Abstract
Sporulation in low-G+C gram-positive bacteria (Firmicutes) is an important survival mechanism that involves up to 150 genes, acting in a highly regulated manner. Many sporulation genes have close homologs in non-sporulating bacteria, including cyanobacteria, proteobacteria and spirochaetes, indicating that their products play a wider biological role. Most of them have been characterized as regulatory proteins or enzymes of peptidoglycan turnover; functions of others remain unknown but they are likely to have a general role in cell division and/or development. We have compiled a list of such widely conserved sporulation and germination proteins with poorly characterized functions, ranked them by the width of their phylogenetic distribution, and performed detailed sequence analysis and, where possible, structural modeling aimed at estimating their potential functions. Here we report the results of sequence analysis of Bacillus subtilis spore germination protein GerM, suggesting that it is a widespread cell development protein, whose function might involve binding to peptidoglycan. GerM consists of two tandem copies of a new domain (designated the GERMN domain) that forms phylum-specific fusions with two other newly described domains, GERMN-associated domains 1 and 2 (GMAD1 and GMAD2). Fold recognition reveals a β-propeller fold for GMAD1, while ab initio modeling suggests that GMAD2 adopts a fibronectin type III fold. SpoVS is predicted to adopt the AlbA archaeal chromatin protein fold, which suggests that it is a DNA-binding protein, most likely a novel transcriptional regulator.
Contact: drigden@liverpool.ac.uk
Supplementary information: Supplementary data are available at ftp://ftp.ncbi.nih.gov/pub/galperin/Sporulation.html
1 INTRODUCTION
Cell division remains one of the least understood processes in the life of the bacterial cell. In stark contrast to the significant progress in the understanding of microbial metabolism and signal transduction brought about by the complete genome sequences, the contribution of genomics to the studies of cell division has been relatively modest. Some cell division proteins are still poorly characterized and the importance of the presence or absence of a certain gene in a given genome cannot be readily interpreted as it is being done for the metabolic enzymes. In addition, cell division involves numerous protein–protein interactions, so mutant phenotypes are fairly complex, making their formal description (e.g. using the Gene Ontology system) almost impossible. Finally, preliminary characterization of such genes in Escherichia coli, Bacillus subtilis or other bacteria usually results in assigning them stable names, e.g. ‘cell division protein FtsN’, which often creates an illusion of at least some understanding and obscures the fact that the functions of these proteins remain enigmatic.
Sporulation in B.subtilis and other low-G+C gram-positive bacteria (Firmicutes) is an important survival mechanism that is related to cell division and involves up to 150 genes, acting in a highly regulated manner (Piggot and Losick, 2002). Mutational analyses and transcriptional profiling revealed the timing of action of each sporulation gene and suggested functions for most of them. In some cases, deduced functions have been verified by structural analysis or direct biochemical experiments, mostly on proteins from B.subtilis. Phylogenetic distribution of B.subtilis sporulation genes is quite complex (Onyenwoke et al., 2004; Wu et al., 2005). Many of them have regulatory roles and appear to be non-essential for spore formation. Accordingly, such genes may be missing in certain bacillar and clostridial genomes. On the other hand, close homologs of several sporulation genes can be found in the genomes of non-sporulating microorganisms (Onyenwoke et al., 2004). Such genes typically encode cell division proteins, e.g. SpoVD (FtsI) or SpoVE (FtsW), enzymes of peptidoglycan turnover, or components of bacterial signaling systems, such as sensory histidine kinases, response regulators, alternative sigma factors and other transcriptional regulators (Piggot and Losick, 2002). However, function of many sporulation genes remains unknown, including some that have wide phylogenetic distribution and are found in a variety of non-sporulating bacteria. It would be reasonable to assume that widespread functionally uncharacterized sporulation genes encode additional components of bacterial division or signal transduction machinery. We have set out to identify such genes and analyze their likely functions using a combination of sequence and structure analysis tools. Here we report the results of domain analysis and structural modeling of two widespread proteins from this group, GerM and SpoVS.
