Summary
ETS gene fusions have been characterized in a majority of prostate cancers, however the key molecular alterations in ETS negative cancers are unclear. Here we used an outlier meta-analysis (meta-COPA) to identify SPINK1 outlier-expression exclusively in a subset of ETS rearrangement negative cancers (~10% of total cases). We validated the mutual exclusivity of SPINK1 expression and ETS fusion status, demonstrated that SPINK1 outlier-expression can be detected non-invasively in urine and observed that SPINK1 outlier-expression is an independent predictor of biochemical recurrence after resection. We identified the aggressive 22RV1 cell line as a SPINK1 outlier-expression model, and demonstrate that SPINK1 knockdown in 22RV1 attenuates invasion, suggesting a functional role in ETS rearrangement negative prostate cancers.
Recently, we developed a bioinformatics approach termed Cancer Outlier Profile Analysis (COPA) to nominate candidate oncogenes from transcriptomic data based on high expression in a subset of cases (“outlier-expression”) (Tomlins et al., 2005). When applied to the Oncomine compendium of tumor profiling studies (www.oncomine.org) (Rhodes et al., 2004), COPA correctly identified several known oncogenes as outliers, such as ERBB2 in breast cancer and PBX1 in leukemia. In addition, COPA identified the ETS family members ERG and ETV1 as high-ranking outliers in multiple prostate cancer profiling studies, leading to the discovery of recurrent gene fusions involving the 5’ untranslated region of the androgen regulated gene TMPRSS2 with ERG, ETV1, ETV4 or ETV5 in prostate cancer cases that over-expressed the respective ETS family member (Helgeson et al., 2008; Tomlins et al., 2006; Tomlins et al., 2005). Recently, we identified additional 5’ fusion partners in cases with ETS family member outlier-expression (Tomlins et al., 2007a).
ETS gene fusions occur in 40–80% of prostate specific antigen (PSA)-screened prostate cancers, leaving 20–60% of prostate cancers in which the key genetic aberration cannot be ascribed to ETS gene fusions. Additionally, we have determined that ETS positive and negative cancers have distinct transcriptional signatures across profiling studies (Tomlins et al., 2007b), suggesting that fusion negative cancers activate unique oncogenes and downstream targets. Here, we attempted to identify such candidate oncogenes through their outlier expression in ETS negative prostate cancers.
The utility of COPA and other strategies to identify outlier genes from microarray data was recently demonstrated in multiple myeloma and breast cancer (Annunziata et al., 2007; Naderi et al., 2007), suggesting that this strategy can be applied across human cancers to identify relevant subtypes. Here, we refined our COPA strategy based on observations from our initial application of COPA. We observed that correctly identified oncogenes, including ERG and ETV1, were typically high-ranking outliers in multiple datasets (Tomlins et al., 2005). This suggests that true candidate oncogenes should demonstrate strong outlier profiles across independent studies and supports the use of a meta-analysis based COPA approach. Thus, in this study, we performed a focused application of COPA to 7 prostate cancer profiling studies (Dhanasekaran et al., 2001; Glinsky et al., 2004; Lapointe et al., 2004; LaTulippe et al., 2002; Vanaja et al., 2003; Welsh et al., 2001; Yu et al., 2004) in the Oncomine database (Rhodes et al., 2004), as described in the Methods, to prioritize candidate oncogenes in ETS negative prostate cancers. Twenty-nine genes were nominated as outliers in at least 3 of the 7 datasets (Table S1), with 11 genes identified as outliers in at least 4 of the 7 datasets (Table 1). Consistent with our previous application of COPA filtered by causal cancer genes (Tomlins et al., 2005), both ERG and ETV1 were high ranking meta-outliers; ERG ranked as the 1st meta-outlier (7 studies) and ETV1 as the 5th meta-outlier (4 studies).
Table 1. Meta-COPA analysis of 7 prostate cancer gene expression profiling datasets in Oncomine.
Genes were ranked by the number of studies in which they scored in the top 100 outliers (ranked by COPA) at any of the three pre-defined percentile cutoffs (75th, 90th, 95th). Genes were further ranked by their average COPA rank (Avg. Rank) in studies where they ranked in the top 100. ETS genes are indicated in bold. Genes showing outlier-expression exclusively in prostate cancer and mutually exclusive outlier expression with ETS genes are indicated in bold italics. Genes showing outlierexpression in benign prostate tissue are indicated by asteriks. Genes without mutual exclusivity with ERG or ETV1 outlier expression are indicated by italics.
| Meta COPA Rank | Gene | # of Studies | Avg. Rank |
|---|---|---|---|
| 1 | ERG | 7 | 19.3 |
| 2 | SPINK1 | 5 | 29.8 |
| 3 | GPR116 | 5 | 46 |
| 4 | ORM1* | 4 | 10 |
| 5 | ETV1 | 4 | 23 |
| 6 | MYL2* | 4 | 26.8 |
| 7 | NEB* | 4 | 27 |
| 8 | TGM4* | 4 | 30.8 |
| 9 | NELL2* | 4 | 33.5 |
| 10 | KRT13* | 4 | 49 |
| 11 | SLC26A4* | 4 | 63.3 |
To identify candidate oncogenes activated in ETS negative prostate cancers, we analyzed the remaining top meta-outliers for two characteristics: 1) over-expression in prostate cancer compared to benign prostate tissue and 2) mutually exclusive over-expression with ERG and ETV1 (as ~95% of cancers with ERG or ETV1 over-expression have detectable ETS fusions (Tomlins et al., 2007a; Tomlins et al., 2005)). Specific examples of meta-outliers that failed one or both criteria are shown in Figure S1 and Table 1. We identified SPINK1 (serine peptidase inhibitor, Kazal type 1), the 2nd ranked meta-outlier, as showing over-expression in prostate cancer compared to benign prostate tissue and mutually exclusive over-expression with ERG and ETV1 across multiple studies. SPINK1 was not measured in one of the studies in the meta-analysis (Lapointe et al., 2004), and ranked in the top 10 in 2 of the remaining 6 studies.
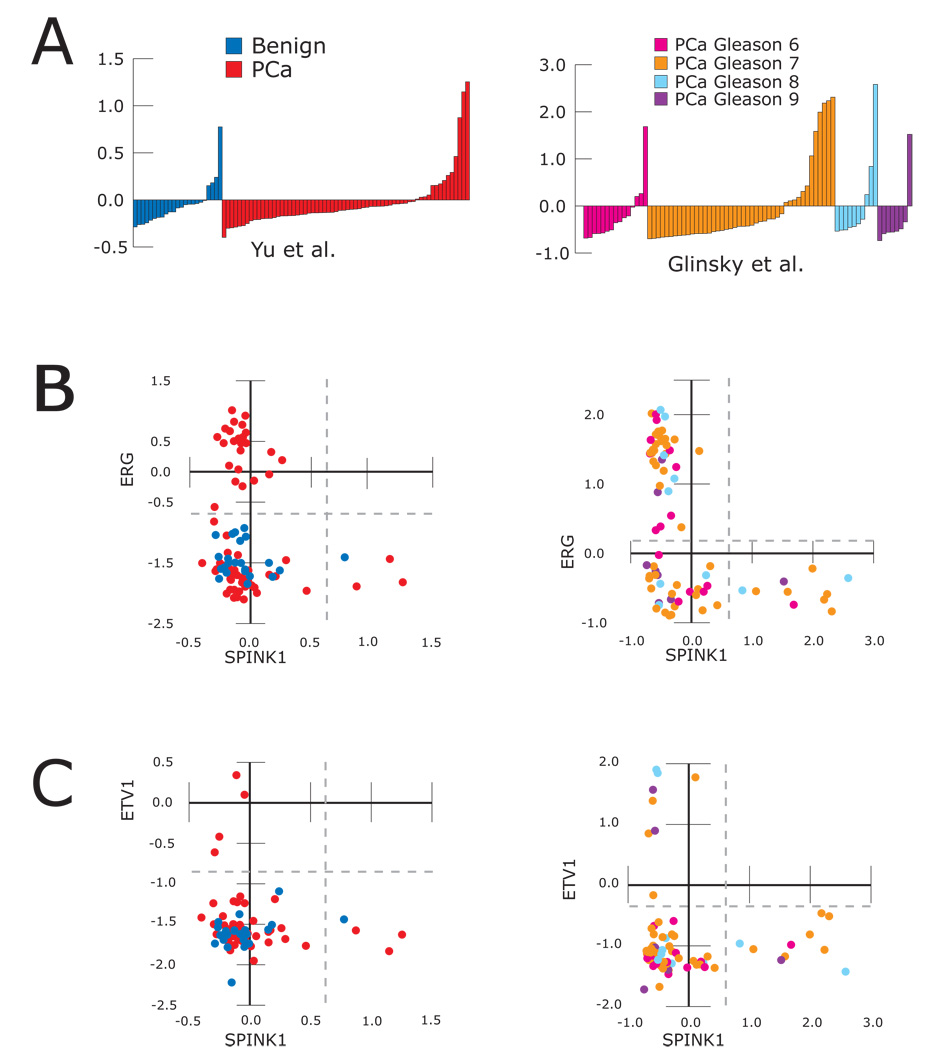
The profile of SPINK1 expression and scatter plots with ERG and ETV1 for two studies (Glinsky et al., 2004; Yu et al., 2004) where SPINK1 was identified as a top 100 outlier are shown in Figure 1. with plots from the other 4 studies in the meta-analysis (Dhanasekaran et al., 2001; LaTulippe et al., 2002; Vanaja et al., 2003; Welsh et al., 2001), shown in Figure S2. Additionally, SPINK1 expression in an additional unpublished prostate cancer profiling study (GSE8218, where SPINK1 was the 3rd ranked outlier at the 90th %) and two multi-cancer studies profiling prostate cancer (((Su et al., 2001) and GSE2109), are also shown in Figure S2. In total, from these nine studies, SPINK1 showed outlier expression (see Methods) in only 4 of 136 (2.9%) benign prostate tissue samples and 56 of 376 (14.9%) clinically localized prostate cancers (two sided Fisher’s exact test, p = 9.5E-7). Remarkably, 372 of 376 profiled clinically localized prostate cancers (98.9%) showed mutually exclusive outlier-expression of SPINK1, ERG, and ETV1, as shown in Figure 1 & Figure S2 and Table S2.
