Abstract
We report the first ever Raman and infrared microspectroscopic images of human cells at different stages of mitosis. These spectroscopic methods monitor the distribution of condensed nuclear chromatin, and other biochemical components, utilizing inherent protein and DNA spectral markers, and, therefore, do not require the use of any stains. In conjunction with previously reported data from the G1, S, and G2 phases of the cell cycle, the complete cell division cycle has now been mapped by spectroscopic methods. Although the results reported here do not offer new insights into the distribution of biochemical components during mitosis, the recognition of cell division without the use of stains, and the possibility of doing so on living cells, may be useful for an automatic, spectroscopic determination of the proliferation rates of cells and tissues. Spectral images were constructed by plotting spectral intensities of DNA or protein versus the coordinates from which spectra were recorded. We found that both Raman and infrared intensities depend on the overall chromatin density variation among the individual subphases of mitosis.
Index Headings: Raman microspectroscopy, Mitosis, Fourier transform infrared, FT-IR microspectroscopy, Cell cycle
INTRODUCTION
Raman1–7 and infrared (IR)8–11 microspectroscopic studies of human cells have only recently become the subject of considerable interest, whereas corresponding efforts on micro-organisms (bacteria and yeast) were first reported over a decade ago.12 For eukaryotic cells, the advantages of vibrational imaging techniques are their noninvasive character, the absence of staining and special sample preparation steps, and the sensitivity of the spectral methods to the information associated with spatial distribution of the chemical components. The overall aim of this work is the establishment of spectral patterns that can be used to determine the metabolic and proliferation status of a cell. We have evidence that proliferation and metabolism affect a cell’s spectral properties as much as the transition from normal to malignant states. Previous infrared microspectroscopic studies have concentrated on the detection of variations of a cell’s biomolecular composition during progression through the cell cycle,13 which precedes mitosis. These studies represent the necessary background information required for an eventual application of vibrational spectroscopic imaging methods to screen cells for the presence of disease that may manifest itself in increased cell proliferation.
Previous Raman microscopic studies have demonstrated that it is possible to visualize cellular components such as the nucleus, nucleoli, and chromatin.2,7 In addition, there have been attempts to correlate Raman and infrared microspectroscopic data with the morphology of cells during the processes of apoptosis and necrosis.3–5,14,15 In this study, we concentrate on the events that take place over the course of one hour, during the cell-cycle phase of mitosis, when the cell physically divides into two daughter cells. We monitor these events by mapping the Raman scattering light intensities of the chromatin during mitosis. These intensities are dependent on the degree of DNA packing in chromatin, which will be described in the third section of the paper. In particular, we report spectral images from cells during the sub-phases of mitosis, using Raman and infrared microspectral methods. These spectroscopic techniques are complementary in that the Raman imaging affords superior spatial and spectral resolution (~1 μm and 1 cm−1, respectively) and spectral discrimination, whereas the infrared technique produces data about 100 times faster and with a much better signal-to-noise ratio (S/N), but lower spatial and spectral resolution (~12 μm and 4 cm−1, respectively).
We employ both techniques to monitor the distribution of cellular components, in particular chromatin, in human epithelial cancer cells during mitosis and compare the results to spectral images acquired from non-mitotic cells. The overall aim of this paper is to develop spectroscopic methods to detect cell proliferation in samples of cultured or exfoliated human cells, and to complete the study of temporal variation of spectral signatures during the entire eukaryotic cell division cycle.
EXPERIMENTAL
Raman Microspectroscopy
Raman spectra and images were collected using a JY-Labram Microscope (Jobin Yvon Inc., Edison, NJ). This instrument is constructed around an Olympus BX30 microscope and incorporates a 300-mm focal length spectrograph with 600 or 1800 lines/mm gratings. A 632.8-nm wavelength HeNe laser, providing about 10 mW of power at the sample, is used for excitation of the Raman spectra. The same 100× objective is used for focusing the laser on a spot size of approximately 1 μm diameter and for collecting the Raman backscattered radiation. Detection of the Raman scattered photons is provided by a charge-coupled device (CCD) detector.
All Raman spectra were recorded at 1 cm−1 spectral resolution. For imaging and single point spectroscopy, two slightly different protocols were used. Raman images were recorded at 30- or 60-second exposure times per pixel, with a fixed grating position. The spatial resolution was 1 μm. To achieve this resolution, the step size of the computer-interfaced microscope stage, which has a spatial accuracy and repeatability of 1 μm, was set at 1 μm, and the laser was focused to a spot size of about 1 μm. The data acquisition time for a single cell image was fifteen hours, on average.
Higher quality, single point Raman spectra of selected cellular regions were acquired using a method first proposed by Kiefer16 to reduce CCD readout patterns. Such a readout pattern is commonly observed in Raman spectra collected via CCD detectors, and manifests itself by a constant level of noise, which does not decrease with increasing number of read-cycles of the detector. However, if a similar spectral range is acquired with slightly different grating position, the readout pattern cancels. In order to correct the spectra for the new grating position, an algorithm was reported17 that recalculates spectra from the linear dispersion of the monochromator, and the physical spacing between detector elements.