2 METHODS
The initial list of B.subtilis sporulation proteins was compiled from published sources (Errington, 2003; Onyenwoke et al., 2004; Piggot and Losick, 2002). Phylogenetic distribution of each protein was judged based on the species lists in the Pfam (Finn et al., 2008), CDD (Marchler-Bauer et al., 2007) and COG (Tatusov et al., 2000) databases, where available, and verified using PSI-BLAST (Altschul et al., 1997) searches, employing an e-value of 0.01, filtered to exclude hits from the Firmicutes. The search results were sorted using the ‘Taxonomy reports’ option in BLAST outputs. Domain composition of the retrieved sequences was analyzed by comparing them against Pfam, CDD and COGs with e-values of below 0.01 taken to represent significant hits.
Possible templates for modeling were sought at the META server, a portal to the several fold recognition methods (Bujnicki et al., 2001) and to the 3D-Jury consensus method, by which scores of over 50 are taken as highly confident (Ginalski et al., 2003). Distant homologies were also sought with HHpred (Soding et al., 2005) using a cut-off of e < 0.01. Secondary structures were predicted using PSI-PRED (Jones, 1999).
Template-based modeling was carried out with MODELLER (Sali and Blundell, 1993). Five variant models were constructed after an initial coordinate randomization step. PROCHECK (Laskowski et al., 1993) was used for their stereochemical evaluation and VERIFY_3D (Lüthy et al., 1992) and PROSA II (Sippl, 1993) employed for model ranking by solvent exposure and residue–residue contacts. ROSETTA was used for ab initio model building using default protocols: 2000 individual models were constructed from 3- and 9-residue segments using Monte Carlo substitution and optimization protocols and clustered based on RMSD calculations (Simons et al., 1997, 1999). I-TASSER (Lee and Skolnick, 2007) ab initio models were obtained from the web server (http://zhang.bioinformatics.ku.edu/I-TASSER/). I-TASSER models and coordinates for centre models of each ROSETTA cluster were submitted to the DALI server (http://www.ebi.ac.uk/dali/; Holm and Sander, 1993) for structural comparison to the Protein Data Bank (PDB). By DALI, Z-scores of <2 are insignificant. Side chains were added to the best ROSETTA model using SCWRL (Canutescu et al., 2003).
PYMOL (http://pymol.sourceforge.net) was used for structure manipulation and visualization as well as display of electrostatic potential surfaces calculated with Adaptive Poisson-Boltzmann Solver (APBS) (Baker et al., 2001). Structural relationships were browsed in the SCOP database (Andreeva et al., 2008).
3 RESULTS
3.1 Sporulation genes in non-sporulating bacteria
Earlier studies identified homologs of B.subtilis sporulation genes in a variety of non-sporulating bacteria both within and outside of the phylum Firmicutes (Onyenwoke et al., 2004; Wu et al., 2005). In order to identify widely conserved sporulation genes, we have looked only for those that have close homologs outside the Firmicute phylum. The resulting list was winnowed down by manually removing known transcriptional regulators, signal transduction proteins, sigma subunits of the RNA polymerase, anti-sigma and anti-anti-sigma factors, enzymes involved in synthesis or hydrolysis of peptidoglycan and previously characterized cell division proteins. Further, detailed analysis of the remaining proteins showed that for some of them functional assignments either had been made or could be made very easily, based on recent experimental data or convincing sequence similarity to experimentally characterized proteins. After these proteins were removed, the final list (Supplementary Table 1) included 20 previously uncharacterized ‘sporulation’ and ‘germination’ proteins that had close homologs in more than three different bacterial phyla. We applied a battery of bioinformatics analyses to yield insights into possible functions and present here results for GerM and SpoVS.
