Abstract
Motivation: Certain chemical substructures are present in many drugs. This has led to the claim of ‘privileged’ substructures which are predisposed to bioactivity. Because bias in screening library construction could explain this phenomenon, the existence of privilege has been controversial.
Results: Using diverse phenotypic assays, we defined bioactivity for multiple compound libraries. Many substructures were associated with bioactivity even after accounting for substructure prevalence in the library, thus validating the privileged substructure concept. Determinations of privilege were confirmed in independent assays and libraries. Our analysis also revealed ‘underprivileged’ substructures and ‘conditional privilege’—rules relating combinations of substructure to bioactivity. Most previously reported substructures have been flat aromatic ring systems. Although we validated such substructures, we also identified three-dimensional privileged substructures. Most privileged substructures display a wide variety of substituents suggesting an entropic mechanism of privilege. Compounds containing privileged substructures had a doubled rate of bioactivity, suggesting practical consequences for pharmaceutical discovery.
Contact: fritz_roth@hms.harvard.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The observation that commercially available drugs have physical properties that distinguish them from other compounds led to the establishment of Lipinski's ‘Rule of 5’ to predict drug absorption and permeation (Lipinski et al., 2001). While the likelihood that any given compound which satisfies this rule will become a drug remains small, the Rule of 5 has been a valuable guide for the design of chemical libraries. The need exists to further enrich chemical libraries with potential drug molecules.
The abundance of certain substructures in commercially available drugs has motivated the search for privileged substructures, i.e. substructures associated with biological activity (DeSimone et al., 2004; Horton et al., 2003). Among the reported privileged substructures are benzodiazepines (Evans et al., 1988) and hydrophobic ring systems (Ariens et al., 1979, Fig. 1A) which are present in drugs active against various protein targets (Andrews and Lloyd, 1982).
Fig. 1.
Reportedly privileged substructures and related compounds. (A) Reportedly privileged substructures. 1,4-benzodiazepin-2-one (1), indole (2), purine (3), spiropiperidine (4) and biphenyl (5) are reported as privileged. The wavy line on spiropiperidine (4) indicates that ring can have variable composition and size. (B) Examples of endogenous molecules containing reportedly privileged substructures. ATP (6) contains purine, the amino acid tryptophan (7) contains indole, and nordiazepam (8), which occurs naturally in mammalian brains, contains 1,4-benzodiazepin-2-one. (C) Examples of drugs with reportedly privileged substructures. Devazepide (9) is a cholecystokinin A antagonist that contains indole and 1,4-benzodiazepin-2-one. Acyclovir (10), used to treat herpes, contains purine. Spiperone (11), a dopamine D2 antagonist, contains spiropiperidine. Diflunisal (12), an anti-inflammatory analgesic, contains biphenyl.
The shape of privileged substructures may be preferred by hydrophobic pockets on protein surfaces (Bondensgaard et al., 2004; Hajduk et al., 2000; McGaughey et al., 1998), or have structural homology to biological substrates (Fig. 1B; Jacobson, 2001; McGaughey et al., 1998). For example, the benzodiazepine scaffold (Evans et al., 1988) (1) in drug compounds (9) (Fig. 1C) is active against opioid receptors and other protein targets (Marsters, 1994; Patchett and Nargund, 2000). This may be explained by structural homology to endogenous biomolecules (8) (Sangameswaran et al., 1986) or to peptide β-turns (Ripka et al., 1993). Similarly, Indole (2) and Purine (3), present in many drug compounds (9, 10) (DeSimone et al., 2004; Dinnell et al., 2001; Heinelt et al., 2001; Jacobson, 2001; Willoughby et al., 2002), are also present in endogenous biomolecules such as tryptophan (7) and ATP (6).
While many of the reported privileged substructures are flat, aromatic ring systems, there are exceptions. For example, spiropiperidines (Klabunde et al., 2002; Patchett and Nargund, 2000; Patchett et al., 1995) (4, 11) and cyclic peptides (seen in cyclosporin A, for example) (Horton et al., 2002) have three-dimensional geometries.
