical center visit for physical measurements 3 years after enrollment. Of the 93,676 postmenopausal women enrolled into the WHI-OS cohort, 82,069 (87.6%) women had no prior history of diabetes or cardiovascular disease. Among these 82,069 nondiabetic participants, incident cases of diabetes were identified based on postbaseline self-report of first-time use of hypoglycemic medication (oral hypoglycemic agents or insulin) during a median follow-up period of 5.9 years (mean = 5.5 years). Following the principle of risk-set sampling (8) for each incident case, controls were selected at random from women who remained free of cardiovascular disease and/or diabetes at the time the case was identified during follow-up. Controls were matched to the cases by age (±2.5 years), racial/ethnic group (white/Caucasian, black/African, Hispanic/Latino, and Asian/Pacific Islander), clinical center (geographic location), time of blood draw (±0.10 h), and length of follow-up. In the present study, 968 cases in white women were randomly chosen and matched with one control each. Of 749 incident cases among ethnic minority women, 366 cases were blacks, 152 were Hispanics, and 98 cases were Asians/Pacific Islanders. The 1:2 matching ratio was used for minorities to strengthen the power in these smaller samples of cases. Our study did not include American Indian or Alaskan Native women because of their limited numbers.
Genotyping
DNA was extracted from the buffy coat fraction of centrifuged blood using the QIAmp Blood Kit (Qiagen, Chatsworth, CA). We genotyped the two previously reported FTO SNPs in intron 1, rs8050136 and rs9939609, in the entire case–control sample consisting of 1,543 cases and 2,132 matched controls in the University of California at Los Angeles Molecular Epidemiology Laboratory (Director: S.L.). The primers and probes were custom designed by the ABI Taqman system (PE Biosystems, Foster City, CA). Following PCR amplification, endpoint fluorescence was read with the Applied Biosystems Primer 7900HT instrument, and genotypes were assigned using SDS2.2.2 Allelic Discrimination Software (Applied Biosystems, Foster City, CA) by two independent technicians blinded to sample identification numbers. A total of 94 duplicate samples were randomly selected and replicated across all plates. Concordance rate was >98.9% for both SNPs in the case–control study. The average genotyping drop-out rates were 1.8% for rs8050136 and 1.6% for rs9939609. Thus, we performed association analysis on 1,517 affected individuals and 2,123 controls.
Quantitative measures
As previously described (10,11), all biochemical assays were carried out by laboratory staff blinded to case/control status. Blood samples from cases and their matched controls were handled identically, shipped in the same batch, and assayed in random order in the same analytical run to reduce systematic bias and interassay variation. The coefficients of variation for each analyte were 1.7% for fasting glucose, 5.7% for fasting insulin, 1.61% for high-sensitivity C-reactive protein, and 6.5% for E-selectin (11). We used the homeostasis model assessment (HOMA) derived from basal fasting glucose and insulin levels to estimate insulin resistance (HOMA-IR) and pancreatic β-cell function (HOMA-B) (10).
Statistical analysis
We first assessed each SNP for the Hardy–Weinberg equilibrium test using the χ2-tests. We tested for heterogeneity of genotype distributions across ethnic groups by the χ2-test. The linkage disequilibrium (LD) for the rs9939609 and rs8050136 pair in each of the four ethnic groups was calculated using both Lewontin’s D’ (12) and r2 (13).
Examination of all baseline continuous variables for sample distribution and outliers was assessed first. To determine the cross-sectional associations between the FTO genotypes and each of BMI, waist circumference, height, and obesity-associated intermediate trait variables at baseline, general linear models were used to compare geometric mean values of each quantitative trait across different genotype groups. All the linear models included matching factors (age, clinical center, and blood draw time) as covariates for each of four ethnic groups. For all the combined groups, weighted mean of linear coefficient estimates of each SNP on continuous outcomes were calculated using inverse-variance (s.e.) weighting of individual results from each ethnic group. All the analyses were performed in both groups of incident diabetes cases and controls, separately. Because all included cases and controls were nondiabetic at baseline and there was also not statistical evidence to suggest that the FTO SNP–obesity association differed by the status of incident diabetes, we subsequently analyzed and presented the results using all the combined samples to further increase statistical power, especially in American Hispanic and Asian/Pacific Islander groups.
We performed unconditional logistic regression to calculate odds ratios (ORs) and 95% confidence intervals (CIs) for each SNP with risk of being overweight (BMI ≥ 25 kg/m2), obese (BMI ≥ 30 kg/m2), and severely obese (BMI ≥ 35 or 40 kg/m2). All analyses were stratified by ethnicity to minimize potential false-positive results due to population stratification. We first made adjustments for matching variables (age, clinical center, and time of blood draw), and then further adjusted for cigarette smoking (never, past, and current), alcohol intake (never, past, and current), hormone replacement therapy (never, past, and current), and physical activity per week at baseline (expressed as total metabolic equivalents (METs); total expenditure of energy from recreational physical activity). Because of the possible association between the FTO genotype and BMI, any obesity index was not included as a covariate in our models. For each SNP, we tested for allelic association with obesity risk under dominant, recessive, and additive models. Following matching strategy in our case–control study, we employed conditional logistic regression to calculate ORs and 95% CIs for allelic association with diabetes risk under three genetic models (dominant, recessive, and additive). To minimize any effect of population stratification on the overall significance of the associations, we performed the DerSimonian and Laird’s random-effects model to synthesize the OR estimate from each ethnic group (14). Likelihood ratio test was used to test the interaction effect between the genotypes and ethnicity on obesity and diabetes risk.