3.2 GerM protein and associated domains
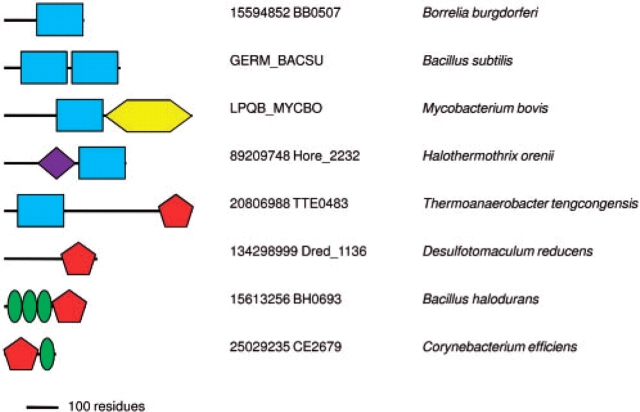
B.subtilis protein GerM has been implicated in both sporulation and spore germination (Sammons et al., 1987; Slynn et al., 1994), suggesting an important role in cell development. According to the COG database, orthologs of GerM (COG5401) are encoded in spirochaetes and cyanobacteria. Indeed, a PSI-BLAST search of complete genome sequences using GERM_BACSU protein (Swiss-Prot accession P39072) as a query and run to convergence retrieved more than 150 proteins (last searched April 1, 2008), encoded in Firmicutes and members of other bacterial phyla, including Actinobacteria, Cyanobacteria, Proteobacteria and Deinococcus–Thermus group. These searches revealed that GerM from B.subtilis and other bacilli contains tandem copies of a ∼100 amino acid-long domain, hereafter called the GERMN domain (Fig. 1 and Supplementary Fig. 1). In other firmicutes, this domain was found mostly in a stand-alone form, whereas Moorella thermoacetica and several other clostridia encoded both a stand-alone and a duplicated version. In a single species in the current database, Halothermothrix orenii, GERMN is fused to the amidase (PF01520) domain which cleaves amide bonds in cell wall peptidoglycans.
Fig. 1.
Domain architectures formed by the GERMN (blue oblongs), GMAD1 (yellow hexagon) and GMAD2 (red pentagon) domains, drawn approximately to scale. The purple diamond is the amidase domain (PF01520) while the green ellipses represent LysM (PF01476) domains. The proteins are listed by their Uniprot identifiers or NCBI gene index (gi) numbers and genome locus tags.
In addition, the GERMN domain was found fused, in a phylum-specific fashion, to two further novel domains (Fig. 1 and Supplementary Figs 2 and 3) named GERMN-associated domains 1 and 2 (GMAD1 and GMAD2). GERMN, GMAD1 and GMAD2 have been deposited with the Pfam database, receiving accession numbers PF10646, PF10647 and PF10648, respectively. The GERMN entry replaces and extends a PfamB entry PB005693 (in Pfam 22.0), which only covered the first two domain architectures shown on Figure 1. In Firmicutes, orthologs of GerM protein, containing duplicated GERMN domain are encoded in spore-forming clostridia and bacilli, but not in non-sporogenous lactobacilli, staphylococci or streptococci. Every sequenced genome of Actinobacteria encodes a GERMN-GMAD1 domain fusion that is referred to as ‘putative lipoprotein LpqB’ although the origin of this name seems obscure. The lpqB gene, nearly always found adjacent to the genes encoding the two-component system MtrAB, has been proposed to modulate the function of these signaling proteins (Hoskisson and Hutchings, 2006). Deletion of MtrA and MtrB in Corynebacterium glutamicum leads to increased susceptibility to cell wall targeting vancomycin and lysozyme as well as to cell elongation (Moker et al., 2004). Thus the MtrAB double mutant appears to have cell envelope and cell division defects. The function of this pair is intriguingly similar to that of YycFG (VicRK) system in the Firmicutes (Aravind et al., 2003).