Infrared Microspectroscopy
Infrared spectra and spectral images were collected using a Perkin-Elmer Spectrum One FTIR spectrometer coupled to a Spectrum Spotlight 300 IR microscope. This totally integrated imaging infrared microspectrometer incorporates a 16 × 1 element (400 × 25 μm2) HgCdTe (MCT) array detector. Specifically designed optics permit 1:1 or 4:1 imaging, resulting in sample areas of 25×25 or 6×6 μm2 to be projected on each detector element. Spectra are collected in rapid scan mode at a maximum rate of about 80 pixels per second. For the single cell spectra reported here, 16 to 64 interferograms were coadded for each pixel at 4 cm−1 spectral resolution. The spatial resolution at 4:1 image magnification is determined mainly by the diffraction limit and was found to be about 12 μm at 1000 cm−1, and significantly better at 3000 cm−1.
Cell Culture and Sample Preparation
Human HeLa cells (cell line CCL-2 ATCC) were grown on sterilized calcium fluoride (CaF2) windows (Wilmad) in 20 mL Dulbecco’s Modified Eagle’s Medium (DMEM) (ATCC), supplemented with100 IU/mL penicillin/streptomycin (ATCC) and 10% Fetal Bovine Serum (FBS, ATCC) and incubated at 37 °C and 5% CO2. The cells were fixed in a 10% phosphate buffered formalin solution (Sigma-Aldrich), washed in distilled water and quick-dried under a compressed air stream.
Fluorescence Staining and Microscopy
After spectral data acquisition, the cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) at a concentration of 0.2 μg/mL at room temperature for three minutes. This minor-groove-binding ligand is used as a fluorescent dye for DNA and chromosomes.18 After staining, the slides were washed three times in Hank’s buffered saline solution (BSS), then washed briefly with Millipore water. The water was quickly removed under a compressed air stream, and the slides were allowed to air dry in the dark. The DAPI fluorophore was visualized via a Nikon Optiphot-2 microscope equipped with a mercury lamp and an episcopic-fluorescence attachment EFD-3, utilizing the appropriate excitation and emission filters and the Plan Apo DIC 60× oil objective and the attached Sony camera. The images were manipulated using Micrografx Picture Publisher version 8 software.
Data Treatment
Raw infrared and Raman data sets were imported into the CytoSpec19 software for baseline correction, smoothing, and uni- and multivariate methods of data analysis. Fourier self-deconvolution, as implemented in the CytoSpec software, was used for spatial resolution enhancement for some of the infrared spectral images.
RESULTS
DNA Packing and Mitosis
The total length of double-stranded DNA in the nucleus of a human cell is about a meter. Since the cell nucleus is only about 6 μm in diameter, the DNA must be greatly condensed. The degree of DNA condensation is expressed as a packing ratio. In the first level of packing organization, DNA winds around histone octamers to form nucleosome fibers, which results in a packing ratio of about 6. The next level of organization results in a solenoid structure with a 30-nm diameter and represents the “average” degree of condensation of an interphase (non-mitotic) cell and corresponds to a packing ratio of 40. During mitosis, the solenoid further condenses to form the metaphase chromosomes in which the packing ratio ranges between 7000 and 10 000.
Mitosis typically lasts less than one hour for most cell types grown in culture and can be divided into several subphases, as summarized: (1) during early prophase, the chromosomes replicated during synthesis (S) phase undergo extensive condensation but are still contained within the nuclear envelope; (2) later in the prophase, the mitotic spindle begins to grow and the nuclear envelope breaks down; (3) in the metaphase, the microtubules begin to interact with the chromosomes, which leads to the alignment of the two complete sets of diploid chromosomes lining up along the “equator” of the parent cell; (4) the anaphase commences with separation of the daughter chromosomes away from the equatorial metaphase plate and toward one of the two spindle pole regions; (5) in the telophase, two separate groups of chromosomes have formed at each pole, and a nuclear envelope begins to appear around each set of chromosomes to form two nuclei that are temporarily in one cell; and (6) furrowing and division of the cell takes place during cytokinesis to yield two distinct daughter cells.