Figure 1. Meta COPA identifies SPINK1 as a mutually exclusive outlier with ERG and ETV1 in prostate cancer.
Meta-COPA analysis of 7 prostate cancer gene expression profiling datasets in Oncomine. The expression of SPINK1 and scatter plots of ERG vs. SPINK1 and ETV1 vs. SPINK1 expression are shown from two studies (Glinsky et al., 2004; Yu et al., 2004) where SPINK1 ranked as a top 100 COPA outlier. A. The expression of SPINK1, in normalized expression units (non-median centered), for all profiled samples including benign prostate tissue (blue) and clinically localized prostate cancer (PCa, red), as well as Gleason pattern 6, 7, 8 or 9 prostate cancer (magenta, orange, light blue and purple, respectively) are shown. B–C. Scatter plots are shown for B) ERG vs. SPINK1 and C) ETV1 vs. SPINK1 for all samples in both studies. Outlier-expression is delineated by the dashed gray lines (See Methods). See Figure S2 for SPINK1 outlier expression in additional prostate cancer profiling studies.
To confirm the outlier expression of SPINK1 exclusively in ETS negative prostate cancers, we measured SPINK1, ERG and ETV1 expression by quantitative PCR (qPCR) in an independent cohort of 10 benign prostate tissues and 61 prostate cancers (54 clinically localized and 7 metastatic samples). While ERG, ETV1, or SPINK1 showed outlier-expression in 25 (41%), 4 (6.5%), and 4 (6.5%) of 61 prostate cancers (clinically localized and metastatic), respectively, no benign prostate tissue samples demonstrated outlier-expression of these genes. Consistent with the above microarray studies, ERG, ETV1 and SPINK1 showed outlier-expression in distinct cancers (Fig S3).
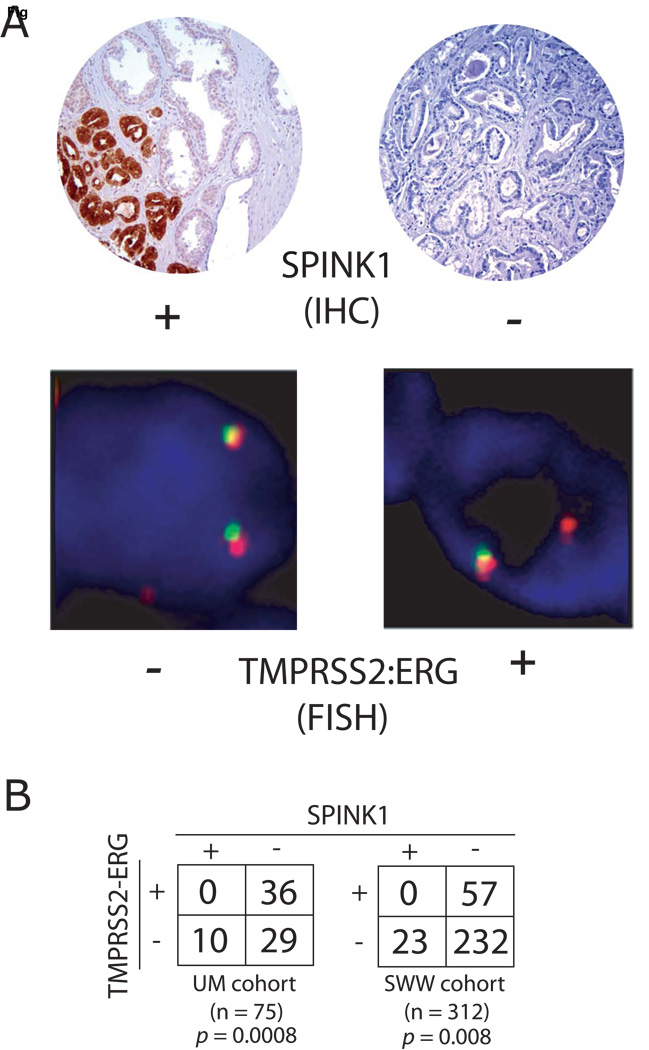
After demonstrating that SPINK1 outlier expression defines a subset of ETS rearrangement negative prostate cancers at the transcript level, we evaluated the expression of SPINK1 protein in prostate cancers. By immunohistochemical (IHC) analysis on tissue microarrays (TMAs), we evaluated SPINK1 expression in two independent cohorts, (University of Michigan (UM) and Swedish Watchful Waiting (SWW)) representing a total of 392 cases of clinically localized prostate cancers. We have previously evaluated both cohorts for TMRPSS2-ERG fusion status by fluorescence in situ hybridization (FISH) (Demichelis et al., 2007; Mehra et al., 2007). In both cohorts, prostate cancer epithelia exhibited either strong or no expression of SPINK1, without intermediate staining as observed for many prostate cancer markers. As shown in Figure 2, in the UM cohort, 10 and 36 of 75 cases were positive for SPINK1 expression (13.3%) and TMRPSS2-ERG fusions (48%), respectively, with all SPINK1 positive cases being TMRPSS2-ERG negative (one sided Fisher’s exact test, p = 0.0008). In the SWW cohort, 23 and 57 of 312 cases were positive for SPINK1 expression (7.4%) and TMRPSS2-ERG fusions (18.3%), respectively, again with all SPINK1 positive cases being TMRPSS2-ERG negative (one sided Fisher’s exact test, p = 0.008).
Figure 2. Confirmation of SPINK1 outlier-expression exclusively in ETS negative prostate cancers.
SPINK1 protein expression was evaluated in two cohorts (University of Michigan (UM) and Swedish Watchful Waiting (SWW)) using immunohistochemsitry (IHC) on tissue microarrays that have previously been evaluated for TMRPSS2-ERG status by fluorescence in situ hybridization (FISH). A. Representative SPINK1 positive and negative cores are shown, along with cells from the same cores negative and positive for TMRPSS2-ERG rearrangement by FISH. A TMRPSS2-ERG rearrangement through intrachromosomal deletion is indicated by loss of one 5’ (green) ERG signal. B. Contingency tables for SPINK1 expression and TMRPSS2-ERG status and p-values for Fisher’s exact tests for both cohorts are indicated.