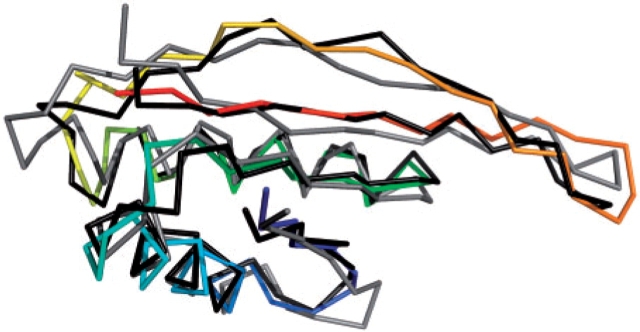
Fig. 2.
Comparison of comparative (blue to red, N- to C-terminus), ROSETTA (grey) and I-TASSER (black) models of SpoVS.
The importance of the GERMN domain is underscored by the fact that it is encoded in all completely sequenced genomes of representatives of such phyla as Spirochaetes and Deinococcus–Thermus, including the relatively small genomes of obligate parasites Borrelia burgdorferi and Treponema pallidum. The GERMN domain is also encoded in many cyanobacterial genomes.
Screening GERMN-containing proteins with PROSITE (Hulo et al., 2008) prokaryotic lipoprotein (PS51257) motif revealed their strong association with this predicted post-translational modification. Single GERMN sequences were predicted lipoproteins in 48/120 cases. The figure for twin GERMN sequences (e.g. GerM) is 33/39, for GERMN-GMAD1 it is 30/36 and for GERMN-GMAD2 it is 6/11. We then considered whether modeling, comparative or ab initio, or domain context could shed light on the functions of these domains. None of the fold recognition methods implemented at the META server (Bujnicki et al., 2001) gave significant results for either the GERMN or the GMAD2 domains. In the case of GERMN, taking a consensus view of secondary structure predictions for different proteins, the fold seems to contain the following principal elements of regular secondary structure—ββααββααββ. The lack of significant hits suggests that GERMN has a novel α+β type fold. Ab initio modeling was unsuccessful for GERMN.
The GMAD2-predicted secondary structure indicates an all-β-fold containing approximately nine β-strands. Several methods rated the immunoglobulin-type fold most highly but with scores not high enough for confident fold assignment. Ab initio modeling with ROSETTA was also unsuccessful, but, suggestively, all five models obtained by ab initio modeling at the I-TASSER server shared the same fold, matched significantly by DALI (Z-scores up to 7.5) to immunoglobulin-type structures. Among these the top scores were for matches to fibronectin type III (FnIII) domains. These have structural roles in animals, but are also present in bacteria, where they are often associated with glycoside hydrolase enzymes (Bork and Doolittle, 1992; Little et al., 1994). Browsing of the current Pfam database (release 22.0) confirms the strong association between this domain and carbohydrate metabolism, as shown by its presence, with few exceptions, alongside catalytic domains that clearly act on carbohydrates. At a molecular level, bacterial FnIII domains act as spacers between catalytic and substrate-binding domains (Toratani et al., 2006) while structurally similar bacterial β-sandwich domains have a direct carbohydrate-binding function (Jee et al., 2002). The GMAD2 domain is found combined with LysM domains in two distinct architectures (Fig. 1). LysM domains were first associated with peptidoglycan binding (Steen et al., 2003), although recent data show that some examples bind chitin or lipochitin (Spaink, 2004; Onaga and Taira, 2008). Specificity for carbohydrate ligand therefore seems to vary between LysM domains.
The GMAD1 domain gave strong hits to β-propeller folds by fold recognition methods at the META server, consistent with earlier data (Hoskisson and Hutchings, 2006). Interestingly, WD40-type 7-bladed propellers consistently achieved the top scores (up to 96 by 3D-Jury). However, the WD motifs themselves are absent and such propellers are, in any case, rare in bacteria and generally contain more blades than the five or six predicted for GMAD1 (Neer et al., 1994). Unfortunately, the β-propeller fold assignment provides few clues as to the function of the GMAD1 domain since β-propellers are involved in a wide variety of binding and catalytic functions. A certain tendency towards sugar binding and metabolism is evident in the functions of smaller 5- and 6-bladed β-propellers of known structure; 2 of the 3 superfamilies of 5-bladed propellers and 4 of the 11 superfamilies of 6-bladed propellers have these functions (Andreeva et al., 2008). However, β-propellers are also known as protein–protein interaction domains, so the interaction mentioned above between LpqB, a GerMN-GMAD1 domain fusion protein and the MtrAB system could be mediated by the GMAD1 propeller. The presence of only a single conserved hydrophilic residue, an Arg, in an alignment of GMAD1 domains, would be more consistent with a passive binding role than with a catalytic function (Koonin and Galperin, 2002).