Infrared and Raman Maps of an Interphase Cell
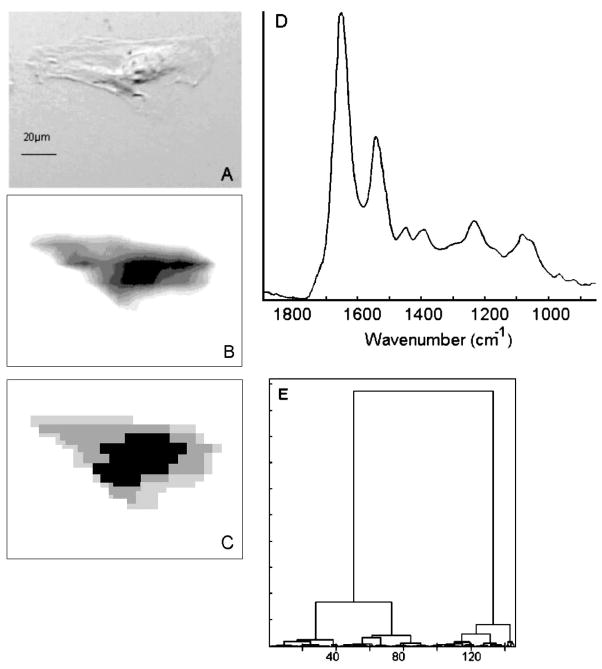
Figure 1 depicts infrared imaging data for a cultured HeLa cell grown on a CaF2 substrate. The nucleus and the nucleoli in the nucleus are discernible in the visual image (Fig. 1A). Figure 1B shows the distribution of the integrated intensity due to the protein amide I vibration over the cellular position. This image shows very large protein spectral intensities from the region of the nucleus. These intensities are nearly an order of magnitude higher than those observed for the cytoplasm. This observation confirms similar results from our and other groups20,21 that the large protein vibrations are due to two factors: the total protein content of the cell is largest in the nucleus, since histone and many other nuclear proteins are localized there. Second, the near-spherical nucleus may present a longer path to the IR beam than the surrounding cytoplasm, which is often only a few micrometers thick. We have recently shown that in dried cells, the nucleus retains a nearly spherical shape, as indicated by its Mie scattering properties.11
Fig. 1.
(A) Visual photomicrograph of unstained HeLa cell, grown on a CaF2 substrate. (B) Infrared spectral image of the cell shown in (A), based on protein absorption intensity. The spatial resolution is about 12 μm at 1000 cm−1 absorption. Light gray hues indicate low intensity, while black indicates highest intensity. (C) Hierarchical Cluster Map of the data set shown in (B). (D) Infrared spectrum of the nuclear (black) region. (E) Dendrogram for the clustering process used to produce (C). The dissimilarity between spectra increases along the Y axis.
The results of cluster analysis22 applied to the data shown in Fig. 1B are displayed in Fig. 1C. Cluster analysis produces images based on spectral similarities. Thus, Fig. 1C represents an image in which the nuclear region (black), the cytoplasm (gray), and the region of the edges of the cell (light gray) can be distinguished. The input data were vector normalized; thus, cluster analysis should detect spectral differences that are independent of the cell thickness. Since Figs. 1B and 1C show different spatial distributions of spectral patterns, one may conclude that the results from the cluster analysis are, indeed, independent of the total intensity and monitor different composition in the cytoplasm and the nucleus. Figure 1E shows the dendrogram of the clustering process. The cluster on the right side (abscissa values 100 to 150) corresponds to the nucleus (black in Fig. 1C), whereas the center cluster (40–100) corresponds to the gray area and the left cluster (0–40) to the light gray area of the cytoplasm. The light gray and gray areas are closely related and are due to different regions of the cytoplasm. The spectra from the nuclear cluster are quite different, as indicated by the merge level in the dendrogram. A spectrum of the nuclear region is shown in Fig. 1D. The intensity in the amide I band is on the order of 0.2 optical density (OD) units. Distinct spectral features due to the symmetric and antisymmetric stretching vibrations of the PO2− are manifested as peaks at ~ 1080 and 1235 cm−1, respectively. In the absence of a distinct peak due to phospholipids at 1740 cm−1, the PO2− vibrations are most likely due to the DNA and RNA. Although a spectral distinction of these two classes of nucleic acids is normally possible, the signals from the cellular nucleus appear to be a superposition of both. This spectrum is quite similar to that reported for the nuclei of cultured cells9 and differs markedly from those of pyknotic nuclei, which show markedly weaker nucleic acid spectral features. Spectra from the cytoplasm (not shown) are significantly less intense and lack the nucleic acid spectral features seen in the nucleus.