We also computed population attributable risk (PAR) of each FTO genotype with a significant association, which indicates the proportion of disease that would not have occurred in the population if the risk genotype was absent or if its disease-predisposing effect could be nullified. PAR was calculated based on case–control studies: (15), where b is the prevalence of the risk genotype in the controls from the source population and the OR is the estimated relative risk of disease in individuals with the risk genotype relative to the referent group.
All reported P values were two-tailed, and statistical significance was defined at the α = 0.05 level. Statistical analyses were performed using SAS statistical package (version 9.0 for window; SAS Institute, Cary, NC).
RESULTS
LD and minor allele frequencies
The observed genotype frequencies for rs8050136 and rs9939609, except for rs8050136 among Hispanic women (P = 0.05), were within Hardy–Weinberg equilibrium among the controls within each of four ethnic groups. Basic anthropometric characteristics and allelic and genotypic frequencies of rs9939609 and rs8050136 are shown in Table 1. The allele frequencies varied significantly by ethnic groups (all P < 0.0001). As reported previously, there was evidence for a high LD region containing all intron 1, exon 2, and part of intron 2 in European whites (Supplementary Figure S1 online). Rs9939609 and rs8050136 are 4,251 bp apart within intron 1 of FTO gene and the values of their pairwise LD metrics were high and were very similar across each of four ethnic groups (D′ =0.91–0.92 and r2=0.74–0.79). The minor allele frequencies (MAFs) for these two SNPs were 0.40 and 0.42 in white women, which were similar to those previously reported in European whites (0.39 for rs9939609 and 0.38 for rs8050136) (refs. 1,2). However, MAFs in Hispanics and Asians/Pacific Islanders were significantly different from those of other ethnic groups. For rs9939609, the minor allele A frequency was similar between whites (0.40) and blacks (0.45) but lower in Hispanics (0.31) and in Asians/Pacific Islanders (0.17). Similar distributions were observed for rs8050136, with MAFs of 0.40 in whites, 0.42 in blacks, 0.30 in Hispanics, and 0.18 in Asians/Pacific Islanders.
Table 1.
Distributions of BMI/waist circumference and two FTO intronic SNPs in our matched case–control study
| Total population | Ethnic groups (case/control) | ||||
|---|---|---|---|---|---|
| White | Black | Hispanic | Asian/Pacific Islander | ||
| N, sample size (case/control) | 1,517/2,123 | 936/935 | 365/749 | 139/276 | 77/163 |
| Age mean (s.d.) (years) | 62.6 (7.03) | 63.9 (6.88) | 61.1 (6.71) | 60.2 (6.77) | 63.8 (7.58) |
| Weight mean (s.d.) (kg) | 77.3 (19.5) | 77.9 (19.1) | 82.1 (19.8) | 72.2 (16.2) | 59.6 (13.2) |
| Height mean (s.d.) (cm) | 161 (7.00) | 162 (6.85) | 163 (6.38) | 157 (6.36) | 155 (6.07) |
| Obesity indices | |||||
| BMI (kg/m2) | |||||
| Mean (s.d.) (kg/m2) | 29.5 (6.79) | 29.5 (6.75) | 30.9 (7.10) | 29.0 (5.78) | 24.7 (4.49) |
| BMI ≥ 25 kg/m2 (n (%)) | 2,620 (72.9) | 1,321 (71.6) | 897 (81.7) | 303 (73.9) | 99 (41.4) |
| BMI ≥ 30 kg/m2 (n (%)) | 1,428 (39.8) | 740 (40.1) | 521 (47.5) | 143 (34.9) | 24 (10.0) |
| BMI ≥ 40 kg/m2 (n (%)) | 288 (8.02) | 147 (7.97) | 123 (11.2) | 16 (3.90) | 2 (0.84) |
| Waist circumference (cm) | |||||
| Mean (s.d.) (cm) | 89.8 (15.5) | 91.2 (16.1) | 91.0 (14.6) | 87.3 (13.9) | 78.2 (11.0) |
| Waist ≥80 cm (n (%)) | 2,622 (72.3) | 1,354 (72.8) | 875 (78.7) | 290 (69.9) | 103 (42.9) |
| Waist ≥88 cm (n (%)) | 1,841 (50.8) | 999 (53.7) | 612 (55.0) | 186 (44.8) | 44 (18.3) |
| Candidate variants | |||||
| rs9939609 | |||||
| Allele frequency (%) | |||||
| Allele T | 59.1/61.5 | 57.7/60.4 | 52.9/55.3 | 73.0/69.3 | 79.9/83.1 |
| Allele A | 40.9/38.5 | 42.3/39.7 | 47.1/44.7 | 27.0/30.7 | 20.1/16.9 |
| Gentoype (%) | |||||
| TT | 37.0/40.0 | 34.6/38.3 | 30.8/31.9 | 54.0/49.8 | 64.9/69.9 |
| TA | 44.2/43.1 | 46.3/44.1 | 44.2/46.9 | 38.1/38.9 | 30.0/26.4 |
| AA | 18.8/17.0 | 19.1/17.6 | 25.0/21.2 | 7.91/11.3 | 5.19/3.68 |
| rs8050136 | |||||
| Allele frequency (%) | |||||
| Allele C | 60.8/62.4 | 58.2/60.2 | 58.4/58.0 | 73.4/70.5 | 80.5/82.0 |
| Allele A | 39.3/37.6 | 41.9/39.8 | 41.6/42.0 | 26.6/29.5 | 19.5/18.0 |
| Gentoype (%) | |||||
| CC | 38.7/40.8 | 34.8/38.0 | 36.4/34.3 | 54.7/52.2 | 67.5/68.3 |
| CA | 44.2/43.2 | 46.8/44.4 | 44.1/47.5 | 37.4/36.6 | 26.0/27.3 |
| AA | 17.1/16.0 | 18.5/17.6 | 19.6/18.3 | 7.91/11.2 | 6.49/4.35 |
SNPs, single-nucleotide polymorphisms.