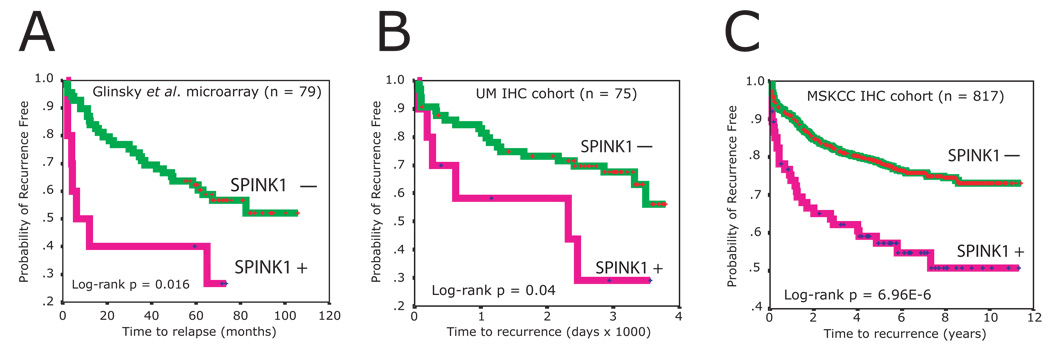
Approximately 25–40% of patients treated by radical prostatectomy for clinically localized prostate cancer will experience disease recurrence, initially indicated by an increase in the serum level of PSA (biochemical recurrence) (Han et al., 2001; Hull et al., 2002). Thus, we next sought to determine if SPINK1 outlier status was associated with biochemical recurrence after surgical resection. We identified two datasets from the evaluated cohorts for which we had access to follow-up biochemical recurrence information and a sufficient number of SPINK1 positive cases (>5). We first examined the Glinsky et al. (Glinsky et al., 2004) gene expression dataset, which contained tumors from 79 patients (with 37 recurrences), 10 of which showed outlier mRNA transcript expression of SPINK1. These patients had a significantly higher risk of recurrence than patients without SPINK1 outlier expression (hazard ratio: 2.65, 95% CI: 1.16–6.07, log rank p = 0.016) by Kaplan-Meier analysis (Fig 3A). Multivariate Cox proportional-hazards regression analysis also revealed that SPINK1 outlier status, independent of Gleason score, lymph node status, surgical margin status, age and pre-operative PSA, was a significant predictor of clinical recurrence of prostate cancer (hazard ratio: 2.5; 95% CI: 1.1–6.0; p = 0.035, Table S3).
Figure 3. SPINK1 outlier expression identifies an aggressive subtype of ETS negative prostate cancers.
Relationship between SPINK1 outlier expression and biochemical recurrence after surgical resection. Kaplan-Meier analyses of outlier SPINK1 expression from the (A) Glinsky et al. DNA microarray dataset (Glinsky et al., 2004) and SPINK1 IHC from the (B) UM and (C) Memorial Sloan Kettering Cancer Center (MSKCC) cohorts and biochemical recurrence after surgical resection are shown.
We next performed the same analysis on the UM cohort (75 cases, 28 recurrences) evaluated for SPINK1 status by IHC. By Kaplan-Meier analysis, SPINK1 positive staining was significantly associated with biochemical recurrence (hazard ratio: 2.49, 95% CI: 1.01–6.18, p = 0.04, Fig 3B). Multivariate Cox proportional-hazards regression analysis again confirmed that SPINK1 status predicted recurrence independently of other clinical parameters (Table S3). With an adjusted hazard ratio of 4.1 (95% CI: 1.4–11.7, p = 0.009), it was the strongest predictor in this model.
As a final validation, we performed IHC for SPINK1 status on an independent cohort of 817 evaluable prostate cancers (200 recurrences) from the Memorial Sloan Kettering Cancer Center (MSKCC). In this MSKCC cohort, with IHC performed independently from the UM and SWW cohorts using a different SPINK1 antibody, 297 of the 817 cases (36%) of cases showed positive SPINK1 immunoreactivity in at least one of three triplicate cores. In addition, staining intensity was more variable than that observed in the UM and SWW cohorts. As the percentage of cases in this cohort with SPINK1 staining (36%) is far greater than the other IHC cohorts (13% and 7%) or the percentage of SPINK1 outlier samples from DNA microarray and qPCR studies (15% and 7%, see Table S4), we defined SPINK1 positive cases in the MSKCC cohort as those with at least one core showing greater than 80% of cells showing positive SPINK1 immunoreactivity, resulting in 75 SPINK positive cases (9%), consistent with the other studies.
By Kaplan-Meier analysis, SPINK1 positive cases in the MSKCC cohort showed significantly shorter time to biochemical recurrence (hazard ratio: 2.32, 95% CI: 1.59–3.39, P = 6.96E-06, Fig 3C). Multivariate Cox proportional-hazards regression analysis again confirmed that SPINK1 outlier status, independent of Gleason score, lymph node status, surgical margin status, seminal vesicle invasion, extracapsular extension and preoperative PSA, was a significant predictor of clinical recurrence (hazard ratio: 2.02; 95% CI: 1.37–2.99; p = 0.0004, Table S3). Clinically, nomograms are commonly used to predict the likelihood of biochemical recurrence after surgical resection by optimally incorporating clinical and pathological parameters. To determine whether the addition of SPINK1 improves a validated nomogram for predicting the 7-year post-prostatectomy probability of biochemical recurrence (Kattan et al., 1999), we assessed the concordance index (Kattan et al., 2003) (the probability that given two randomly selected patients, the patient with the worse outcome is indeed predicted to have a worse outcome) of the nomogram and the nomogram plus SPINK1 status. The bootstrap-corrected concordance index was minimally improved in all three datasets by the addition of SPINK1 status to the nomogram (Glinsky et al: 0.772 vs 0.762, UM IHC: 0.698 vs 0.676 and MSKCC: 0.775 vs. 0.765). Thus, while SPINK1 does not dramatically add to the predictive ability of an optimized multivariate model, we demonstrate by analyzing 971 cancers from three independent cohorts that SPINK1 outlier status identifies an aggressive subset of prostate cancers.
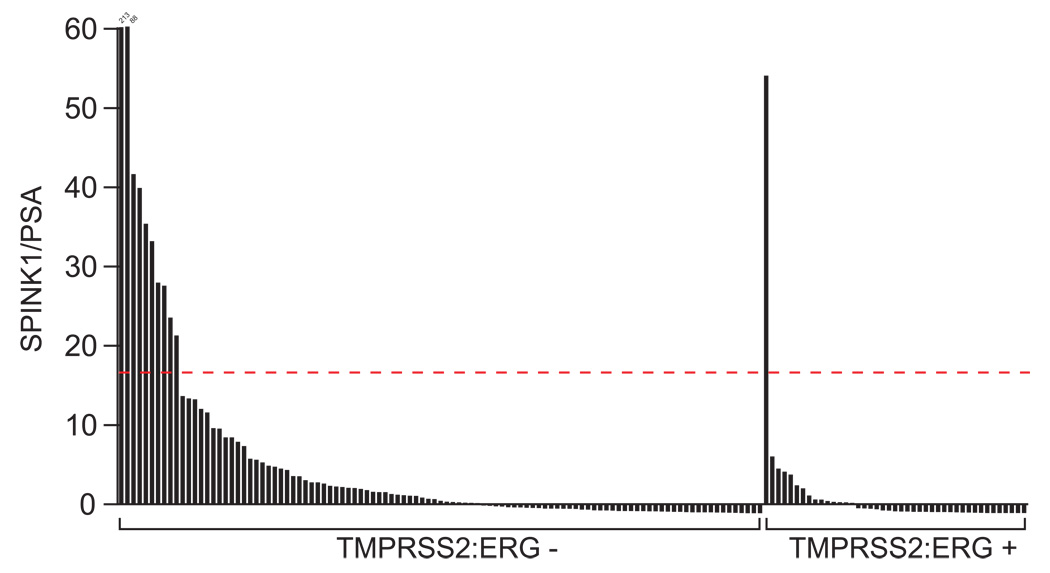
We next sought to determine if outlier expression of SPINK1 could be detected non-invasively. As increased serum levels of SPINK1 occur in multiple malignancies (Paju and Stenman, 2006; Stenman, 2002), and 44% of patients with prostate cancer are reported to have elevated serum levels of SPINK1 (Paju et al., 2007), we sought to identify a more specific assay to identify patients with tumors showing SPINK1 outlier expression. We have recently described the detection of TMRPSS2-ERG fusion transcripts in the urine of men with prostate cancer (Laxman et al., 2006), and this assay allows us to more directly assess transcripts contributed by prostatic cells. Thus, we assessed SPINK1 expression from a cohort of 148 urine samples collected from men with prostate cancer that we have characterized as TMRPSS2-ERG positive (43) or negative (105). As expected, SPINK1 expression was higher in TMPRSS2:ERG negative vs. positive samples (Mann-Whitney U test, p = 5E-5), and 21 of the 22 samples with the highest SPINK1 expression were TMPRSS2:ERG negative. Using the same method to identify SPINK1 outlier samples as described for our tissue qPCR cohort, 1 of the 43 TMRPSS2-ERG positive samples (2.3%) showed SPINK1 outlier-expression, 10 of the 105 TMRPSS2-ERG negative samples (10%) showed SPINK1 outlier-expression (Fig 4) (Fisher’s exact test, p = 0.12). In addition, compared to urine collected from 96 men presenting for evaluation of prostate cancer with negative needle biopsies, SPINK1 expression is a significant predictor of prostate cancer in both univariate and multivariate analyses, and no negative samples show SPINK1 outlier-expression (Laxman et al., In press).