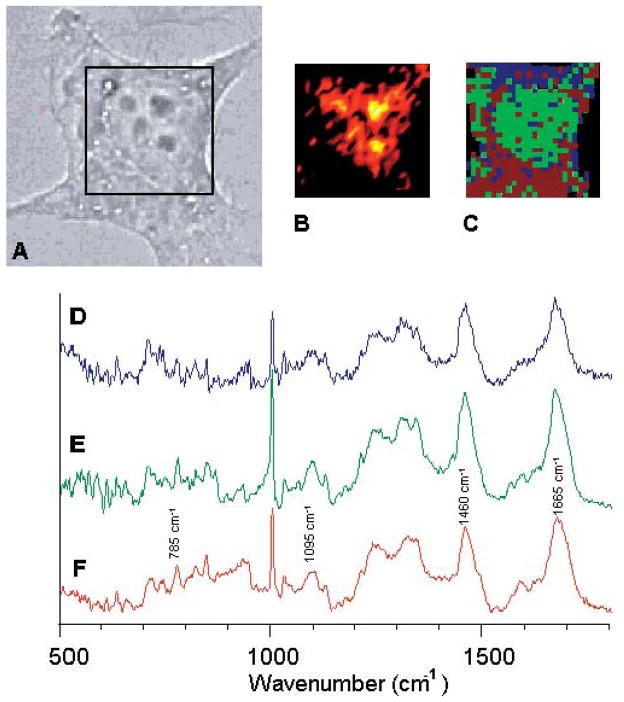
Figure 2 shows Raman images and Raman spectra of a HeLa cell. Under the higher magnification and optical quality of the Raman microscope, the nucleus and the nucleoli in the nucleus and the surrounding cytoplasm are easily detected. Thus, it is possible to record and compare the Raman spectra from these distinct areas. Figure 2 depicts spectra collected from areas about 1 × 1 μm2 in size, from the cytoplasm (Fig. 2D), nucleus (Fig. 2E) and nucleolus (Fig. 2F). The spectrum collected from the cytoplasm is dominated by protein features. Although spectra of the cytoplasm typically are much noisier than those of the nucleus, the amide I protein C=O stretching vibration around 1650 cm−1, the methyl and methylene deformations around 1460 cm−1, and the protein amide III (N–H/C–H) deformation vibration modes between 1285 and 1350 cm−1 are clearly discernible in Fig. 2D. The phenylalanine ring breathing vibration occurs as a sharp band at 1004 cm−1. In addition, there may be small RNA signals (from messenger RNA, ribosomal RNA, and transfer RNA) in the cytoplasm.
Fig. 2.
(A) Visual photomicrograph of an unstained HeLa cell, grown on a CaF2 substrate. (B) Raman spectral image of the region indicated by the black square, based on the Raman scattered intensity at 785 cm−1. Black indicates low intensity, to red, to white (highest intensity). (C) Pseudo-color map of same cell, based on hierarchical cluster analysis. (D, E, F) Single point Raman spectra from within the cytoplasm, the nucleus, and the nucleoli, respectively.
In the nucleus, additional weak bands associated with nucleic acid vibrations are observed, as represented in Fig. 2E. The guanine/adenine C=C ring stretching modes appear at 1575 cm−1, the phosphodiester backbone stretching mode at 1095 cm−1, and the uracil/thymine ring deformation modes around 785 cm−1. The spectra from the cytoplasm and nucleus (Figs. 2D and 2E) are dominated by protein spectra, with only weak nucleic acid features. Since the DNA in the nucleus of interphase cells is less condensed, as compared to M-phase DNA, it shows very weak Raman signals. This aspect will be discussed below in more detail. Figure 2F shows a spectrum from within the nucleoli. Here, the nucleic acid features are more pronounced than in the other regions of the nucleus, since the nucleic acid concentration in the nucleolus is very high.
A univariate map of the intensity of the nucleic acid peak at 785 cm−1 (Fig. 2B) reproduces the location of the nucleoli, but does not clearly delineate the nucleus. This may be due to the fact, as discussed above, that RNA signatures may be found throughout the nucleoli, the nucleus, and the cytoplasm, and that in an interphase cell, the DNA is less condensed and produces low Raman signals.
However, cluster analysis of the same data set produces a map, shown in Fig. 2C, that partially delineates the nucleus and cytoplasm. Cluster analysis works best for large data sets and spectral data of very high S/N ratio. Although both of these conditions are fulfilled only marginally in this data set, the cluster analysis recognizes the two main cellular features, the cytoplasm and nucleus, much better than the analysis of single wavelength intensities. However, cluster analysis does not delineate the nucleoli features from the nucleolus as shown in Fig. 2B.
Infrared and Raman Maps of Mitotic Cells
We now turn to the discussion of the mitotic cells. We have previously reported cells in the other phases of the cell cycle (G1, S, and G2) by infrared microspectroscopy.10,13 However, these three stages cannot be distinguished visually; thus, staining procedures were employed to determine the cell’s exact stage in the cell cycle. Mitotic cells, on the other hand, can be distinguished from interphase cells by microscopic inspection of the live cells growing on a substrate in culture. Cells exhibiting mitotic figures were selected and fixed and spectral maps acquired. The spectral maps reported below for different stages of mitosis (i.e., prophase, metaphase, anaphase, and telophase) do not represent a time course of mitosis of one individual cell, but time points in mitosis of several different cells. Each Raman spectral data set required 12 to 24 hours of data acquisition time.