Associations of SNPs with continuous quantitative obesity traits
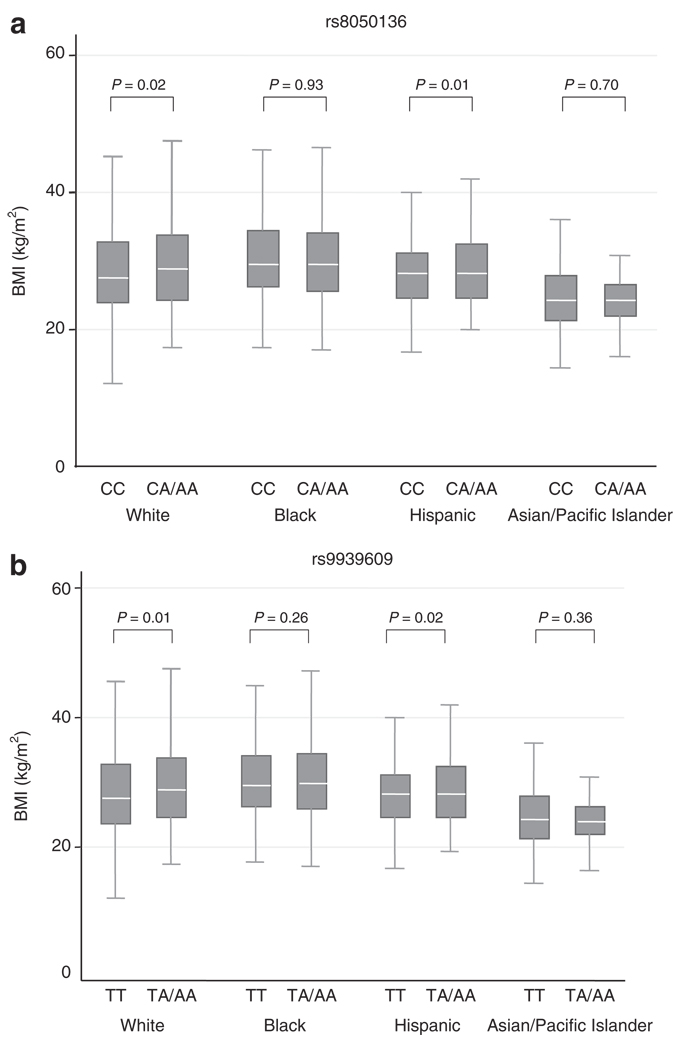
Both rs9939609 and rs8050136 showed consistent and significant associations with BMI and waist circumference among white and Hispanic women, but not in black and Asian/Pacific Islander women (Table 2 and Table 3 and Figure 1). For rs9939609, homozygous carriers of the risk-allele A had higher BMI than the heterozygous carriers while heterozygous carriers showed a similar increment in BMI compared with noncarriers in American white and Hispanic women (Table 2). Hence, the association of rs9939609 with BMI was highly significant and consistent with an additive effect. On average, each additional copy of the A allele of rs9939609 was associated with a BMI increment of 0.53 kg/m2 (95% CI: 0.10–0.96; P =0.01) for whites and of 1.25 kg/m2 (95% CI: 0.43–2.07; P =0.003) for Hispanics. Overall, the risk-allele A at rs9939609 was significantly associated with a mean of 0.45 kg/m2 increase in BMI (95% CI: 0.16–0.74; P =0.004), which is similar to the effect observed by Frayling et al. (0.40 kg/m2) (1). Similar trends were noted for rs8050136 (Table 2). Both SNPs were similarly associated with waist circumference and body weight but not height (data not shown).
Table 2.
Associations of FTO rs9939609 and rs8050136 genotypes with BMI and waist circumference as continuous traits
| Groups | FTO rs9939609 | ||||
|---|---|---|---|---|---|
| BMI (mean ± s.d.) (kg/m2) | Linear effect on BMIa | ||||
| Genotype | |||||
| T/T | T/A | A/A | β Coefficient (s.e.) | P values | |
| White | 29.0 ± 6.52 (n = 672) | 29.7 ± 6.92 (n = 826) | 30.0 ± 6.77 (n = 339) | 0.53 (0.22) | 0.01 |
| Black | 30.6 ± 6.56 (n = 343) | 31.2 ± 7.42 (n = 504) | 30.9 ± 7.21 (n = 245) | 0.19 (0.29) | 0.52 |
| Hispanic | 28.3 ± 4.92 (n = 208) | 29.3 ± 5.94 (n = 159) | 31.2 ± 8.12 (n = 42) | 1.25 (0.42) | 0.003 |
| Asian/Pacific Islander | 24.9 ± 4.68 (n = 163) | 24.1 ± 4.17 (n = 66) | 25.6 ± 3.35 (n = 10) | −0.44 (0.52) | 0.40 |
| All groups combinedb | 28.8 ± 6.33 (n = 1,386) | 29.9 ± 7.05 (n = 1,555) | 30.3 ± 7.02 (n = 636) | 0.45 (0.15) | 0.004 |
| Groups | Waist circumference (mean ± s.d.) (cm) | Linear effect on waist circumferencea | |||
|---|---|---|---|---|---|
| Genotype | |||||
| T/T | T/A | A/A | β Coefficient (s.e.) | P values | |
| White | 89.8 ± 15.6 (n = 676) | 91.9 ± 16.5 (n = 837) | 92.3 ± 16.2 (n = 339) | 1.39 (0.52) | 0.008 |
| Black | 90.7 ± 13.5 (n = 348) | 91.1 ± 14.9 (n = 509) | 91.2 ± 15.5 (n = 249) | 0.23 (0.60) | 0.70 |
| Hispanic | 85.9 ± 13.8 (n = 212) | 88.1 ± 13.5 (n = 160) | 91.7 ± 14.9 (n = 42) | 2.57 (1.02) | 0.01 |
| Asian/Pacific Islander | 78.4 ± 11.3 (n = 164) | 77.5 ± 11.2 (n = 66) | 78.9 ± 6.21 (n = 10) | −0.70 (1.29) | 0.59 |
| All groups combinedb | 88.1 ± 14.9 (n = 1,400) | 90.6 ± 15.8 (n = 1,572) | 91.6 ± 15.8 (n = 640) | 0.97 (0.35) | 0.006 |
| Groups | FTO rs8050136 | ||||
|---|---|---|---|---|---|
| BMI (mean ± s.d.) (kg/m2) | Linear effect on BMIa | ||||
| Genotype | |||||
| C/C | C/A | A/A | β Coefficient (s.e.) | P values | |