Intriguingly, a direct connection can be made with PSI-BLAST between a GERMN query and Streptococcus pneumoniae Wzd, a component of the capsule polysaccharide export machinery (Aanensen et al., 2007). The match appears in the third iteration in the form of an ungapped 56 residue alignment with a bit score of 43 and an e-value of 0.006. As yet, no Wzd structure exists with which to assess the significance of this match.
3.3 SpoVS protein
Mutations in B.subtilis spoVS gene block sporulation at Stage V but allow the spoIIBspoVG double mutant to bypass the sporulation block at Stage II (Resnekov et al., 1995). According to the Pfam database, proteins with the SpoVS domain (PF04232), in addition to firmicutes, are encoded in members of the bacterial phyla Chloroflexi, Thermotogae and Deinococcus–Thermus. This is consistent with results of PSI-BLAST searches, which detect orthologs of B.subtilis SpoVS (SP5S_BACSU, Swiss-Prot accession P45693) in every completely sequenced genome of these phyla. In firmicutes, SpoVS is found in sporulating bacilli and clostridia, but not in non-sporulating lactobacilli, listeria, staphylococci or streptococci.
PSI-BLAST searches failed to reveal any distant homologs of SpoVS. At the META server, the single match scoring above the 3D-Jury confidence threshold of 50 (Ginalski et al., 2003) was between Thermotoga maritima SpoVS and Sulfolobus solfataricus Alba (PDB code 1h0x; Wardleworth et al., 2002) with a score of 53. An excellent match between predicted secondary structure for SpoVS and the actual secondary structure of Alba was seen (Supplementary Fig. 4). In order to assess the compatibility of the SpoVS sequences and the Alba fold in more detail, modeling was carried out. We modeled the T.maritima protein TM1059 which, among those we tested, achieved the best 3D-Jury score. The S.solfataricus Alba structure was used as template. Although there are no experimental data regarding the SpoVS oligomeric state we supposed that it might exist, like Alba and likely its most closely related families of relatives (Aravind et al., 2003), as a dimer, a hypothesis supported by the presence in the dimer model of a large hydrophobic interface between the subunits. The modeling showed that indels between SpoVS and Alba could be readily accommodated and the final model has favorable VERIFY_3D and PROSA II profiles and an optimal pG value (Sanchez and Sali, 1998) of 1.0.
Ab initio modeling of B.subtilis SpoVS lent further support for its structural correspondence with Alba. The top cluster of 2000 ROSETTA models contained many more models, 268, than the next most populated cluster (81) indicative of likely success. Indeed, the top cluster centre gave a highly significant Z-score of 9.3 by DALI, far in excess of the 3.3–5.6 achieved by other cluster centres. The top cluster centre matched S.solfataricus Alba and corresponded to an alignment of 81 residues with a Cα RMSD of 2.2 Å. For the top I-TASSER model the corresponding figures were Z-score of 13.2 for an alignment of 84 residues with a Cα RMSD of 1.5 Å.
As shown in Figure 2, the top ROSETTA and I-TASSER models of B.subtilis SpoVS and the homology model of T.maritima TM1059 are remarkably similar. We interpret the ability of ab initio modeling to produce the Alba fold as strong evidence that this is indeed the correct fold assignment for the SpoVS family.