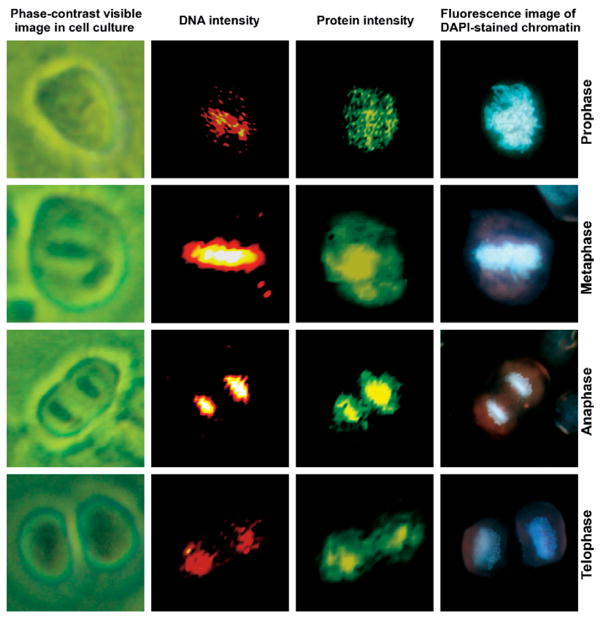
Figure 3 shows HeLa cells undergoing the typical stages of mitosis: prophase, metaphase, anaphase, and telophase. The left column of images shows the cells as seen in cell culture under an inverted phase contrast microscope. The second column shows maps of the nucleic acid band intensity at 785 cm−1, normalized with respect to the protein peak, whereas the third column depicts the protein amide I intensity (1655 cm−1). The protein intensity delineates the microtubules and the dense histone-packed chromatin. The far right column shows, for comparison, the fluorescence image of the DAPI-stained cells, demonstrating the density and distribution of the condensed chromatin within the cell at each of the phases of mitosis. The DNA-based Raman and DAPI fluorescence images show a high degree of agreement, indicating that it is possible to use Raman spectral imaging instead of fluorescence staining methods to visualize the DNA condensation during mitosis.
Fig. 3.
Photomicrographs and Raman and fluorescence (DAPI stain) images of HeLa cells during various phases of mitosis. (Top row) prophase, (second row) metaphase, (third row) anaphase, and (bottom row) late telophase. All images are collected at 40× magnification. (Column 1) Phase-contrast images of live cells in culture. (Column 2) Raman scattering intensity plots for the DNA scattering intensities. The colors range from black (low intensity), to red, to white (high intensity). (Column 3) Raman scattering intensity plots for the protein scattering intensities. The colors range from black (low intensity), to green, to yellow (high intensity). (Column 4) Fluorescence images of the DAPI-stained cells.
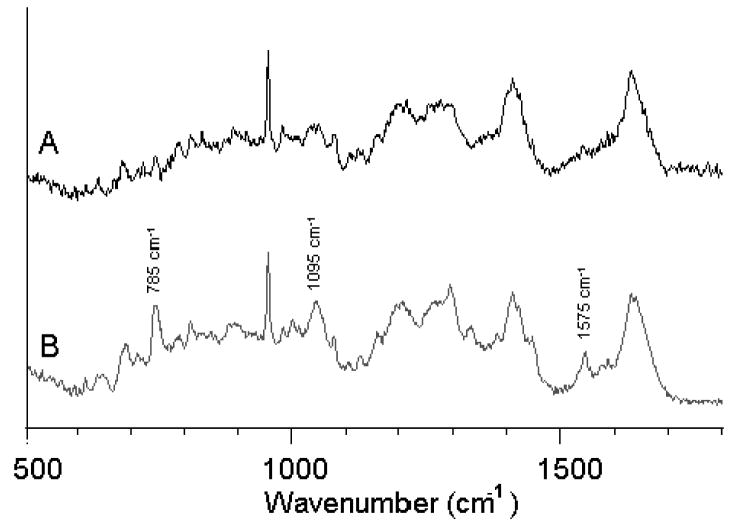
The chromosomal condensation during metaphase and anaphase, shown in the second and third rows, respectively, is manifested by a large intensity increase of the DNA-related peaks. Figure 4 shows the increased spectral features of DNA in metaphase and anaphase. Trace A represents the region of the cytoplasm whereas Trace B represents the spectra of the chromatin during anaphase. Since the laser beam is probing the chromatin in Fig. 4B, we may be certain that the nucleic acid signatures are mostly due to DNA/histone complexes. Furthermore, the DNA is highly condensed, providing much higher signals as compared to the interphase cell in Fig. 2.
Fig. 4.
Raman spectra of (A) cytoplasm and (B) DNA/chromatin complex during the anaphase of mitosis. Major bands due to DNA are indicated by their wavenumbers.
The bottom row of images in Fig. 3 was taken for a cell in telophase. In contrast to the images of the cells in anaphase and metaphase, the DNA signal is reduced, since the DNA distribution has already become relatively diffuse during this stage of mitosis. However, the protein signals clearly indicate the presence of two new nuclei forming.
In Fig. 3 (right column) the corresponding fluorescence images, using DAPI stain, are depicted. There is, obviously, excellent agreement in the position of the chromatin when Raman DNA and DAPI stains are plotted. However, the Raman data permit images to be constructed from the Raman scattering signals inherent in the cell, such as the protein/DNA distribution. This detection requires the addition of stains for fluorescence imaging.