| White | 29.0 ± 6.52 (n = 666) | 29.7 ± 6.87 (n = 833) | 29.9 ± 6.87 (n = 332) | 0.49 (0.22) | 0.02 |
| Black | 31.0 ± 6.94 (n = 380) | 30.8 ± 7.17 (n = 506) | 31.3 ± 7.30 (n = 205) | 0.09 (0.30) | 0.76 |
| Hispanic | 28.3 ± 4.97 (n = 216) | 29.4 ± 5.92 (n = 152) | 30.6 ± 8.27 (n = 42) | 1.12 (0.42) | 0.009 |
| Asian/Pacific Islander | 24.8 ± 4.72 (n = 161) | 24.4 ± 4.19 (n = 64) | 25.3 ± 3.30 (n = 12) | −0.21 (0.51) | 0.69 |
| All groups combinedb | 28.9 ± 6.49 (n = 1,423) | 29.8 ± 6.90 (n = 1,555) | 30.4 ± 7.13 (n = 591) | 0.43 (0.11) | 0.0002 |
| Groups | Waist circumference (Mean ± s.d.) (cm) | Linear effect on waist circumferencea | |||
|---|---|---|---|---|---|
| Genotype | |||||
| C/C | C/A | A/A | β Coefficient (s.e.) | P values | |
| White | 89.9 ± 15.3 (n = 671) | 91.8 ± 16.6 (n = 843) | 92.1 ± 16.4 (n = 332) | 1.24 (0.52) | 0.02 |
| Black | 91.3 ± 13.8 (n = 386) | 90.4 ± 14.8 (n = 512) | 91.9 ± 15.9 (n = 207) | 0.10 (0.62) | 0.87 |
| Hispanic | 86.0 ± 13.7 (n = 220) | 88.5 ± 13.5 (n = 153) | 89.8 ± 15.8 (n = 42) | 2.06 (1.02) | 0.04 |
| Asian/Pacific Islander | 78.4 ± 11.4 (n = 162) | 77.8 ± 11.1 (n = 64) | 77.8 ± 7.55 (n = 12) | −0.75 (1.27) | 0.56 |
| All groups combinedb | 88.4 ± 14.8 (n = 1,439) | 90.5 ± 15.8 (n = 1,572) | 91.6 ± 16.1 (n = 593) | 0.81 (0.36) | 0.02 |
Results in boldface were statistically significant (P < 0.05).
Linear regression models were adjusted for matching factors (age, clinical center, and time of blood draw).
In all groups combined, crude mean ± s.d. were shown for each genotype and weighted mean linear coefficients (s.e.) for linear effects.
Table 3.
OR (95% CI) for the associations between two FTO SNPs and risk of being overweight and/or obese as compared to being normal weight (BMI < 25 kg/m2)
| Ethnic groups | Pooled estimatesa | ||||
|---|---|---|---|---|---|
| White | Black | Hispanic | Asian/Pacific Islander | ||
| rs9939609b | |||||
| BMI ≥ 25 kg/m2 | |||||
| Recessive model: | 1.32 (1.00–1.73) | 0.84 (0.59–1.20) | 1.14 (0.54–2.43) | 2.17 (0.58–8.06) | 1.13 (0.83–1.54) |
| TT+TA vs. AA | P = 0.05 | P = 0.33 | P = 0.73 | P = 0.25 | P = 0.45 |
| Dominant model: | 1.23 (1.00–1.52) | 0.90 (0.64–1.26) | 0.94 (0.60–1.47) | 0.58 (0.33–1.04) | 0.95 (0.71–1.26) |
| TT vs. TA+AA | P = 0.05 | P = 0.54 | P = 0.77 | P = 0.07 | P = 0.71 |
| Additive model: | 1.19 (1.03–1.37) | 0.90 (0.73–1.12) | 0.99 (0.71–1.38) | 0.76 (0.47–1.24) | 1.00 (0.83–1.21) |
| TT vs. TA vs. AA | P = 0.02 | P = 0.35 | P = 0.95 | P = 0.27 | P = 1.00 |
| BMI ≥ 30 kg/m2 | |||||
| Recessive model: | 1.30 (0.96–1.75) | 0.90 (0.61–1.32) | 1.67 (0.74–3.75) | 1.43 (0.15–13.8) | 1.17 (0.93–1.47) |
| TT+TA vs. AA | P = 0.09 | P = 0.58 | P = 0.22 | P = 0.76 | P = 0.18 |
| Dominant model: | 1.32 (1.04–1.67) | 0.95 (0.67–1.37) | 1.28 (0.76–2.14) | 0.57 (0.21–1.56) | 1.13 (0.88–1.46) |
| TT vs. TA+AA | P = 0.02 | P = 0.80 | P = 0.35 | P = 0.28 | P = 0.33 |
| Additive model: | 1.22 (1.04–1.44) | 0.95 (0.76–1.18) | 1.28 (0.88–1.87) | 0.68 (0.28–1.65) | 1.09 (0.88–1.33) |
| TT vs. TA vs. AA | P = 0.01 | P = 0.63 | P = 0.20 | P = 0.39 | P = 0.44 |
| BMI ≥ 35 kg/m2c | |||||