The Alba fold is strongly linked with the broad function of nucleic acid binding. Although Alba itself binds DNA, homologs recognizable by iterative database searching include several families of RNA binding proteins (Aravind et al., 2003). Furthermore, Alba's IF3-C fold is found in many other families lacking sequence similarity but recognizable at the structural level. As reported (Aravind et al., 2003), and verified in the current SCOP database, this fold is strongly associated with nucleic acid binding.
The comparative SpoVS model has the significant dipole characteristic of nucleic acid binding proteins (Szilagyi and Skolnick, 2006). The positively charged face (see Supplementary Fig. 5), also seen for the ab initio models (data not shown) corresponds to a similarly positively charged surface in the Alba dimer (Wardleworth et al., 2002) that has been convincingly modeled as forming its interface with duplex DNA.
4 CONCLUSIONS
Our census (Supplementary Table 1) and the work of others (Errington, 2003; Onyenwoke et al., 2004; Piggot and Losick, 2002); show that many ‘sporulation’ and ‘germination’ proteins are in fact of broad phyletic distribution. Furthermore, their annotation as being involved in these processes often obscures a real lack of knowledge of their molecular functions. Here, we have attempted in-depth sequence analysis of some of them. Three novel domains are described which will contribute to extending Pfam coverage towards its upper limit (Sammut et al., 2008).
The strong theme emerging from domain context analysis of GERMN (Fig. 1) and the available structure predictions is carbohydrate binding. We therefore propose that the GERMN domain interacts with bacterial carbohydrate. With the separate implication of the GERMN domain in cell envelope and division for GerM (Sammons et al., 1987; Slynn et al., 1994) and LpqB (Moker et al., 2004), likely carbohydrate targets would be peptidoglycan and/or capsule polysaccharide. The former is supported by associations with the amidase and LysM domains (Fig. 1), while the PSI-BLAST link from GERMN to Wzd hints at the latter possibility.
Experimental data regarding SpoVS are apparently limited to two reports (Resnekov et al., 1995; Perez et al., 2006). In the first, mutation of the spoVS gene halted sporulation at Stage V and reduced expression of two σK-directed genes, cotA and gerE (Resnekov et al., 1995). On the other hand, spoVS mutation increases σD-directed gene expression, cell separation and autolysis (Perez et al., 2006). These sigma factors direct RNA polymerase to transcribe certain sets of genes and, along with other DNA-binding regulators such as GerE and SpoIIID, form complex networks that control sporulation (Eichenberger et al., 2004). Along with our structure-based prediction, these data suggest that SpoVS is a further member of these networks and, by binding to specific DNA sites, influences the transcriptional profile of the cell during the onset of sporulation.
Supplementary Material
ACKNOWLEDGEMENTS
Funding: M.Y.G. was supported by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health. Funding to pay the Open Access publication charges for this article was provided by the NIH Intramural Research Program.
Conflict of Interest: none declared.