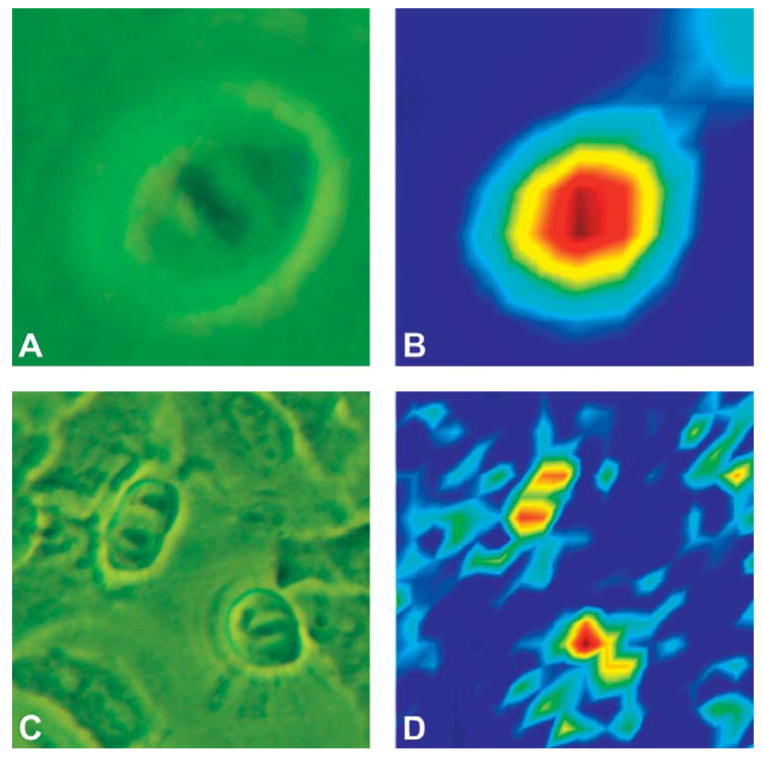
Figure 5 shows the results of efforts to monitor the mitotic stages via infrared imaging. The cell shown in Fig. 5 is much smaller than the one shown in Fig. 1, and is nearly spherical in shape. Although the S/N ratio of the infrared spectra is more than an order of magnitude higher than that of the Raman spectra, and the data acquisition times are nearly 100 times faster, the infrared images reveal very little detail in the mid-infrared (800–1800 cm−1, or 12.5 to 5.6 μm) spectral region about the distribution of condensed chromatin. In Fig. 5B, just a hint of the chromatin features in the nucleus can be detected. This is, of course, due to the fact that the spatial resolution of the infrared spectral images is diffraction limited. This aspect will be discussed in the next section. However, in the C–H stretching region, between 2800 and about 3100 cm−1, or 3.6 to 3.2 μm, the spatial resolution of infrared spectral images is improved by nearly a factor of two. Thus, mild resolution enhancement (see below) of an image of a cell in the anaphase, obtained by displaying the intensity at 2924 cm−1, reveals the newly separated chromosomes, as shown in Fig. 5D. These results represent the best spatially resolved single cell spectra available from non-synchrotron based measurements.
Fig. 5.
Phase-contrast images of HeLa cells in (A) metaphase and (C) anaphase. (B) Infrared spectral map based on the amide I absorption intensities, of the cell shown in (A). In the center of the nucleus, the largest intensity correlates with the position of the chromatin. Dark blue indicates low intensity, and yellow to red indicate higher and highest intensity, respectively. (D) Resolution-enhanced infrared spectral map based on the 2925 cm−1 absorption region of the cell shown in (C). The two chromatin regions of the cell in anaphase are detectable. The color scheme is as in (B).
DISCUSSION
One aspect of the research in the authors’ laboratories deals with the characterization of spectral patterns of cells in response to cell division, cell differentiation, cell death, drug treatment, and onset of disease. In the process of our research, we have collected individual Raman and infrared spectra of tens of thousands of cells. In this paper, we present spectral maps of the distribution of the cell’s major biochemical components during mitosis. Since the original submission of our manuscript, a paper has appeared in the literature23 that reports the changes in Raman spectral features of yeast cells during the cell division cycle and mitosis. We have reported the changes of infrared spectra of somatic cells during the cell cycle in the past,10 and present in this paper the first spectral maps of somatic cells during mitosis. Since the nucleus of these somatic cells is much bigger than the nucleus of yeast cells, clear spectral maps during the subphases of mitosis could be observed.
The exact sequence of events during mitosis is, of course, well understood from light microscopy and fluorescence microscopy studies. Spectral imaging methodology, however, affords the advantage of monitoring the distribution of biochemical components, using only their inherent vibrational fingerprints without employing any stains or other commonly used probes. Furthermore, spectral recognition of mitotic cells is necessary for an application of vibrational imaging methodology to aid in the detection of cell proliferation, for example, in samples of exfoliated or exponentially growing cultured cells.