Figure 4. SPINK1 outlier expression can be detected non-invasively in urine.
Non-invasive detection of SPINK1 outlier-expression in men with TMRPSS2-ERG negative prostate cancer. Total RNA was isolated from the urine of 148 men with prostate cancer and assessed for TMRPSS2-ERG and SPINK1 expression by quantitative PCR. Samples above the dashed red line show SPINK1 outlier expression (See Methods).
SPINK1 encodes a 56 amino acid secreted peptide, also known as PSTI or TATI. Originally isolated from bovine pancreas and human pancreatic juice, its normal function is thought to be the inhibition of serine proteases such as trypsin (Greene et al., 1976; Haverback et al., 1960; Kazal et al., 1948; Paju and Stenman, 2006). SPINK1 levels are strongly elevated during inflammation and pancreatitis (Paju and Stenman, 2006). Like the pancreas, the prostate gland also secretes a variety of serine proteases, most notably the kallikrein enzyme prostate specific antigen (PSA), but also trypsin, the expression of which is increased in prostate cancer (Bjartell et al., 2005). Thus, SPINK1 outlier expression may have a role in modulating the activity of cancer-related proteases. Additionally, SPINK1 has been reported to stimulate DNA synthesis in rat pancreatic cancer cells and human fibroblasts, suggesting additional roles in oncogenesis(Freeman et al., 1990; Ogawa et al., 1985). SPINK1 mRNA and protein have been detected in a variety of benign and cancerous tissues, and recently its expression in prostate and prostate cancer has been described (Paju et al., 2007; Paju and Stenman, 2006; Stenman, 2002). It is notable that SPINK is also overexpressed in other cancers and elevated serum level is an independent prognostic sign in many of these (reviewed in 21 and 22).
To investigate a functional role for SPINK1 in prostate cancer, we generated adenoviruses expressing SPINK1 and infected the benign immortalized prostate epithelial cell line RWPE to generate RWPE-SPINK1 cells. Over-expression of SPINK1 had no significant effect on the proliferation or invasion of RWPE cells (Fig 5A–B). As SPINK1 over-expression had no effect on benign prostate cells, we hypothesized that SPINK over-expression may occur later in prostate cancer progression in the presence of coexisting genetic lesions, consistent with its association with aggressive prostate cancer.
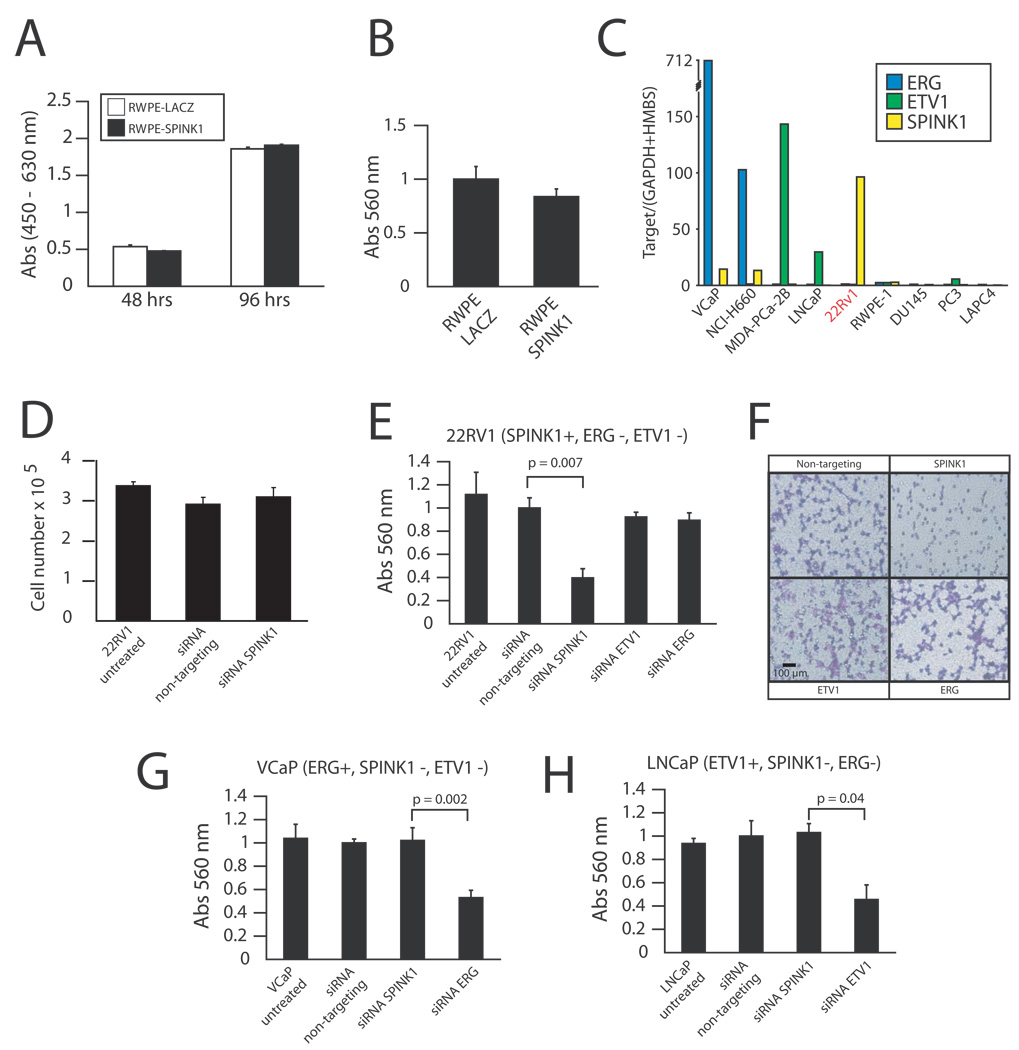
Figure 5. Knockdown of SPINK1 in 22RV1 prostate cancer cells attenuates invasiveness.
A–B. To recapitulate the outlier-expression of SPINK1, we generated adenoviruses expressing SPINK1 or LACZ (control). The benign immortalized prostate cell line RWPE was infected with SPINK1 or LACZ adenovirus as indicated and assayed for (A) proliferation or (B) invasion through a modified basement membrane. C. As these results suggest that SPINK1 may require co-existing genetic lesions to function in prostate cancer, we assayed prostate cancer cell lines by qPCR for SPINK1 (yellow), ERG (blue) and ETV1 (green) outlier expression. D–F. SPINK1 mediates invasiveness in 22RV1 cells. To investigate the role of SPINK1 in the outlier-expressing cell line 22RV1, cells were treated with transfection reagent alone (untreated), or transfected with non-targeting or siRNA against SPINK1, ETV1 or ERG as indicated. Cells were assayed for (D) proliferation and (E) invasion. Photomicrographs of invaded cells treated with the indicated siRNA are shown in F. G–H. VCaP (TMPRSS2-ERG positive) and LNCaP (ETV1 rearrangement positive) prostate cancer cell lines as indicated were treated with transfection reagent alone (untreated), or transfected with non-targeting or siRNA against SPINK1, ETV1 or ERG as indicated and assayed for invasion. For all proliferation and invasion experiments, mean (n = 3) + S.E. are shown, and P-values < 0.05 are given.