The activity of privileged substructures is distinct from that of promiscuous inhibitors which act by molecular aggregation and inhibit proteins non-selectively (McGovern et al., 2002). Many compounds containing privileged substructures bind proteins selectively, as revealed by NMR studies of biphenyl (5), for example (Fig. 1A; Hajduk et al., 2000), also present in the drug diflunisal (12) (Fig. 1C).
Privileged substructures remain controversial because their abundance in drug compounds may be a trivial consequence of their abundance in chemical libraries (DeSimone et al., 2004). Numerous computational analyses have identified privileged substructures abundant in bioactive compounds (Bemis and Murcko, 1996; Lewell et al., 1998; Nilsson et al., 2001; Sheridan, 2003; Wagener and van Geerestein, 2000), without considering bias towards some substructures in compound library construction. In contrast, others have employed decision trees (Rusinko et al., 1999, 2002; van Rhee, 2003; Young and Hawkins, 1995) to identify substructures that discriminate activity from inactivity within a given collection of compounds. The decision tree estimates the conditional probability of activity given the combination of substructures present (or absent) in a compound, while accounting for the abundance of substructures within the library. Although these studies identified some enriched substructures, the activity for which enrichment was shown was defined narrowly according to a single biological assay or specific class of protein targets (Muller, 2003).
To assess reportedly privileged substructures and to identify new ones, we evaluated diverse high-throughput phenotypic assays applied to a commercially available chemical library. ‘Active’ compounds were defined as those showing activity in at least one of these assays. Decision trees were then used to identify substructures that best discriminated active from inactive compounds. Once identified, each of these discriminating substructures were tested statistically for enriched activity and compared with the privileged substructures reported in the literature. In addition, substructures were independently assessed for their ability to broadly enrich for compound activity across different assays of different chemical libraries. Strikingly, we found that top-ranked lead compounds yielded more than twice the rate of bioactives in many cases.
2 METHODS
2.1 Assay data
We examined 24 cell-based phenotypic assays (including four assays of zebrafish embryos) applied at the Harvard Institute of Chemistry and Cell Biology to the 16 320-compound Chembridge Diverse Set E library. A compound was deemed positive for a particular assay if it was reported to induce a visually detectable phenotype (qualitative scoring facilitated by automated microscopy) or if it achieved a quantitative score in the top 1% for an assay. Alternatively, we defined positives for quantitative scores according to a factor of three departure of assay signal from baseline. After pooling the data, a compound was considered active if it was positive in at least one assay (18.4% of all compounds) and inactive otherwise. A similar procedure was used to identify active compounds among a 37 330-member National Cancer Institute (NCI) chemical library tested for growth inhibition in 70 cancer cell lines. Compounds in the NCI library scoring in the top 3% of at least one assay were identified as active (11.4% of all compounds); the more liberal assay threshold of 3% was used because compounds meeting this threshold were active below the reported Chembridge screening concentration of 10 μM. Because most of the compounds in the Chembridge library had molecular weights below 500, only compounds with a molecular weight below 500 were examined in the NCI library. In addition, we studied an assay of the Chembridge Microformat library measuring inhibition of histone acetylation and two assays of the Chembridge Diverse Set E library (not included above) measuring arginine methyltransferase inhibition (Cheng et al., 2004) and selective killing of Neu-overexpressing cells (Fantin et al., 2002).
2.2 Fragmentation of compounds
The structure of each compound was converted to a SMILES string using Daylight's mol2smi algorithm. A collection of 4860 unique substructures were generated by fragmenting each compound SMILES in the Chembridge Diverse Set E library using the Daylight SMARTS and SMIRKS toolkits and one of six fragmentation strategies, including RECAP (Csizmadia, 2000; Lewell et al., 1998, see Supplementary Material) which employs retrosynthetic analysis and tends to produce substructures that would be useful in guiding medicinal chemistry optimization and combinatorial library design. Each fragmentation strategy was applied exhaustively using a series of virtual chemical reactions (represented by SMIRKS strings). The resulting substructures were represented as SMARTS strings (symbols representing the non-hydrogen wild-card ‘[!#1]’ were used where appropriate). The generated substructures were then pooled and those appearing at least five times in the library were retained, yielding a non-redundant set of 4860 chemical substructures. We note the analogy of this strategy to previous graph mining methods (Cook and Holder, 2006; Nijssen and Kok, 2004; Rosenkranz and Klopman, 1990). The Daylight SMARTS toolkit was applied to generate an array of 1's and 0's indicating the presence or absence of each substructure in each molecule in each library.