| Recessive model: | 1.42 (0.99–2.03) | 0.89 (0.57–1.40) | 2.30 (0.88–6.06) | — | 1.28 (0.82–1.98) |
| TT+TA vs. AA | P = 0.06 | P = 0.62 | P = 0.09 | P = 0.27 | |
| Dominant model: | 1.61 (1.20–2.16) | 1.18 (0.77–1.80) | 1.34 (0.68–2.61) | 0.21 (0.02–2.09) | 1.44 (1.15–1.81) |
| TT vs. TA+AA | P = 0.002 | P = 0.45 | P = 0.40 | P = 0.18 | P = 0.002 |
| Additive model: | 1.37 (1.13–1.67) | 1.03 (0.79–1.34) | 1.42 (0.88–2.30) | 0.23 (0.03–1.94) | 1.24 (1.01–1.53) |
| TT vs. TA vs. AA | P = 0.002 | P = 0.86 | P = 0.15 | P = 0.18 | P = 0.04 |
| rs8050136b | |||||
| BMI ≥ 25 kg/m2 | |||||
| Recessive model: | 1.25 (0.95–1.65) | 1.02 (0.69–1.51) | 1.00 (0.48–2.07) | 1.42 (0.43–4.63) | 1.16 (0.94–1.44) |
| CC+CA vs. AA | P = 0.11 | P = 0.93 | P = 0.99 | P = 0.56 | P = 0.17 |
| Dominant model: | 1.15 (0.93–1.41) | 0.86 (0.62–1.19) | 0.96 (0.61–1.51) | 0.69 (0.39–1.23) | 0.98 (0.80–1.20) |
| CC vs. CA+AA | P = 0.21 | P = 0.35 | P = 0.87 | P = 0.2059 | P = 0.81 |
| Additive model: | 1.13 (0.98–1.31) | 0.94 (0.76–1.16) | 0.98 (0.70–1.36) | 0.96 (0.75–1.23) | 1.04 (0.94–1.15) |
| CC vs. CA vs. AA | P = 0.09 | P = 0.56 | P = 0.90 | P = 0.42 | P = 0.47 |
| BMI ≥ 30 kg/m2 | |||||
| Recessive model: | 1.28 (0.95–1.72) | 1.17 (0.77–1.78) | 1.28 (0.57–2.88) | 1.08 (0.12–9.72) | 1.24 (0.99–1.57) |
| CC+CA vs. AA | P = 0.11 | P = 0.46 | P = 0.55 | P = 0.95 | P = 0.06 |
| Dominant model: | 1.26 (0.99–1.59) | 0.86 (0.61–1.23) | 1.31 (0.78–2.18) | 0.62 (0.23–1.71) | 1.08 (0.83–1.41) |
| CC vs. CA+AA | P = 0.06 | P = 0.41 | P = 0.31 | P = 0.36 | P = 0.57 |
| Additive model: | 1.19 (1.02–1.40) | 0.99 (0.79–1.24) | 1.22 (0.84–1.78) | 0.72 (0.30–1.70) | 1.12 (0.99–1.27) |
| CC vs. CA vs. AA | P = 0.03 | P = 0.90 | P = 0.30 | P = 0.45 | P = 0.07 |
| BMI ≥ 35 kg/m2c | |||||
| Recessive model: | 1.37 (0.96–1.96) | 1.12 (0.69–1.81) | 2.01 (0.78–5.18) | — | 1.33 (1.01–1.74) |
| CC+CA vs. AA | P = 0.08 | P = 0.64 | P = 0.15 | P = 0.04 | |
| Dominant model: | 1.17 (1.04–1.31) | 0.89 (0.59–1.34) | 1.38 (0.71–2.68) | 0.24 (0.03–2.41) | 1.15 (1.03–1.29) |
| CC vs. CA+AA | P = 0.007 | P = 0.57 | P = 0.35 | P = 0.23 | P = 0.01 |
| Additive model: | 1.32 (1.09–1.61) | 0.99 (0.76–1.29) | 1.39 (0.87–2.22) | 0.26 (0.03–2.16) | 1.20 (0.97–1.48) |
| CC vs. CA vs. AA | P = 0.006 | P = 0.91 | P = 0.17 | P = 0.21 | P = 0.09 |
Results in boldface were statistically significant (P < 0.05).
CI, confidence interval; OR, odds ratio; SNPs, single-nucleotide polymorphisms.
Random-effect meta-analysis to estimate the pooled RR estimates for all case–control samples.
Unconditional logistic regression adjusted for matching factors including age, clinical center, and time of blood draw.
Pooled estimates for risk of having a BMI ≥35 kg/m2 did not include the unstable estimates for Asian/Pacific Islander group due to sparse sample numbers.
Figure 1.
Distributions of BMI in four ethnic groups according to FTO genotypes (a) rs8050136 and (b) rs9939609. Box plots illustrate median, 25th, and 75th percentile values for BMI. Differences between the heterozygous carriers of the risk alleles and the homozygotes of the common allele were tested using student t-tests.