Thus we analyzed a panel of prostate cancer cell lines to identify an appropriate in vitro model for SPINK1 outlier-expression. We identified marked over-expression of SPINK1 exclusively in the 22RV1 cell line (Fig 5C), consistent with previous work that reported high expression in this cell line (Paju et al., 2007). The aggressive 22RV1 prostate cancer cell line was derived from a human prostate carcinoma xenograft that was serially propagated in nude mice after castration-induced regression and relapse of the parental, androgen-dependent CWR22 xenograft (Sramkoski et al., 1999). Importantly, 22RV1 does not over-express ERG or ETV1 (Fig 5C), similar to clinical SPINK1 outlier cases, supporting its use as a cell line model of SPINK1 outlier expression. To assess the function of SPINK1 in 22RV1, we utilized siRNA knockdown. While SPINK1 knockdown had no affect on 22RV1 proliferation (Fig 5D), SPINK1 knockdown markedly attenuated the invasiveness of 22RV1 cells through a modified basement membrane (Fig 5E–F). Similar results were obtained with two additional siRNA duplexes targeting SPINK1 (Fig S4).
Consistent with the mutually exclusive over-expression of ERG, ETV1 and SPINK1, siRNA knockdown of ERG or ETV1 in 22RV1 had no effect on invasion, while SPINK1 knockdown had no effect on the invasiveness of VCaP (TMPRSS2-ERG +, SPINK1 −) or LNCaP (ETV1 rearrangement +, SPINK1 −) (Fig 5G–H). Importantly, siRNA knockdown of ERG in VCaP and ETV1 in LNCaP similarly attenuated invasion (Fig 5G–H) without affecting proliferation (Tomlins et al., 2007a; Tomlins et al., In press). Additionally, microarray analysis of 22RV1-si SPINK1 cells revealed only limited transcriptional effects (76 features over-expressed, 14 features under-expressed, Table S5 and Fig S5), suggesting that SPINK1 knockdown directly affects cellular invasiveness. Together, these results support a role for SPINK1 in prostate cancer invasion, consistent with its over-expression in aggressive prostate cancers.
The outlier expression of SPINK1 in a subset of prostate cancers suggests that SPINK1 expression may be activated by a unique molecular event, similar to TMPRSS2:ETS positive prostate cancers. However, FISH studies using locus/control and 5’/3’ split probes demonstrated no evidence of amplification or gross rearrangements, respectively, in samples with SPINK1 over-expression (data not shown). Additionally, sequencing of the SPINK1 coding region identified no mutations in samples with SPINK1 outlier-expression (data not shown). Thus, SPINK1 may be activated by increased transcription, possibly through promoter mutations affecting regulatory elements. Alternatively, SPINK1 may be activated by a unique upstream genetic event. However, few genes show consistent correlation with SPINK1 across data sets (Fig S6), suggesting that SPINK1 would be an exclusive downstream target. It is also possible that SPINK1 may be down-regulated in TMPRSS2:ETS positive cancers, however we would expect high SPINK1 expression in benign prostatic epithelium and the in vitro data described above supports a role for SPINK1 over-expression in prostate cancer progression.
Future studies will be directed at determining the mechanism by which SPINK1 is over-expressed in TMPRSS2:ETS negative prostate cancers and whether determination of SPINK1 in serum is of diagnostic and prognostic use.
Although conflicting reports of TMPRSS2:ETS fusion status and aggressiveness have been reported, recent large cohort studies have shown that TMPRSS2:ERG fusion positive prostate cancers harboring an intrachromosomal deletion between the TMPRSS2 and ERG loci on chromosome 21 are associated with aggressiveness (Attard et al., 2008; Demichelis et al., 2007; Lapointe et al., 2007; Mehra et al., 2007; Nam et al., 2007a; Nam et al., 2007b; Perner et al., 2006; Rajput et al., 2007; Wang et al., 2006; Winnes et al., 2007; Yoshimoto et al., 2006). Supporting this hypothesis, in a cohort of patients undergoing rapid autopsy after death from hormone refractory metastatic prostate cancer, we found that all TMPRSS2:ERG fusion positive patients were deletion positive (R Mehra et al., unpublished observations). These “deletion positive” TMPRSS2:ERG positive cases, representing ~25% of all prostate cancers, likely account for the association of TMPRSS2:ERG positivity with aggressiveness. In this report, we identify SPINK1 positive samples as defining an aggressive subset of TMRPSS2:ETS negative prostate cancers (~10% of all prostate cancers). Future studies will be needed to identify the molecular mechanisms, including response or resistance to current therapies, that drive the aggressiveness of TMRPSS2:ERG deletion positive and SPINK1 positive prostate cancers.
In conclusion, using a combination of in silico bioinformatics analysis coupled with independent experimental validation, we analyzed data on ~1,800 prostate cancers, demonstrating the consistent outlier-expression of SPINK1 in TMPRSS2:ETS negative prostate cancers (Table S4). We provide evidence that SPINK1 outlier-expression defines an aggressive molecular sub-type of prostate cancer (~10% of cases) not attributable to known gene fusion events. We hypothesize that the molecular lesion(s) that initially drive ETS negative tumors, which are presently unclear, may predispose to activation of SPINK1 expression later in prostate cancer progression. Additionally, SPINK1 positive tumors may arise from a different prostate progenitor cell type than ETS positive tumors, and SPINK1 expression may be a marker of this cell type. We demonstrate that SPINK1 may be monitored non-invasively in urine and thus could serve to complement gene-fusion based urine testing for prostate cancer. Additionally, we demonstrate the utility of 22RV1 as a cell line model for SPINK1 outlier expression. Finally, we extend the utility of our original COPA approach by using a meta-COPA strategy to nominate candidate oncogenes in specific cancer types.
Materials and Methods
Cancer Outlier Profile Analysis (COPA) and outlier analysis
COPA analysis was performed on 7 prostate cancer gene expression data sets (Dhanasekaran et al., 2001; Glinsky et al., 2004; Lapointe et al., 2004; LaTulippe et al., 2002; Vanaja et al., 2003; Welsh et al., 2001; Yu et al., 2004) in Oncomine 3.0 (www.oncomine.org) as described (Tomlins et al., 2005). 1) For each data set considering all samples, gene expression values are median-centered per gene, setting each gene’s median expression value to zero. 2) The median absolute deviation (MAD) is calculated per gene and scaled to 1 by dividing each gene expression value by its MAD. Of note, median and MAD are used for transformation as opposed to mean and standard deviation so that outlier expression values do not unduly influence the distribution estimates, and are thus preserved post-normalization.3) For each gene in each dataset, COPA scores are computed as the 75th, 90th and 95th percentile of ascending transformed gene expression values. Thus, each gene in each dataset has 3 COPA scores, one at each percentile cutoff, representing the degree of over-expression in decreasing subsets of cases. 4) In each dataset, all genes are rank-ordered by the 3 COPA scores, generating 3 rank-ordered lists of genes per dataset. 5) For each dataset, we defined outlier genes as those that ranked in the top 100 COPA scores in any one of the 3 rank-ordered lists. 6) To identify “metaoutlier” genes, we ranked genes by the number of datasets where the gene was identified as an outlier gene. Genes identified as outliers in the same number of studies were further ranked by their average outlier rank across those studies. This process is summarized in Figure S7. Data sets can be accessed in Oncomine by searching for “[author last name]_prostate”, e.g “Yu_prostate”.
SPINK1 expression was also interrogated in prostate cancer specimens from two multi-cancer profiling studies ((Su et al., 2001) and the International Genomics Consortium’s expO dataset (GSE2109)), and the Yang et al. “Gene expression data from prostate cancer samples” dataset (GSE8218). The two multi-cancer studies were not included in the meta-analysis, as prostate cancer samples comprised a minority of the profiled samples, and GSE8218 was not available at the time the meta-analysis was performed.