2.3 Decision trees
In order to find the substructures most associated with biological activity, decision trees (Rusinko et al., 1999, 2002; van Rhee, 2003; Young and Hawkins, 1995) were used to partition compounds in the Chembridge Diverse Set E library based on the presence or absence of highly discriminating substructures chosen from the set of 4860 substructures. The most discriminating substructures were identified based on mutual information between substructure presence and compound activity. Compounds were then partitioned into subgroups depending on the presence or absence of a given discriminating substructure, and those subgroups were further partitioned (recursively) based on additional discriminating substructures. To avoid overfitting, discriminating substructures were chosen using the Bayesian information criterion (BIC) (Friedman and Goldszmidt, 1996; King et al., 2003, see Supplementary Material). The final partitions (‘leaf nodes’ in the decision tree) represent sets of compounds with (or without) specific substructures that are enriched or depleted in activity relative to other compounds in the library.
2.4 Significance of substructure activity
Each discriminating substructure selected by the decision tree was tested for association with activity within each individual assay and using the definition of activity based on multiple assays. The cumulative hypergeometric test of association was used (Klekota et al., 2005, see Supplementary Material). For each substructure, the number of individual assays yielding significant (P < 0.01) associations (either positive or negative) was counted.
2.5 Ability of the decision tree to enrich for bioactivity
The ability of the decision tree to enrich for compound activity in various assays and compound libraries was determined. Compounds assigned to a given leaf in the decision tree are assigned an activity score equal to the fraction of active compounds observed in that leaf (to avoid small sample effects on estimated proportions, one pseudocount was distributed according to the overall fraction of active compounds within the library). The tree was also used to assign leaf nodes and corresponding activity scores to compounds not used in tree construction. Rates of bioactivity among compounds ranked by the decision tree were compared with randomly permutated compound rankings. The number of active compounds in the Chembridge Diverse Set E, the Chembridge Microformat and the NCI libraries recovered by ranking according to activity scores was compared with randomly ranked lists.
3 RESULTS
3.1 Using substructures to group compounds according to bioactivity
We examined 24 cell-based phenotypic assays applied to the 16 320-compound Chembridge Diverse Set E library. These assays encompass a variety of chemical-induced phenotypes including mitotic arrest, endocytosis inhibition and histone acetylation (Boyce et al., 2005; Feng et al., 2003; Haggarty et al., 2000, 2003; Mayer et al., 1999; Nieland et al., 2002; Yarrow et al., 2003, 2005). This set of assays was selected from an original set of 85 assays, excluding assays with phenotypes attributable to compound fluorescence, toxicity (cell-death), non-specific transcriptional upregulation or inhibition of luciferase (a commonly used assay reporter). Because false positive ‘promiscuous inhibitors’ form molecular aggregates that are less membrane permeable (McGovern et al., 2002), we used only cell-based assays.
We focused primarily on the Chembridge Diverse Set E library, since it is not highly biased towards particular protein target classes. We confirmed its diversity, showing that the average pairwise Tanimoto coefficient—a measure of chemical substructure similarity—is 0.2 within this library. This is well below the threshold of 0.85 that is widely used to classify compounds as similar.