Associations of SNPs with dichotomously defined obesity traits
We also attempted to replicate the associations of FTO SNPs with the risk of being overweight (BMI ≥ 25 kg/m2) and obese (BMI ≥ 30 kg/m2) as reported previously (1,16). In white women, we found a significant association of the risk-allele A at rs9939609 with overweight and obesity under an additive model and a dominant model. The ORs per risk-allele A at rs9939609 were 1.19 for overweight (95% CI: 1.03–1.37; P =0.02), 1.22 for obesity class I (BMI ≥ 30 kg/m2) (95% CI: 1.04–1.44; P =0.01), and 1.37 for obesity class II (BMI ≥ 35 kg/m2) (95% CI: 1.13–1.67; P =0.002) in white women. Results for rs8050136 in whites and Hispanics were similar to those for rs9939609 but not significant for overweight (Table 3). Further adjustment for cigarette smoking, alcohol intake, hormone replacement therapy, and physical activity attenuated slightly these results. In the combined dataset, our random-effect pooled OR estimates replicated the significant associations of rs9939609 (OR = 1.44; 95% CI: 1.15–1.81; P =0.002) and rs8050136 (OR=1.15; 95% CI: 1.03–1.29; P =0.01) with obesity class II under a dominant model.
Associations of FTO SNPs with diabetes risk
As showed in Table 4, we found no evidence of any increased diabetes risk associated with these two FTO SNP obesity-related risk alleles in all four ethnic groups regardless of BMI adjustment. Although we found a tendency toward a recessive model association for whites, blacks, and Asians/Pacific islanders, all large ORs included 1.00 and were not statistically significant. To increase the statistical power and minimize population stratification, we used meta-analysis to combine individual OR estimates rather than individual data simply combined from each ethnic group. Overall, we observed no evidence of significant associations of either rs8050136 or rs9939609 with diabetes risk in all groups combined (Table 4).
Table 4.
OR (95% CI) for the associations of two FTO SNPs with risk of diabetes mellitus
| Ethnic groups | Pooled estimatesa |
||||
|---|---|---|---|---|---|
| White | Black | Hispanic | Asian/Pacific Islander | ||
| rs9939609 | |||||
| Matching-adjusted modelb | |||||
| Recessive model: TT+TA vs. AA | 1.14 (0.89–1.44) | 1.20 (0.89–1.63) | 0.59 (0.27–1.29) | 1.33 (0.38–4.73) | 1.12 (0.94–1.35) |
| Dominant model: TT vs. TA+AA | 1.18 (0.97–1.43) | 0.99 (0.75–1.32) | 0.83 (0.53–1.28) | 1.23 (0.67–2.23) | 1.09 (0.94–1.26) |
| Additive model: TT vs. TA vs. AA | 1.11 (0.98–1.27) | 1.06 (0.89–1.28) | 0.82 (0.59–1.13) | 1.20 (0.73–1.96) | 1.07 (0.96–1.18) |
| Multivariate-adjusted modelc | |||||
| Recessive model: TT+TA vs. AA | 1.12 (0.87–1.45) | 1.17 (0.84–1.61) | 0.36 (0.13–0.96) | 2.12 (0.54–8.23) | 1.06 (0.75–1.49) |
| Dominant model: TT vs. TA+AA | 1.09 (0.88–1.35) | 0.98 (0.72–1.33) | 0.77 (0.47–1.26) | 1.56 (0.81–3.02) | 1.04 (0.88–1.24) |
| Additive model: TT vs. TA vs. AA | 1.07 (0.93–1.23) | 1.05 (0.86–1.27) | 0.72 (0.49–1.05) | 1.53 (0.88–2.66) | 1.03 (0.87–1.23) |
| rs8050136 | |||||
| Matching-adjusted modelb | |||||
| Recessive model: CC+CA vs. AA | 1.09 (0.86–1.39) | 1.12 (0.81–1.56) | 0.67 (0.31–1.41) | 1.39 (0.41–4.66) | 1.07 (0.89–1.29) |
| Dominant model: CC vs. CA+AA | 1.14 (0.94–1.39) | 0.89 (0.68–1.16) | 0.89 (0.58–1.36) | 1.00 (0.56–1.80) | 1.02 (0.89–1.18) |
| Additive model: CC vs. CA vs. AA | 1.09 (0.95–1.24) | 0.98 (0.82–1.18) | 0.87 (0.63–1.19) | 1.05 (0.65–1.69) | 1.03 (0.93–1.14) |
| Multivariate-adjusted modelc | |||||
| Recessive model: CC+CA vs. AA | 1.06 (0.82–1.37) | 1.15 (0.81–1.63) | 0.49 (0.21–1.17) | 1.93 (0.53–7.04) | 1.04 (0.79–1.37) |
| Dominant model: CC vs. CA+AA | 1.07 (0.87–1.33) | 0.88 (0.66–1.17) | 0.89 (0.55–1.44) | 1.17 (0.62–2.19) | 1.00 (0.85–1.16) |
| Additive model: CC vs. CA vs. AA | 1.05 (0.91–1.21) | 0.98 (0.81–1.19) | 0.82 (0.57–1.17) | 1.22 (0.73–2.04) | 1.01 (0.91–1.13) |
CI, confidence interval; OR, odds ratio; SNPs, single-nucleotide polymorphisms.
Random-effect meta-analysis to estimate the pooled OR estimates for all case–control samples.
Conditional logistic regression adjusted for matching factors including age, clinical center, and time of blood draw; all the P values are >0.05.
Multivarite-adjusted model additionally adjusted for cigarette smoking, alcohol intake, postmenopausal hormone use, and exercise; all the P values are >0.05.
