Abstract
Spidermonkey is a new component of the Datamonkey suite of phylogenetic tools that provides methods for detecting coevolving sites from a multiple alignment of homologous nucleotide or amino acid sequences. It reconstructs the substitution history of the alignment by maximum likelihood-based phylogenetic methods, and then analyzes the joint distribution of substitution events using Bayesian graphical models to identify significant associations among sites.
Availability: Spidermonkey is publicly available both as a web application at http://www.data-monkey.org and as a stand-alone component of the phylogenetic software package HyPhy, which is freely distributed on the web (http://www.hyphy.org) as precompiled binaries and open source.
Contact: afpoon@ucsd.edu
1 INTRODUCTION
Detection of coevolving residues in a protein by the comparative analysis of homologous gene sequences is an important source of evidence for the functional and/or structural characterization of proteins. Similarly, comparative analysis of non-coding nucleotide sequences can reveal secondary structure, e.g. stem-loops in ribosomal RNAs. By failing to address the evolutionary nature of sequence variation, however, such methods are susceptible to spurious associations between sites due to identity by descent (Felsenstein, 1985). Additionally, pairwise association tests cannot capture higher order interactions and do not provide a means for compiling the ‘big picture’ from a list of significant pairs. Spidermonkey provides an easy-to-use web interface to a framework for detecting coevolving sites from coding and non-coding nucleotide or protein sequences, which combines phylogenetic and machine learning techniques to address these issues (Poon et al., 2007).
2 METHODS
The history of substitution events is inferred from an alignment using standard phylogenetic methods. If a tree is not uploaded with the alignment, then one is estimated using the neighbor-joining method (Saitou and Nei, 1987). A substitution model corresponding to the user-defined data type (nucleotide/codon/protein) is fitted to these data by maximum likelihood and the inferred ancestral sequences are used to map substitution events to branches in the tree (Kosakovsky Pond and Frost, 2005c). Replicate sets of ancestral sequences can be resampled from the posterior probability distribution and analyzed in parallel. For codon data, only non-synonymous substitutions are retained for further analysis. Invariant sites are automatically excluded in all cases. Correlated patterns of substitutions in the tree implies coevolution among sites. The joint distribution of substitutions in the tree is encoded as a binary state matrix, in which each row corresponds to a unique branch and each column to a site in the alignment, and is analyzed using Bayesian graphical models (BGMs).
A BGM is a compact representation of a joint probability distribution in which each node represents a distinct random variable (Pearl, 1988). An edge originating from ‘parent’ node P and terminating in ‘child’ node C postulates a conditional dependence between the corresponding sites, i.e. C is ‘influenced’ by P. We use the order-MCMC algorithm (Friedman and Koller, 2003) to infer the configuration of edges in the graph that best explains the data. Due to limited computing resources, we restrict BGM analyses on Spidermonkey to 150 sequences and 1000 nodes if k=1 or 75 nodes if k=2, where k is the maximum number of parents per node. Spidermonkey executes a single MCMC run with a burn-in period of 104 steps followed by 105 steps, sampled at regular intervals of 103 steps. We have found these default settings to provide sufficient conditions for convergence and sampling.
