Abstract
Summary: The web server MetalDetector classifies histidine residues in proteins into one of two states (free or metal bound) and cysteines into one of three states (free, metal bound or disulfide bridged). A decision tree integrates predictions from two previously developed methods (DISULFIND and Metal Ligand Predictor). Cross-validated performance assessment indicates that our server predicts disulfide bonding state at 88.6% precision and 85.1% recall, while it identifies cysteines and histidines in transition metal-binding sites at 79.9% precision and 76.8% recall, and at 60.8% precision and 40.7% recall, respectively.
Availability: Freely available at http://metaldetector.dsi.unifi.it
Contact: metaldetector@dsi.unifi.it
Supplementary Information: Details and data can be found at http://metaldetector.dsi.unifi.it/help.php
1 INTRODUCTION
Metal-binding proteins play critical catalytic, regulatory and structural roles in the cell. They are implicated in heavy metal toxicity, in processes such as apoptosis (Formigari et al., 2007) and aging (Mocchegiani et al., 2006), as well as in numerous diseases, including Alzheimer (Crouch et al., 2007), Parkinson (Santamaria et al., 2007) and AIDS (Diamond and Bushman, 2006). Their identification and characterization can contribute toward a better understanding of these phenomena. Here, we introduce a web server that takes the protein sequence as input and outputs predictions of transition-metal binding for cysteine and histidine residues; for cysteines it also predicts disulfide bonding bridges.
2 METALDETECTOR: INTEGRATING METAL LIGAND PREDICTOR AND DISULFIND
We previously developed a method, Metal Ligand Predictor (MLP; Passerini et al., 2006), which predicts transition-metal binding for cysteines and histidines from sequence information alone. The method classifies cysteines into one of three states: free (F), disulfide bridged (D) metal bound (M) and histidines into one of two states (F or M). The main purpose of MetalDetector is to make the predictor available online as a web application. When in the process of developing a server for MLP, however, we observed some inconsistencies with DISULFIND (Ceroni et al., 2006), a server we previously made available for predicting the disulfide bonding state of cysteines and their disulfide connectivity. In particular, on the same test set used in (Passerini et al., 2006), conflicting cysteine classifications by the two predictors involved 761 out of 9187 cases (i.e. 8.3%). Two types of inconsistency may arise: (1) MLP predicts D and DISULFIND predicts F (554 cases), and (2) MLP predicts F or M and DISULFIND predicts D (207 cases). MetalDetector integrates MLP and DISULFIND and tries to resolve their inconsistencies.
3 CONCEPT
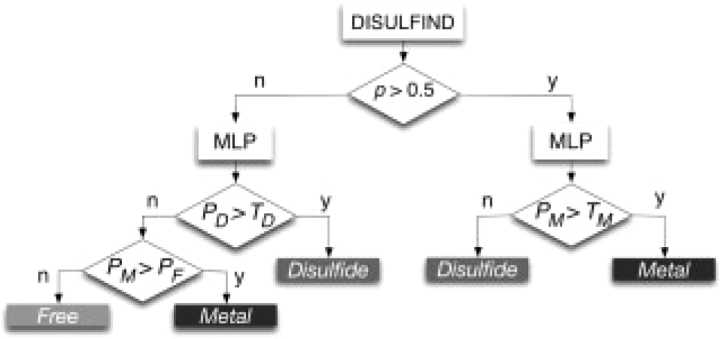
When a protein sequence is submitted to MetalDetector, both constituent methods, MLP and DISULFIND, are queried. For histidines, the results are just read off MLP. For cysteines, the output of MetalDetector is determined by a decision tree architecture (Fig. 1). We start with the output of DISULFIND that classifies all cysteines as either F or D. For the same residues, MLP provides probabilities for classes F, D and M (PF, PD, PM). For a given cysteine, if DISULFIND predicts class F, we apply a simple threshold TD to the PD output of MLP. If PD>TD, MetalDetector will predict class D, else the cysteine will be predicted to be either in class F (if PF>PM), or M (if PF<PM). We apply a similar threshold TM when DISULFIND predicts D. If the output PM of MLP exceeds TM, the cysteine will be assigned to class M, otherwise to class D. Changing the thresholds TD and TM enables the user to decide how much trust to put in each of the constituent predictors. For example, if TD=TM=1, disulfide bridges are only predicted by DISULFIND, while lowering both thresholds increases the weight for MLP. Prior knowledge about the protein may therefore help users to find a metal bound/disulfide bound/free cysteine. At the end of the decision process, a finite state automation (Passerini et al., 2006) constrains the number of disulfide predictions to be even (inter-chain bridges are ignored). In case of an odd number of disulfide predictions, it relabels a single cysteine from free or metal bonded to disulfide bonded or vice versa, depending on which relabeling produces the least reduction in likelihood. The probabilities used by the automaton come either from DISULFIND, or from MLP, depending on which predictor has made the final prediction on each residue. MetalDetector also outputs predicted disulfide connectivity by calling the second stage of DISULFIND.
Fig. 1.
Decision-tree architecture for cysteine bonding state predictions.