Associations of FTO SNPs with obesity-related metabolic traits
For obesity-related metabolic traits including systolic blood pressure, diastolic blood pressure, plasma fasting insulin, glucose, HOMA-IR, HOMA-B, high-sensitivity C-reactive protein, and E-selectin, there were no statistically significant differences between different genotypes at both rs8050136 and rs9939609 (data not shown). Neither were there any significant differences in these well-established quantitative metabolic parameters under recessive and dominant models.
PAR estimation for obesity
The PAR is defined as the proportion of disease cases in a population that would be prevented if the risk allele were monomorphic for the protective allele in either the heterozygote carriers or the homozygote carriers of the risk allele. As compared with the noncarriers, heterozygous and homozygous carriers of the risk-allele A at rs9939609 had PAR of 12% and 5.0% for white women, respectively; 12% and 7.0% for Hispanic women, respectively; and 5.1% and 2.8% for overall population, respectively. Similar results were noted for rs8050136.
DISCUSSION
Motivated by the recent intriguing discovery of the associations of the FTO genotypes with BMI in a genome-wide association study of type 2 diabetes in European populations (1), we hypothesized that these same SNPs may also play a role in the development of obesity and diabetes in US populations. On the basis of 1,543 diabetes cases and 2,132 matched controls from a multiethnic cohort of American postmenopausal women, we observed significant associations of the two common SNPs, rs9939609 and rs8050136, in intron 1 of FTO gene with obesity in white and Hispanic women but not in black and Asian/Pacific Islander women. None of the SNPs showed any significant associations with type 2 diabetes risk in any of the four ethnic groups. Our findings of the FTO-obesity relation in American white women were remarkably similar to those that were previously reported in European children and adults both in terms of direction and magnitude of these associations (2,16).
As we are entering the era of “whole-genome association study” in which numerous false-positive results are to be expected, replication in diverse ethnic populations becomes very important in the field of population genetics for complex diseases (17,18). A MAF of 0.40 was found at both rs9939609 and rs8050136 in American white women, which were comparable with those reported in Europeans (0.38–0.39) (refs. 1,2). Consistent with those reported by the HapMap, we observed diverse frequencies of the risk alleles in FTO gene ranged from 0.17 to 0.45 across various ethnic groups. In HapMap, frequencies of the minor allele A at rs9939609 and rs8050136 were 0.45 in CEPH Europeans, 0.47–0.52 in Yorubans (Sub-Saharan African), and 0.12/0.17 in Chinese/Japanese.
Consistent with previous findings in European populations (1,16), we observed significant and consistent associations between these two FTO intronic SNPs and obesity assessed using BMI and waist circumference, and we also did not find any significant associations of the same SNPs with height. Considering obesity as a discrete trait based on various thresholds of BMI, an additive association of each FTO SNP with obesity risk was also replicated in white women. Although the MAFs of rs9939609 (0.31) and rs8050136 (0.30) were less frequent in Hispanics, their associations with BMI and waist circumference were much stronger than white women under an additive model. Given the relatively high frequency of the risk allele at the FTO SNP, they may confer a relatively large PAR for obesity in American white and Hispanic populations. The findings of ethnicity-specific FTO–obesity associations may most likely reflect different haplotype block structure within the FTO gene between whites and other nonwhite populations. This notion is also supported by recent studies that failed to replicate the association of FTO and obesity in several non-white populations such as African Americans (4), Oceanic populations (7), and Chinese population (6). Further investigation using fine-mapping studies in nonwhite and genetically homogenous populations is necessary to confirm our findings of ethnicity differences.
It should be noted that, although the relations of rs9939609 and rs8050136 SNPs in intron 1 with obesity and diabetes risk were the strongest and most reproducible in the European populations (1,2,16), the causal variant has not yet been found. The finding of an extensive LD region (>30 kb) across the FTO gene intron 1 region makes it difficult to discern which of these intronic SNPs within intron 1 is the best single SNP surrogate to fully capture genetic variability for this region. It is plausible that intron 1 of the FTO gene may contain a regulatory element that is important in regulating both FTO mRNA splicing and expression. Intriguingly, a recent study found that FTO rs8050136 SNP is located in a putative transcriptional factor (Cutl-like 1, CUTL1) binding site and the rare allele A has been associated with lower CUTL1 binding to the DNA fragments and lower FTO expression levels (19). In the absence of conclusive evidence, the molecular consequences of genetic variation at the FTO locus remain to be determined. We also cannot rule out the possibility that these FTO intronic SNPs may be highly linked to an as yet unknown causal variant located in the vicinity of the FTO gene, but outside the intron 1 LD block. Dina et al. recently sequenced exon 2 and the exon/intron junctions in 363 severely obese individuals, but failed to identify any coding sequence variant leading to amino acid substitutions (16), indicating that potentially causal variants, if any, would be very rare even in obese adults. Additional function studies will be needed to investigate the regulation mechanisms underlying FTO mRNA expression, splicing, and degradation and the consequences on protein translation, stability, activity, and tissue-specific expression patterns.
The precise mechanism underlying the effect of FTO protein on body fat regulation remains largely unknown. Recent studies suggest that FTO is a member of the nonheme dioxygenase super family and functions as a DNA demethylase (5). The relatively high expression of FTO in hypothalamus could point to a critical role of FTO in neuroregulation of energy metabolism via the hypothalamic–pituitary–adrenal axis (1,5,19). There is some evidence, although limited, to suggest that FTO may, at least in part, regulate body fat mass through adipocyte lipolysis (20) or cerebrocortical insulin response (21). However, further studies are necessary to elucidate whether FTO is a true obesity-predisposing gene or a potential candidate gene for obesity-associated metabolic disorders.