REFERENCES
- Aanensen DM, et al. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 2007;189:7856–7876. doi: 10.1128/JB.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST – a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, et al. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl Acad. Sci. USA. 1992;89:8990–8994. doi: 10.1073/pnas.89.19.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM, et al. Structure prediction meta server. Bioinformatics. 2001;17:750–751. doi: 10.1093/bioinformatics/17.8.750. [DOI] [PubMed] [Google Scholar]
- Canutescu AA, et al. A graph-theory algorithm for rapid protein side-chain prediction. Protein Sci. 2003;12:2001–2014. doi: 10.1110/ps.03154503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat. Rev. Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginalski K, et al. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–1018. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Hoskisson PA, Hutchings MI. MtrAB-LpqB: a conserved three-component system in actinobacteria? Trends Microbiol. 2006;14:444–449. doi: 10.1016/j.tim.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Hulo N, et al. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee JG, et al. Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 chitinase A1. J. Biol. Chem. 2002;277:1388–1397. doi: 10.1074/jbc.M109726200. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. Sequence-Evolution-Function. Boston, MA: Kluwer Academic; 2003. [PubMed] [Google Scholar]
- Laskowski RA, et al. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- Lee SY, Skolnick J. Development and benchmarking of TASSER(iter) for the iterative improvement of protein structure predictions. Proteins. 2007;68:39–47. doi: 10.1002/prot.21440. [DOI] [PubMed] [Google Scholar]
- Little E, et al. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J. Mol. Evol. 1994;39:631–643. doi: 10.1007/BF00160409. [DOI] [PubMed] [Google Scholar]
- Lüthy R, et al. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moker N, et al. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 2004;54:420–438. doi: 10.1111/j.1365-2958.2004.04249.x. [DOI] [PubMed] [Google Scholar]
- Neer EJ, et al. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Onaga S, Taira T. A new type of plant chitinase containing LysM domains from a fern (Pteris ryukyuensis): roles of LysM domains in chitin binding and antifungal activity. Glycobiology. 2008;18:414–423. doi: 10.1093/glycob/cwn018. [DOI] [PubMed] [Google Scholar]
- Onyenwoke RU, et al. Sporulation genes in members of the low G+C Gramtype- positive phylogenetic branch (Firmicutes) Arch. Microbiol. 2004;182:182–192. doi: 10.1007/s00203-004-0696-y. [DOI] [PubMed] [Google Scholar]
- Perez AR, et al. Suppression of engulfment defects in Bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 2006;188:1159–1164. doi: 10.1128/JB.188.3.1159-1164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot PJ, Losick R. Sporulation genes and intercompartmental regulation. In: Sonenshein AL, et al., editors. Bacillus subtilis and its Closest Relatives: from Genes to Cells. Washington, D.C.: ASM Press; 2002. pp. 483–518. [Google Scholar]
- Resnekov O, et al. Identification and characterization of sporulation gene spoVS from Bacillus subtilis. J. Bacteriol. 1995;177:5628–5635. doi: 10.1128/jb.177.19.5628-5635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sammons RL, et al. Genetical and molecular studies on gerM, a new developmental locus of Bacillus subtilis. J. Gen. Microbiol. 1987;133:3299–3312. doi: 10.1099/00221287-133-12-3299. [DOI] [PubMed] [Google Scholar]
- Sammut SJ, et al. Pfam 10 years on: 10,000 families and still growing. Brief. Bioinform. 2008;9:210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Sali A. Large-scale protein structure modeling of the Saccharomyces cerevisiae genome. Proc. Natl Acad. Sci. USA. 1998;95:13597–13602. doi: 10.1073/pnas.95.23.13597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons KT, et al. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- Simons KT, et al. Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins. 1999;34:82–95. doi: 10.1002/(sici)1097-0134(19990101)34:1<82::aid-prot7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Slynn GM, et al. Molecular genetical and phenotypical analysis of the gerM spore germination gene of Bacillus subtilis 168. FEMS Microbiol. Lett. 1994;121:315–320. doi: 10.1111/j.1574-6968.1994.tb07119.x. [DOI] [PubMed] [Google Scholar]
- Soding J, et al. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP. Specific recognition of bacteria by plant LysM domain receptor kinases. Trends Microbiol. 2004;12:201–204. doi: 10.1016/j.tim.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Steen A, et al. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Szilágyi A, Skolnick J. Efficient prediction of nucleic acid binding function from low-resolution protein structures. J. Mol. Biol. 2006;358:922–933. doi: 10.1016/j.jmb.2006.02.053. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, et al. TheCOGdatabase: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toratani T, et al. Structure of full-length bacterial chitinase containing two fibronectin type III domains revealed by small angle X-ray scattering. Biochem. Biophys. Res. Commun. 2006;348:814–818. doi: 10.1016/j.bbrc.2006.07.096. [DOI] [PubMed] [Google Scholar]
- Wardleworth BN, et al. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler ME, Hoch JA. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 2008;190:2645–2648. doi: 10.1128/JB.01682-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, et al. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 2005;1:e65. doi: 10.1371/journal.pgen.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.