In this paper, we demonstrate that the recognition of mitosis using methods of vibrational spectral imaging is possible, although the instrumentation utilized may not have been optimal for this purpose. In Raman microspectroscopy, the high spatial resolution reveals details of the spatial distribution of biochemical components within the cell and nucleus. The use of somewhat higher laser power, and more efficient optical components, will reduce the data acquisition time and improve the signal quality to such an extent that this methodology may be useful for screening applications of cellular samples.
The Raman images reveal the chromatin distribution for the prophase, metaphase, and anaphase cells in exquisite detail. This chromatin distribution parallels the distribution observed via fluorescence (DAPI) staining. However, the chromatin exhibits low Raman signals in interphase cells, as well as for the late-telophase cells. We explain this by the fact that in an interphase cell, the protein–DNA chromatin complexes are packed with a packing ratio of 40, as compared to a metaphase chromosome with a packing ratio of 10 000. Thus, the chromatin may be too diffuse to yield a detectable Raman scattering signal in the volume probed in Raman microspectroscopy, which is on the order of 1 μm2, or about 10−12 g. It appears that there exists a complementary relationship between Raman and infrared microspectroscopy with regard to the detectability of DNA. We have shown before that highly condensed DNA, such as in a pyknotic nucleus, cannot be observed in infrared microspectroscopy, since the absorbance of chromatin may be 20–50 OD units.8,11,13 By contrast, it appears that Raman signals are most prominent for most highly condensed DNA.
Although the S/N ratio reported here is substantially better than that in the report by Huang,23 we refrain from interpreting the Raman spectra in terms of biochemical changes. In situations of much higher S/N, for example, in synchrotron-based Fourier transform infrared (FT-IR) spectral maps of cells, we have demonstrated that spectral changes can be associated with selective removal of biochemical components.24 In spectra of lower S/N ratios, data interpretation is best carried out by methods of multivariate statistics, as shown above.
Infrared microspectral images can be collected much more readily than Raman images. However, as indicated, the spatial resolution, or the smallest spot size from which data can be collected, is restricted by the diffraction limit. This limit is about 10 μm at 1000 cm−1, 6 μm for the protein amide I vibration (1650 cm−1), and about 3 μm in the C–H stretching vibrations at ~2950 cm−1. The Perkin-Elmer Spotlight 300 Microspectrometer system was calibrated, using standard resolution targets, and was found to produce an actual spatial resolution of 12 μm at 1000 cm−1. Thus, the details of the nuclear structure during mitosis, which are readily observed in the Raman images, cannot be reproduced at a comparable level of detail in the infrared spectral maps, although hints of the chromatin protein intensities of metaphase and telophase cells can just be discerned in the images.
The use of resolution-enhancing techniques, such as Fourier self-deconvolution (FSD),25 can improve the spatial resolution beyond the diffraction limit. These methods utilize the principle that spectral or spatial resolution is contained in the high-frequency part of the Fourier spectrum. FSD is carried out by transforming a three-dimensional (X,Y, and spectral intensity dimensions) data set into Fourier space, and multiplying the X and Y dimensions by a resolution-enhancing function. Such a resolution-enhancing function is generally an exponential function that amplifies the high-frequency components, albeit at an increase of the noise level, reducing the S/N ratio. After transforming the data set back into spectral and XY space, an image with enhanced spatial resolution is obtained. The map of a cell in anaphase, obtained after Fourier self-deconvolution, is shown in Fig. 5D. This figure shows the newly divided chromatin of the daughter cells as monitored in the C–H stretching vibrations of all nuclear constituents. As pointed out before, the spectral region of the C–H stretching modes offers a higher spatial resolution due to the shorter wavelength. The spatial resolution was further enhanced by FSD, which produces increased noise level around the cells. This method works best if data are collected at an image size smaller than the diffraction limit (over-sampling) and can improve the spatial resolution by about a factor of two. In the data presented in Fig. 5D, the increased spatial resolution is due to the lower diffraction limit in the C–H stretching vibrations, along with the resolution enhancement, and reveals for the first time the chromatin in the daughter cells in infrared spectral maps.
CONCLUSION
We report, for the first time, detailed spectral maps collected via infrared and Raman microspectroscopy of cells during mitosis and correlate these maps to images collected using widely used and accepted stains for cellular chromatin; however, the Raman and infrared imaging modalities provide data that utilize the inherent spectral fingerprint of cellular components to visualize the reorganization of chromatin during mitosis. The information provided in this paper will be required for any applications of vibrational imaging methods to detect cell proliferation in cultured or exfoliated cells.
Acknowledgments
Partial support of this research from grants GM 60654 and CA 090346 from the National Institutes of Health is gratefully acknowledged. A Research Centers in Minority Institutions award RR-03037 from the NCRR of the NIH, which supports the infrastructure of the Chemistry Department at Hunter, is also acknowledged.