The new method deals efficiently with inconsistencies: at the default thresholds TD=0.76 and TM=0.65, there are 274 non-consistent predictions, 191 of type (1) and 83 of type (2) (a reduction from 8.3% inconsistencies to 3.0%). For these 274 residues, the predictions of MetalDetector are identical to those of MLP in 256 cases and better than those of DISULFIND 56 and 75% of these cases, for inconsistencies of type (1) and type (2), respectively. A paired t-test revealed that MetalDetector is significantly better than MLP in terms of accuracy (P<0.01). MetalDetector also significantly outperforms both DISULFIND and MLP on the two-classes problem D versus M/F (P<0.01), while there is no significant difference between MLP and DISULFIND. Thus, the new method provides better performance and succeeds in achieving our stated goal, which was to make available a metal-binding state predictor that would largely agree with DISULFIND on disulfide bonding state. In Tables 1 and 2, we report the best results achieved by MetalDetector considering both cysteine and histidine predictions using default thresholds. The corresponding protein-level accuracy Qp is 77% as in (Passerini et al., 2006). Sample predictions are shown in Figure 2.
Table 1.
Comparison of precision (P), recall (R) and disulfide bonding state accuracy (A) on the test set used in (Passerini et al., 2006)
| MLP |
MetalDetector |
DISULFIND | ||||||
|---|---|---|---|---|---|---|---|---|
| Cys | His | All | Cys | His | All | Cys | ||
| Metal | P | 79.7 | 60.8 | 73.3 | 79.9 | 60.8 | 73.5 | – |
| R | 74.9 | 40.7 | 60.5 | 76.8 | 40.7 | 61.6 | – | |
| Disulfide | P | 86.4 | – | 86.4 | 88.6 | – | 88.6 | 88.4 |
| R | 87.0 | – | 87.0 | 85.1 | – | 85.1 | 82.7 | |
| D versus M/F | A | 88.8 | – | 88.8 | 90.0 | – | 90.0 | 89.1 |
All values are in percentage.
Table 2.
Contingency matrix of MetalDetector for TD and TM default values, including histidine predictions
| Metal | Disulfide | Free | |
|---|---|---|---|
| Metal | 993 | 117 | 501 |
| Disulfide | 77 | 3024 | 451 |
| Free | 281 | 273 | 17130 |
| Precision (%) | 73.5 | 88.6 | 94.7 |
| Recall (%) | 61.6 | 85.1 | 96.9 |
Fig. 2.
Sample predictions, inconsistencies highlighted in boldface. Top: MetalDetector (MD) corrects the first wrong D assignment of DISULFIND thanks to MLP prediction, but cannot correct MLP's missed metal. Bottom: MD corrects the wrong D assignments of MLP thanks to DISULFIND predictions. In all cases, where MLP predicts M and DISULFIND predicts F (highlighted in lowercase), MD picks the right choice from MLP.
4 SERVER
Three preset working points can be chosen from the web interface. They correspond to high metal accuracy (default, TD=0.76 and TM=0.65), high metal−precision (TD=0.5, TM=1), and high metal recall (TD=1, TM=0.5) for the metal class. In the case of histidines, the decision threshold is 0.5. Precision/recall for the disulfide class are 83.1/88.7 and 90.1/82.0 at the high metal precision and high metal recall working points, respectively.
ACKNOWLEDGEMENTS
Funding: M.P. and B.R. were supported by the grants R01-GM079767, R01-LM07329, and U54-GM75026 from the National Institutes of Health (NIH) in the USA.
Conflict of Interest: none declared.
REFERENCES
- Ceroni A, et al. Disulfind: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006;34(Web Server issue):W177–81. doi: 10.1093/nar/gkl266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch PJ, et al. The modulation of metal bio-availability as a therapeutic strategy for the treatment of alzheimer's disease. FEBS J. 2007:3775–3783. doi: 10.1111/j.1742-4658.2007.05918.x. [DOI] [PubMed] [Google Scholar]
- Diamond TL, Bushman FD. Role of metal ions in catalysis by hiv integrase analyzed using a quantitative pcr disintegration assay. Nucleic Acids Res. 2006:6116–6125. doi: 10.1093/nar/gkl862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formigari A, et al. Zinc, antioxidant systems and metallothionein in metal mediated-apoptosis: biochemical and cytochemical aspects. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2007:443–459. doi: 10.1016/j.cbpc.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Mocchegiani E, et al. Zinc homeostasis in aging: two elusive faces of the same ‘metal’. Rejuvenation Res. 2006:351–354. doi: 10.1089/rej.2006.9.351. [DOI] [PubMed] [Google Scholar]
- Passerini A, et al. Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins. 2006:305–316. doi: 10.1002/prot.21135. [DOI] [PubMed] [Google Scholar]
- Santamaria AB, et al. State-of-the-science review: Does manganese exposure during welding pose a neurological risk? J. Toxicol. Environ. Health B Crit. Rev. 2007:417–465. doi: 10.1080/15287390600975004. [DOI] [PubMed] [Google Scholar]