3 IMPLEMENTATION
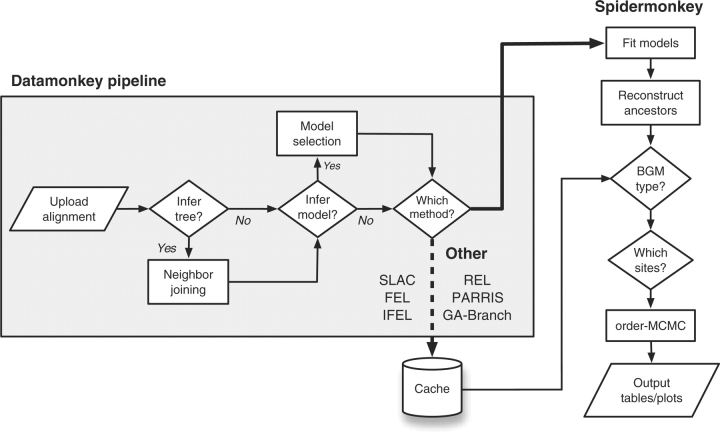
A web interface was constructed using custom Perl CGI and HyPhy batch language scripts (Kosakovsky Pond et al., 2005) and tested on the web browsers Safari, Firefox, Konqueror and Internet Explorer; and the computing platforms Mac OS X, Red Hat Linux, Windows XP Professional for 32- and 64-bit architectures and Windows 2003 Server. Presently, Spidermonkey is hosted on a Linux cluster comprising 20 quad-processor computing nodes. Its functionality is also available as a prepackaged analysis in HyPhy, which can be downloaded and run on local machines. Preprocessing of uploaded alignments (supporting NEXUS, PHYLIP, MEGA and FASTA formats), estimation of tree topology and MPI-enabled model selection and nucleotide and codon model fitting are handled using modified pre-existing scripts in the Datamonkey system (Kosakovsky Pond and Frost, 2005a; Fig. 1). The alignment, tree and analysis results are cached on our server for up to 96 h and can be retrieved from a temporary webpage with a randomized identifier.
Fig. 1.
Flowchart diagram of the Spidermonkey pipeline. Abbreviations: SLAC=single likelihood ancestor counting; FEL=fixed effects likelihood; IFEL=internal FEL; REL=random effects likelihood (Kosakovsky Pond and Frost, 2005c); PARRIS=a partitioning approach for robust inference of selection (Scheffler et al., 2006); GA-Branch=genetic algorithm for detecting branch-specific selection (Kosakovsky Pond and Frost, 2005b).
The inferred distribution of substitutions in the tree is transferred to the Spidermonkey BGM scripts (Fig. 1). The subset of sites to be analyzed as a BGM can be arbitrary or determined by a user-defined threshold in the following statistics on substitutions per site: (1) raw count; (2) percentage of branches affected or (3) information entropy. The analysis reports edges with marginal posterior probabilities exceeding a default cutoff of 0.5, which may be reset to a user-defined value. A visualization of the graph (Gansner and North, 2000) can be exported in PNG, Postscript or PDF formats.
4 DISCUSSION
The availability of rapid algorithms using phylogenetic methods for detecting coevolving sites from sequence data is a critical resource for the accurate exploratory analysis of biological variation. Spidermonkey is a key component update of our Datamonkey suite of bioinformatic tools providing intuitive web access to cutting-edge methods for detecting coevolving sites.
ACKNOWLEDGEMENTS
We thank Selene Zarate and León Martínez-Castilla for their assistance in beta-testing.
Funding: This work was supported by grants AI43638, AI47745 and AI57167 from the National Institutes of Health, and by University of California San Diego Centers for AIDS Research / National Institute of Allergy and Infectious Disease (NIAID) developmental awards AI36214 to S.D.W.F. and S.L.K.P. F.I.L. received financial support from the Wellcome Trust.
Conflict of Interest: none declared.
REFERENCES
- Felsenstein J. Phylogenies and the comparative method. Am. Nat. 1985;125:1–15. [Google Scholar]
- Friedman N, Koller D. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 2003;50:95–125. [Google Scholar]
- Gansner E, North S. Software: Practice and Experience. Chichester, New York: John Wiley & Sons, Ltd.; 2000. An open graph visualization system and its applications to software engineering. [Google Scholar]
- Kosakovsky Pond SL, Frost S.DW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005a;21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost S.DW. A genetic algorithm approach to detecting lineage-specific variation in selection pressure. Mol. Biol. Evol. 2005b;22:478–485. doi: 10.1093/molbev/msi031. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost S.DW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005c;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, et al. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Pearl J. Probabilistic reasoning in intelligent systems: networks of plausible inference. San Mateo, CA: Morgan Kaufmann Publishers; 1988. p. 552. [Google Scholar]
- Poon A.FY, et al. An evolutionary-network model reveals stratified interactions in the V3 loop of the HIV-1 envelope. PLoS Comput. Biol. 2007;3:e231. doi: 10.1371/journal.pcbi.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scheffler K, et al. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]