Our null findings of the association between the same FTO variants and type 2 diabetes risk in each of the four ethnic samples were in stark contrast to the significant positive association previously reported in Europeans. First, it might be due to false negative due to insufficient statistical power, especially if the true OR is <1.15 in each individual ethnic group. However, the overall results from our matched studies (1:2 for minority groups) were well powered (>80%) to detect an OR of ≥1.15 for an additive allele with a frequency of 30 and 70%. Second, the observation that the genetic effects of FTO on BMI varied across different populations may reflect potential gene–gene or gene–environmental interactions. We did not find any significant interactions between FTO variants and traditional diabetes risk, including age, smoking, alcohol intake, and exercise. Nor did we find significant effect modification by BMI, insulin resistance index, and inflammatory biomarker levels for diabetes risk. Third, the absence of association between FTO variants and intermediate metabolic traits indicate that these FTO variants did not contribute to a clinically meaningful significant risk of type 2 diabetes in postmenopausal women with older ages. It also raises the possibility that the FTO may likely be an early predisposition gene for increased weight gain and has less influence on the progression from weight gain to obesity-associated metabolic disorders. Fourth, selection bias might exist in previous studies and inherently tended to overestimate the effect sizes for FTO and diabetes. Of note, it is unclear whether most controls used in individual studies were a random representative sample from the same study population from where cases were identified. Although a meta-analysis has been increasingly applied in genetic association studies to increase statistical power to lead to very narrow CIs with very small P values even for a modest-effect size, any meta-analysis results take no account of inherent selection bias in individual studies and should be interpreted cautiously. Finally, it is worth mentioning that population stratification is a concern in any association studies from genetically different populations. To minimize any potential bias caused by genetic heterogeneity within each ethnic group in the present study, we analyzed the data within each of four ethnicity groups and then used meta-analytic approach to pool the final estimates rather than simply combining all individual data together. In addition, the direction and magnitudes of our association results in American whites are very similar to those previously reported in other white populations. Thus, it is less likely that the presence of population stratification would substantially explain our findings.
In conclusion, our results in a large multiethnic case–control study of postmenopausal women confirmed the previously reported associations between two common SNPs in the intron 1 of FTO and obesity risk in both white and Hispanic women, indicating a possible ethnicity-specific relation of FTO to obesity. The FTO gene’s lack of associations with diabetes endpoint and with intermediate metabolic traits indicates that variants of FTO are unlikely to confer clinically significant risk of type 2 diabetes. Further confirmation in future large-scale prospective studies, especially among American minority populations, however, is warranted.
Supplementary Material
ACKNOWLEDGMENTS
The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts N01WH2210, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. This ancillary study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01 grant DK062290 from the National Institutes of Health. Y.S. is supported by a grant (K01-DK078846) from the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institutes of Health. We would like to acknowledge all WHI centers and their principal investigators for their participation in this study. We are indebted to all dedicated and committed participants of the WHI-OS. A list of WHI investigators is available in the Supplementary Data.
Footnotes
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at http://www.nature.com/oby
DISCLOSURE
The authors declare no conflict of interest.
REFERENCES
- 1.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Wu Y, Loos RJ, et al. Variants in FTO gene are not associated with obesity in a Chinese Han population. Diabetes. 2007;57:264–268. doi: 10.2337/db07-1130. [DOI] [PubMed] [Google Scholar]
- 7.Ohashi J, Naka I, Kimura R, et al. FTO polymorphisms in oceanic populations. J Hum Genet. 2007;52:1031–1035. doi: 10.1007/s10038-007-0198-2. [DOI] [PubMed] [Google Scholar]
- 8.Anderson GL, Manson J, Wallace R, et al. Implementation of the Women’s Health Initiative study design. Ann Epidemiol. 2003;13(Suppl 9):S5–S17. doi: 10.1016/s1047-2797(03)00043-7. [DOI] [PubMed] [Google Scholar]
- 9.Design of the Women’s Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 10.Song Y, Manson JE, Tinker L, et al. Insulin sensitivity and insulin secretion determined by the homeostasis model assessment (HOMA) predict risk of diabetes mellitus in a multi-ethnic cohort of women: the Women Health Initiative Observational Study. Diabetes Care. 2007;30:1747–1752. doi: 10.2337/dc07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song Y, Manson JE, Tinker L, et al. Circulating levels of endothelial adhesion molecules and risk of diabetes mellitus in an ethnically diverse cohort of women. Diabetes. 2007;56:1898–1904. doi: 10.2337/db07-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewontin RC. The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics. 1964;49:49–67. doi: 10.1093/genetics/49.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard JK, Przeworski M. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 2001;69:1–14. doi: 10.1086/321275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird NM. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Rothman KJ, Greenland S. Measures of Effect and Measures of Association. Philadelphia, PA: Lippincott Williams & Wilkins; 1998. [Google Scholar]
- 16.Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 17.Daly MJ, Altshuler D. Partners in crime. Nat Genet. 2005;37:337–338. doi: 10.1038/ng0405-337. [DOI] [PubMed] [Google Scholar]
- 18.Todd JA. Statistical false positive or true disease pathway? Nat Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 19.Stratigopoulos G, Padilla SL, Leduc CA, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294:R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahlen K, Sjolin E, Hoffstedt J. The common rs9939609 gene variant of the fat mass- and obesity-associated gene FTO is related to fat cell lipolysis. J Lipid Res. 2008;49:607–611. doi: 10.1194/jlr.M700448-JLR200. [DOI] [PubMed] [Google Scholar]
- 21.Kloting N, Schleinitz D, Ruschke K, et al. Inverse relationship between obesity and FTO gene expression in visceral adipose tissue in humans. Diabetologia. 2008;51:641–647. doi: 10.1007/s00125-008-0928-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.