References
- 1.Esposito AP, Talley CE, Huser T, Hollars CW, Schaldach CM, Lane SM. Appl Spectrosc. 2003;57:868. doi: 10.1366/000370203322102979. [DOI] [PubMed] [Google Scholar]
- 2.Uzunbajakava N, Lenferink A, Kraan Y, Willekens B, Vrensen G, Greve J, Otto C. Biopolymers. 2003;72:1. doi: 10.1002/bip.10246. [DOI] [PubMed] [Google Scholar]
- 3.Verrier S, Notingher I, Polak JM, Hench LL. Biopolymers. 2004;74:157. doi: 10.1002/bip.20063. [DOI] [PubMed] [Google Scholar]
- 4.Notingher I, Verrier S, Haque S, Polak JM, Hench LL. Biopolymers. 2003;72:230. doi: 10.1002/bip.10378. [DOI] [PubMed] [Google Scholar]
- 5.Uzunbajakava N, Lenferink A, Kraan Y, Volokhina E, Vrensen G, Greve J, Otto C. Biophys J. 2003;84:3968. doi: 10.1016/S0006-3495(03)75124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uzunbajakava N, Otto C. Opt Lett. 2003;28:2073. doi: 10.1364/ol.28.002073. [DOI] [PubMed] [Google Scholar]
- 7.Otto C, Greve J. Internet J Vib Spectrosc. 1999;2(Section 4) [Google Scholar]
- 8.Pacifico A, Chiriboga L, Lasch P, Diem M. Vib Spectrosc. 2003;32:107. [Google Scholar]
- 9.Diem M, Chiriboga L, Lasch P, Pacifico A. Biopolymers. 2002;67:349. doi: 10.1002/bip.10109. [DOI] [PubMed] [Google Scholar]
- 10.Boydston-White S, Chernenko T, Regina A, Miljkovic M, Matthäus C, Diem M. Vib Spectrosc. 2005;38:169. [Google Scholar]
- 11.Mohlenhoff B, Romeo M, Wood BR, Diem M. Biophys J. 2005;88:3635. doi: 10.1529/biophysj.104.057950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maquelin K, Choo-Smith L-P, Kirschner C, Ngo-Thi NA, Naumann D, Pupples GJ. Vibrational Spectroscopic Studies of Microorganisms. In: Chalmers JM, Griffiths PR, editors. Handbook of Vibrational Spectroscopy. John Wiley and Sons; Chichester, UK: 2002. [Google Scholar]
- 13.Boydston-White S, Gopen T, Houser S, Bargonetti J, Diem M. Biospectroscopy. 1999;5:219. doi: 10.1002/(SICI)1520-6343(1999)5:4<219::AID-BSPY2>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 14.Jamin N, Miller L, Moncuit J, Fridman WH, Dumas P, Teillaud JL. Biopolym Biospectrosc. 2003;72:366. doi: 10.1002/bip.10435. [DOI] [PubMed] [Google Scholar]
- 15.Liu KZ, Jia L, Kelsey SM, Newland AC, Mantsch HH. Apoptosis. 2001;6:267. doi: 10.1023/a:1011383408381. [DOI] [PubMed] [Google Scholar]
- 16.Deckert V, Kiefer W. Appl Spectrosc. 1992;46:322. [Google Scholar]
- 17.Diem M, Adar F, Grayzel R. Computer Enhanced Spectrosc. 1986;3:29. [Google Scholar]
- 18.Kapuscinski J. Biotechnol Histochem. 1995;70:220. doi: 10.3109/10520299509108199. [DOI] [PubMed] [Google Scholar]
- 19.www.Cytospec.com.
- 20.Gazi E, Dwyer J, Lockyer NP, Miyan J, Gardner P, Hart CA, Brown MD, Clarke NW. Vib Spectrosc. 2005;38:193. doi: 10.1002/bip.20167. [DOI] [PubMed] [Google Scholar]
- 21.Lasch P, Pacifico A, Diem M. Biopolymers. 2002;67:335. doi: 10.1002/bip.10095. [DOI] [PubMed] [Google Scholar]
- 22.Wood BR, Chiriboga L, Yee H, Quinn MA, McNaughton D, Diem M. Gynecol Oncol. 2004;93:59. doi: 10.1016/j.ygyno.2003.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang YS, Karashima T, Yamamoto M, Hamaguchi HO. Biochemistry. 2005;44:10009. doi: 10.1021/bi050179w. [DOI] [PubMed] [Google Scholar]
- 24.Diem M, Romeo M, Matthäus C, Miljkovic M, Miller L, Lasch P. Infrared Phys Technol. 2004;45:331. [Google Scholar]
- 25.Kaupinnen JK, Moffatt DJ, Mantsch HH, Cameron DG. Appl Spectrosc. 1981;35:271. [Google Scholar]