Individual samples showing outlier-expression in each data set were identified by a two step process that recreates the visual process of identifying the natural “gap” between non-outlier and outlier sample populations. First, Oncomine generated gene expression values (ERG, ETV1 and SPINK1) for all prostate samples in each data set (non-COPA transformed, excluding metastatic prostate cancer) were median centered. Next, for each gene, all samples were rank ordered in ascending order and the difference between each rank ordered sample and the preceding sample was calculated. In each data set, ERG showed two distributions of expression separated by a natural gap in expression levels. This visual gap for each data set was quantified after ordering the samples as just described and ranged from 0.22 to 1.0 (median 0.63) normalized expression units. This same method was then applied to define ETV1 outlier expression, with the natural gap for ETV1 populations ranging from 0.25 to 2.1 (median 0.48), except for the GSE2109 study, which showed no ETV1 outlier population. SPINK1 populations showed a similar distribution in all datasets, with the natural gap ranging from 0.27 to 1.3 (median 0.41). Hence, formally described, the first sample with a positive median centered value and greater than 0.22 normalized expression unit difference compared to the preceding sample marked the transition to the outlier population for all genes in each dataset (Fig S8). Specific reporters used and the number of SPINK1, ERG, ETV1 outliers for each data set are shown in Table S2. Outlier expression in quantitative PCR (qPCR) samples (tissue and urine) was determined similarly, except normalized expression values for each target gene were log transformed before median centering and rank ordering. Metastatic prostate cancer samples were also included in the qPCR tissue cohort.
Samples
Tissues used for qPCR were from the radical prostatectomy series at the University of Michigan and from the Rapid Autopsy Program, which are both part of University of Michigan Prostate Cancer Specialized Program of Research Excellence (S.P.O.R.E.) Tissue Core. For combined fluorescence in situ hybridization (FISH) and immunohistochemsitry (IHC) evaluation, the University of Michigan (UM) cohort consisted of samples from the radical prostatectomy series. The Swedish Watchful Waiting (SWW) cohort consisted of samples from a Swedish population-based cohort of men with localized prostate cancer diagnosed incidentally by trans-urethral resection of the prostate for symptomatic benign prostatic hyperplasia as described (Andren et al., 2006; Johansson et al., 2004). The Memorial Sloan Kettering Cancer Center (MSKCC) cohort consisted of patients with localized or locally advanced prostate cancer that were treated by radical prostatectomy at MSKCC between 1985 and 2003. All samples were obtained with IRB approval from the respective institutions (UM, MSKCC or Örebro Medical Centre (for the SWW cohort). The prostate cancer cell line 22-RV1 was provided by Jill Macoska (University of Michigan).
Quantitative PCR (qPCR) from tissue samples
qPCR was performed using SYBR Green dye on an Applied Biosystems 7300 Real Time PCR system (Applied Biosystems, Foster City, CA) essentially as described(Tomlins et al., 2006; Tomlins et al., 2005). Briefly, total RNA was isolated from tissues using Trizol (Invitrogen, Carlsbad, CA). RNA was quantified using a ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE) and 3–5 µg of total RNA was reverse transcribed into cDNA using SuperScript III (Invitrogen) in the presence of random primers. All qPCR reactions were performed with Power SYBR Green Master Mix (Applied Biosystems) and 25 ng of both the forward and reverse primer using the manufacturer’s recommended thermocycling conditions. For each experiment, threshold levels were set during the exponential phase of the qPCR reaction using Sequence Detection Software version 1.2.2 (Applied Biosystems). The amount of ERG, ETV1 and SPINK1 relative to the average of the housekeeping genes GAPDH and HMBS for each sample was determined using the comparative threshold cycle (Ct) method (according to the Applied Biosystems User Bulletin #2, http://docs.appliedbiosystems.com/pebiodocs/04303859.pdf). All oligonucleotide primers were synthesized by Integrated DNA Technologies (Coralville, IA). GAPDH and HMBS, and ERG (exon5_6) and ETV1 (exon6_7) primers were as described (Tomlins et al., 2005). Sequences for SPINK1 are as follows:
SPINK1_f- CAAAAATCTGGGCCTTGCTGAGAAC
SPINK1_r- AGGCCTCGCGGTGACCTGAT
Approximately equal efficiencies of the primers were confirmed using serial dilutions of pooled prostate cancer cDNA in order to use the comparative Ct method. All reactions were subjected to melt curve analysis.
Immunohistochemsitry (IHC) and fluorescence in situ hybridization (FISH)
IHC for the University of Michigan (UM) and Swedish Watchful Waiting (SWW) cohorts was performed using a mouse monoclonal antibody against SPINK1 (H00006690-M01, Abnova, Taipei City, Taiwan) on tissue microarrays (TMA) containing cores from 75 (UM) and 312 (SWW) evaluable cases of localized prostate cancer. Cases with staining in any cancerous epithelial cells were deemed positive (median 40%, range 1–90%). Previously, we have evaluated cases on these tissue microarrays for TMRPSS2-ERG fusion status by FISH using break apart ERG assays as previously described (Demichelis et al., 2007; Mehra et al., 2007; Tomlins et al., 2005). A one-sided Fisher’s exact test was used to evaluate the relationship between SPINK1 and fusion status, as these studies were performed with the prior hypothesis that there was an inverse correlation between SPINK1 expression and fusion status.
MSKCC Immunohistochemistry
IHC for the MSKCC cohort was performed using an in house mouse monoclonal antibody against SPINK1 (code 6E8 (Osman et al., 1993)) on tissue microarrays containing triplicate cores from 817 evaluable cases of localized prostate cancer. The percentage of positive tumor cells in each core was estimated and assigned values of 0%, 5%, or multiples of 10%. The intensity of the expression was assigned a value of 0, 1, 2, or 3. Triplicate cores from each specimen were scored separately and the presence of tumorous tissue in at least two interpretable cores was required to include a case for analysis. We considered cases as SPINK1 positive if any of the three cores showed >80% of cancerous cells showing positive SPINK1 immunoreactivity (intensity 1–3).
Outcome Analyses
For Kaplan-Meier analysis of the Glinsky et al. (Glinsky et al., 2004) and UM datasets, biochemical recurrence was defined as a 0.2 ng/ml increase in PSA or recurrence of disease after prostatectomy, such as development of metastatic cancer, if biochemical recurrence information was not available. For the MSKCC cohort, only biochemical recurrence, defined as PSA > 0.2 ng/ml after surgical resection with a second confirmatory PSA-measurement > 0.2 ng/ml, was considered, as all patients with a clinical failure had previously had a biochemical recurrence. For outcome analysis from the Glinsky et al. dataset, samples positive for outlier expression of SPINK1 were defined as described above. For the IHC analysis of the UM and MSKCC cohorts, positive cases were defined as described above. Kaplan-Meier analysis and multivariate Cox proportional-hazards regression were then used to examine the association of SPINK1 with biochemical PSA recurrence. To predict the probability of disease recurrence, we used the Kattan 7-year post-operative nomogram(Kattan et al., 1999), and the concordance index of the nomogram and the nomogram plus SPINK1 status was evaluated using 1000 times bootstrapping as described (Kattan et al., 2003).
Urine based detection of SPINK1 expression
The collection of urine, isolation of RNA, RNA amplification and qPCR for TMRPSS2-ERG from men with prostate cancer was as described (Laxman et al., In press; Laxman et al., 2006). Briefly, 25 ng of isolated RNA was amplified using TransPlex Whole Transcriptome Amplification (WTA) kit (Rubicon Genomics, Ann Arbor, MI) according to the manufacturer’s instructions. For each qPCR reaction, 10ng of WTA amplified cDNA was used as template. 2× Power SYBR Green Master Mix (Applied Biosystems, Foster City, CA) and 25 ng of both the forward and reverse primers were used for SPINK1, ERG (primers as described above) and PSA (Laxman et al., 2006). For all experiments, the same threshold and baseline was set using Sequence Detection Software version 1.2.2 (Applied Biosystems). All samples with a threshold cycle (Ct) value greater than 26 for PSA were excluded to remove samples with insufficient prostate cell recovery. Samples were considered TMRPSS2-ERG positive if both ERG and TMRPSS2-ERG assays showed Ct values less than 37. The amount of SPINK1 relative to PSA was determined for each sample using the comparative Ct method. Outlier samples were identified as described above. One-sided Fisher’s exact test and Mann Whitney U tests were used to evaluate the relationship between SPINK1 and TMPRSS2-ERG status, as this study was performed with the prior hypothesis that there was an inverse correlation between SPINK1 expression and fusion status.
In vitro over-expression of SPINK1
cDNA of SPINK1 (NM_003122.2), as present in a clinical prostate cancer specimen over-expressing SPINK1, was amplified by RT-PCR using the following primers, with the forward primer including a consensus Kozak sequence (start and stop codons underlined):
SPINK1_full-f: ACCACCATGAAGGTAACAGGCATCTTTCTT
SPINK1_full-r: TCAGCAAGGCCCAGATTTTTGA
The cDNA product was TOPO cloned into the Gateway entry vector pCR8/GW/TOPO (Invitrogen), yielding pCR8-SPINK1. To generate adenoviral constructs, pCR8-SPINK1 was recombined with pAD/CMV/V5 (Invitrogen) using LR Clonase II (Invitrogen). Control pAD/CMV/LACZ clones were obtained from Invitrogen. Adenoviruses were generated by the University of Michigan Vector Core. The benign immortalized prostate cell line RWPE was infected with SPINK1 or LACZ adenoviruses, generating RWPE-SPINK1 and RWPE-LACZ for transient over-expression.
Proliferation assay
Proliferation for RWPE-LACZ and RWPE-SPINK1 cells was measured by a colorimetric assay based on the cleavage of the tetrazolium salt WST-1 by mitochondrial dehydrogenases (cell proliferation reagent WST1, Roche Diagnostics, Mannheim, Germany) at the indicated time points in triplicate. Cell counts for 22RV1 cells were estimated by trypsinizing cells and analysis by Coulter counter (Beckman Coulter, Fullerton, CA) at 72 hours in triplicate.
Invasion assays
For invasion assays, RWPE-SPINK1 and –LACZ cells (48 hours after infection with adenoviruses), or 22RV1 cells were used. Equal numbers of the indicated cells were seeded onto the basement membrane matrix (EC matrix, Chemicon, Temecula, CA) present in the insert of a 24 well culture plate, with fetal bovine serum added to the lower chamber as a chemoattractant. After 48 hours, non-invading cells and EC matrix were removed by a cotton swab. Invaded cells were stained with crystal violet and photographed. The inserts were treated with 10% acetic acid and absorbance was measured at 560nm.
SPINK1 knockdown
For siRNA knockdown of SPINK1 in 22RV1 cells, the individual siRNAs composing the Dharmacon SMARTpool against SPINK1 (LQ-019724-00, Chicago, IL) were tested for SPINK1 knockdown by qPCR, and the most effective single siRNA (J-019724-07) was used for further experiments. siCONTROL Non-Targeting siRNA #1 (D-001210-01) or siRNA against SPINK1 was transfected into 22RV1 cells using Oligofectamine (Invitrogen). After 24 hours we carried out a second identical transfection and cells were harvested 24 hours later for RNA isolation, invasion assays or proliferation assays as described above. Invasion experiments using two other siRNA directed against SPINK1 (J-019724-05 and J-019724-06, SPINK1-b and –c, respectively) were also performed (Fig S4).
Expression Profiling
Expression profiling was performed using the Agilent Whole Human Genome Oligo Microarray (Santa Clara, CA). Total RNA isolated using Trizol was purified using the Qiagen RNAeasy Micro kit (Valencia, CA). One µg of total RNA was converted to cRNA and labeled according to the manufacturer’s protocol (Agilent). Hybridizations were performed for 16 hrs at 65°C, and arrays were scanned on an Agilent DNA microarray scanner. Images were analyzed and data extracted using Agilent Feature Extraction Software 9.1.3.1, with linear and lowess normalization performed for each array. For 22RV1-si SPINK1 hybridizations, the reference was 22RV1 cells infected with non-targeting siRNA. Duplicate hybridizations were performed with duplicate dye flips, for a total of four arrays. Over and under-expressed signatures were generated by filtering to include only features with significant differential expression (PValueLogRatio < 0.01) in all hybridizations and Cy5/Cy3 ratios (LogRatio) > or < 1 (unlogged) in all hybridizations, after correction for the dye flip.
Electronic availability of the Data
The 22RV1 expression profiling data is available from GEO, accession number GSE11132.
Supplementary Material
Acknowledgements
The authors thank Lei Wang, Anjana Menon, Xiaojun Jing and Elise Nilsson for excellent technical support, and Jill Macoska for the 22RV1 cell line. Supported in part by the Department of Defense ( A.M.C.), the National Institutes of Health (U54 DA021519-01A1 to A.M.C., Prostate SPORE P50CA69568 to A.M.C., and R.B.S.), the Early Detection Research Network (UO1 CA111275-01, UO1 CA113913 to A.M.C. and J.T.W.) and the Prostate Cancer Foundation (S.A.T.). S.A.T. is supported by the GPC Biotech Young Investigator Award from the Prostate Cancer Foundation. A.M.C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation. S.A.T. and D.R.R. are Fellows of the Medical Scientist Training Program. Supported in part by P50-CA92629 SPORE from the National Cancer Institute (P.K. and M.A.R.), Translational cancer research fellowship Award No TCR/05/009/2005 from the International Union Against Cancer (UICC), European Union 6th Framework contract LSHC-CT-2004-503011 (P-Mark), the Swedish Cancer Society (Projects 4294 and 3555); the Research Fund and the Cancer Research Fund of Malmö University Hospital; the Faculty of Medicine, Lund University; the Maud and Birger Gustavsson Foundation; the Gunnar Nilsson Cancer Foundation, Finnish Cancer Foundation, Sigrid Juselius Foundation, Finnish Academy of Sciences, Helsinki University Central Hospital, and University of Helsinki.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Significance
While ETS rearrangements play a role in a majority of prostate cancers, little is known about molecular alterations driving ETS gene fusion negative cancers. In this study, we identified SPINK1 outlier-expression exclusively in a subset of ETS negative cancers. SPINK1 is associated with prostate cancer aggressiveness and can be detected non-invasively in urine. Furthermore, SPINK1 mediates invasion in a prostate cancer cell line with outlier-expression. The mechanism of SPINK1 outlier-expression remains to be characterized and is not explained by chromosomal rearrangement, deletion, or amplification. Thus, SPINK1 is a biomarker specific to a subset of aggressive ETS negative prostate cancers. Our study also demonstrates the utility of a meta-outlier strategy to identify cancer subtypes.
References
- Andren O, Fall K, Franzen L, Andersson SO, Johansson JE, Rubin MA. How well does the Gleason score predict prostate cancer death? A 20-year followup of a population based cohort in Sweden. J Urol. 2006;175:1337–1340. doi: 10.1016/S0022-5347(05)00734-2. [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, et al. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. doi: 10.1038/sj.onc.1210640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjartell A, Paju A, Zhang WM, Gadaleanu V, Hansson J, Landberg G, Stenman UH. Expression of tumor-associated trypsinogens (TAT-1 and TAT-2) in prostate cancer. Prostate. 2005;64:29–39. doi: 10.1002/pros.20236. [DOI] [PubMed] [Google Scholar]
- Demichelis F, Fall K, Perner S, Andren O, Schmidt F, Setlur SR, Hoshida Y, Mosquera JM, Pawitan Y, Lee C, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007 doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Freeman TC, Curry BJ, Calam J, Woodburn JR. Pancreatic secretory trypsin inhibitor stimulates the growth of rat pancreatic carcinoma cells. Gastroenterology. 1990;99:1414–1420. doi: 10.1016/0016-5085(90)91170-b. [DOI] [PubMed] [Google Scholar]
- Glinsky GV, Glinskii AB, Stephenson AJ, Hoffman RM, Gerald WL. Gene expression profiling predicts clinical outcome of prostate cancer. J Clin Invest. 2004;113:913–923. doi: 10.1172/JCI20032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LJ, Pubols MH, Bartelt DC. Human pancreatic secretory trypsin inhibitor. Methods Enzymol. 1976;45:813–825. doi: 10.1016/s0076-6879(76)45075-9. [DOI] [PubMed] [Google Scholar]
- Han M, Partin AW, Pound CR, Epstein JI, Walsh PC. Long-term biochemical disease-free and cancer-specific survival following anatomic radical retropubic prostatectomy The 15-year Johns Hopkins experience. The Urologic clinics of North America. 2001;28:555–565. doi: 10.1016/s0094-0143(05)70163-4. [DOI] [PubMed] [Google Scholar]
- Haverback BJ, Dyce B, Bundy H, Edmondson HA. Trypsin, trypsinogen and trypsin inhibitor in human pancreatic juice. Am J Med. 1960;29:421–433. doi: 10.1016/0002-9343(60)90038-3. [DOI] [PubMed] [Google Scholar]
- Helgeson BE, Tomlins SA, Shah N, Laxman B, Cao Q, Prensner JR, Cao X, Singla N, Montie JE, Varambally S, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- Hull GW, Rabbani F, Abbas F, Wheeler TM, Kattan MW, Scardino PT. Cancer control with radical prostatectomy alone in 1,000 consecutive patients. J Urol. 2002;167:528–534. doi: 10.1016/S0022-5347(01)69079-7. [DOI] [PubMed] [Google Scholar]
- Johansson JE, Andren O, Andersson SO, Dickman PW, Holmberg L, Magnuson A, Adami HO. Natural history of early, localized prostate cancer. Jama. 2004;291:2713–2719. doi: 10.1001/jama.291.22.2713. [DOI] [PubMed] [Google Scholar]
- Kattan MW, Shariat SF, Andrews B, Zhu K, Canto E, Matsumoto K, Muramoto M, Scardino PT, Ohori M, Wheeler TM, Slawin KM. The addition of interleukin-6 soluble receptor and transforming growth factor beta1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J Clin Oncol. 2003;21:3573–3579. doi: 10.1200/JCO.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- Kazal LA, Spicer DS, Brahinsky RA. Isolation of a Crystalline Trypsin Inhibitor-Anticoagulant Protein from Pancreas. Journal of the American Chemical Society. 1948;70:3034–3040. doi: 10.1021/ja01189a060. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Kim YH, Miller MA, Li C, Kaygusuz G, van de Rijn M, Huntsman DG, Brooks JD, Pollack JR. A variant TMPRSS2 isoform and ERG fusion product in prostate cancer with implications for molecular diagnosis. Mod Pathol. 2007 doi: 10.1038/modpathol.3800759. [DOI] [PubMed] [Google Scholar]
- Lapointe J, Li C, Higgins JP, van de Rijn M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim U, et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci U S A. 2004;101:811–816. doi: 10.1073/pnas.0304146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- Laxman B, Morris DS, Yu J, Siddiqui J, Cao J, Mehra R, Lonigro RJ, Tsodikov A, Wei JT, Tomlins SA, Chinnaiyan AM. A First Generation Multiplex Biomarker Analysis of Urine for the Early Detection of Prostate Cancer Cancer Res In Press. doi: 10.1158/0008-5472.CAN-07-3224. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laxman B, Tomlins SA, Mehra R, Morris DS, Wang L, Helgeson BE, Shah RB, Rubin MA, Wei JT, Chinnaiyan AM. Noninvasive detection of TMPRSS2:ERG fusion transcripts in the urine of men with prostate cancer. Neoplasia. 2006;8:885–888. doi: 10.1593/neo.06625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra R, Tomlins SA, Shen R, Nadeem O, Wang L, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM, Shah RB. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007 doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- Naderi A, Teschendorff AE, Beigel J, Cariati M, Ellis IO, Brenton JD, Caldas C. BEX2 is overexpressed in a subset of primary breast cancers and mediates nerve growth factor/nuclear factor-kappaB inhibition of apoptosis in breast cancer cell lines. Cancer Res. 2007;67:6725–6736. doi: 10.1158/0008-5472.CAN-06-4394. [DOI] [PubMed] [Google Scholar]
- Nam RK, Sugar L, Wang Z, Yang W, Kitching R, Klotz LH, Venkateswaran V, Narod SA, Seth A. Expression of TMPRSS2 ERG Gene Fusion in Prostate Cancer Cells is an Important Prognostic Factor for Cancer Progression. Cancer Biol Ther. 2007a;6 doi: 10.4161/cbt.6.1.3489. [DOI] [PubMed] [Google Scholar]
- Nam RK, Sugar L, Yang W, Srivastava S, Klotz LH, Yang LY, Stanimirovic A, Encioiu E, Neill M, Loblaw DA, et al. Expression of the TMPRSS2:ERG fusion gene predicts cancer recurrence after surgery for localised prostate cancer. Br J Cancer. 2007b;97:1690–1695. doi: 10.1038/sj.bjc.6604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Tsushima T, Ohba Y, Ogawa N, Tanaka S, Ishida M, Mori T. Stimulation of DNA synthesis in human fibroblasts by human pancreatic secretory trypsin inhibitor. Research communications in chemical pathology and pharmacology. 1985;50:155–158. [PubMed] [Google Scholar]
- Osman S, Turpeinen U, Itkonen O, Stenman UH. Optimization of a time-resolved immunofluorometric assay for tumor-associated trypsin inhibitor (TATI) using the streptavidin-biotin system. J Immunol Methods. 1993;161:97–106. doi: 10.1016/0022-1759(93)90201-h. [DOI] [PubMed] [Google Scholar]
- Paju A, Hotakainen K, Cao Y, Laurila T, Gadaleanu V, Hemminki A, Stenman UH, Bjartell A. Increased Expression of Tumor-Associated Trypsin Inhibitor, TATI, in Prostate Cancer and in Androgen-Independent 22Rv1 Cells. Eur Urol. 2007 doi: 10.1016/j.eururo.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Paju A, Stenman UH. Biochemistry and clinical role of trypsinogens and pancreatic secretory trypsin inhibitor. Crit Rev Clin Lab Sci. 2006;43:103–142. doi: 10.1080/10408360500523852. [DOI] [PubMed] [Google Scholar]
- Perner S, Demichelis F, Beroukhim R, Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD, Pienta KG, et al. TMPRSS2:ERG Fusion-Associated Deletions Provide Insight into the Heterogeneity of Prostate Cancer. Cancer Res. 2006;66:8337–8341. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- Rajput AB, Miller MA, De Luca A, Boyd N, Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007 doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sramkoski RM, Pretlow TG, 2nd, Giaconia JM, Pretlow TP, Schwartz S, Sy MS, Marengo SR, Rhim JS, Zhang D, Jacobberger JW. A new human prostate carcinoma cell line, 22Rv1. In vitro cellular & developmental biology. 1999;35:403–409. doi: 10.1007/s11626-999-0115-4. [DOI] [PubMed] [Google Scholar]
- Stenman UH. Tumor-associated trypsin inhibitor. Clin Chem. 2002;48:1206–1209. [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007a;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. The Role of the TMPRSS2-ERG Gene Fusion in Prostate Cancer Neoplasia In Press. doi: 10.1593/neo.07822. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM, Kalyana-Sundaram S, Wei JT, Rubin MA, Pienta KJ, et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007b;39:41–51. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chinnaiyan AM. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Vanaja DK, Cheville JC, Iturria SJ, Young CY. Transcriptional silencing of zinc finger protein 185 identified by expression profiling is associated with prostate cancer progression. Cancer Res. 2003;63:3877–3882. [PubMed] [Google Scholar]
- Wang J, Cai Y, Ren C, Ittmann M. Expression of Variant TMPRSS2/ERG Fusion Messenger RNAs Is Associated with Aggressive Prostate Cancer. Cancer Res. 2006;66:8347–8351. doi: 10.1158/0008-5472.CAN-06-1966. [DOI] [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- Winnes M, Lissbrant E, Damber JE, Stenman G. Molecular genetic analyses of the TMPRSS2-ERG and TMPRSS2-ETV1 gene fusions in 50 cases of prostate cancer. Oncology reports. 2007;17:1033–1036. [PubMed] [Google Scholar]
- Yoshimoto M, Joshua AM, Chilton-Macneill S, Bayani J, Selvarajah S, Evans AJ, Zielenska M, Squire JA. Three-Color FISH Analysis of TMPRSS2/ERG Fusions in Prostate Cancer Indicates That Genomic Microdeletion of Chromosome 21 Is Associated with Rearrangement. Neoplasia. 2006;8:465–469. doi: 10.1593/neo.06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.