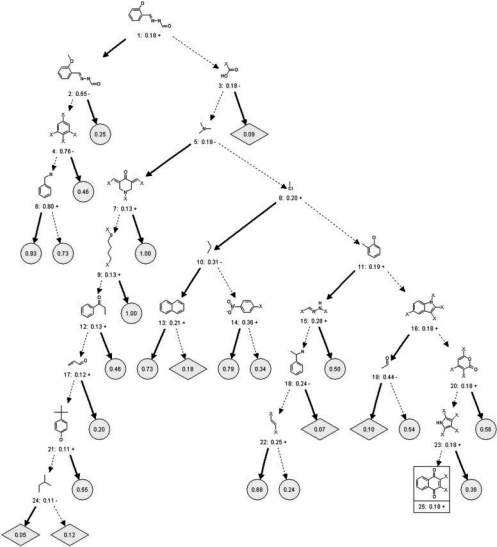
The set of 16 320 Chembridge Diverse Set E compounds was partitioned using a decision tree (Fig. 2). In this tree, the ‘root’ node corresponds to the set of all compounds. Compounds were successively divided into ever smaller subsets according to the presence or absence of ‘discriminating’ substructures. The ‘X’ symbol indicates a non-hydrogen atom and all hydrogen atoms in ‘X’-containing substructures (whether shown or implied) must be exactly matched. All other substructures have unspecified patterns of hydrogen and non-hydrogen atom substitution. At each node, the discriminating substructure that was used to divide the corresponding set of compounds was chosen using an unbiased information-theoretic criterion (see Section 2). Each compound was ultimately classified into one of 44 ‘leaf nodes’—compound subsets that are enriched or depleted in biological activity relative to the rest of the library—based on the presence or absence of 43 ‘discriminating’ substructures (Fig. 2).
Fig. 2.
Discriminating substructures identified by the decision tree. The substructures were selected by the decision tree to discriminate active and inactive compounds in the Chembridge library. The ‘X’ symbol indicates a non-hydrogen atom, and hydrogen atoms (whether shown or implied) in ‘X’-containing substructures must be matched. All other substructures have unspecified patterns of hydrogen and non-hydrogen atom substitution. The symbols ‘+’ and ‘-’ indicate whether or not the node substructure is associated with an increase or decrease in compound activity relative to its parent node in the tree. Bold arrows pointing away from a substructure indicate its presence and dotted arrows indicate its absence. The substructure composition of each leaf (blue circle or red diamond) is constrained by the intersection of statements about the presence or absence of substructures traced from the tree root (node 1) to each leaf. The nodes containing the substructures are numbered and the fraction of active compounds is listed in each node and leaf. Leaves shown as blue circles are enriched in activity and leaves shown as red diamonds are depleted in activity relative to the entire library (18.4% of the library is active as indicated by the tree root, node 1). For space considerations, a subtree stemming from node 25 has been excluded (indicated by an enclosing box; see Supplementary Fig. S1 for this subtree). Supplementary Table S1 details the prevalence of selected substructures within the library as well as their enrichment in bioactivity when considered individually (without respect to the presence of any other substructure).
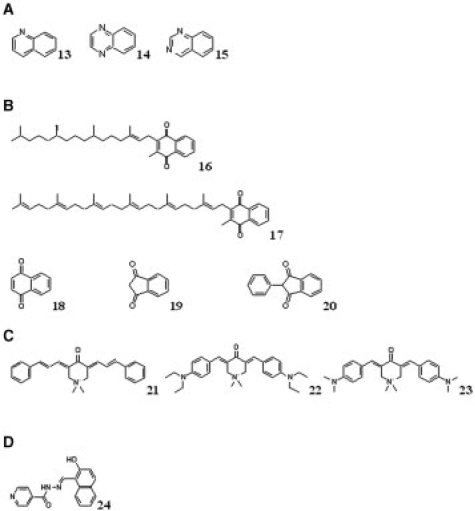
Discriminating substructures selected by the decision tree (Fig. 2 and Supplementary Fig. S1) include many which were reported as privileged (DeSimone et al., 2004; Horton et al., 2003). For example, indole is associated with an increase in biological activity in the Chembridge library assays among compounds lacking the substructures shown at nodes 1, 3, 5, 8 and 11. Interestingly, the selected indole substructure (node 16) had multiple non-hydrogen atoms (‘X’) attached to it, supporting previous intuition that privileged substructures may represent molecular scaffolds enriched for favorable binding entropy rather than enthalpy or complementary charge (Bondensgaard et al., 2004; Hajduk et al., 2000; Jacobson, 2001; McGaughey et al., 1998). Other potential scaffolds with multiple non-hydrogen substituents were also associated with activity: these include pyrrole (substructure at node 23) and benzene (substructures at nodes 32 and 39), which are components of indole and certain amino acids. Quinoline (13) (Fig. 3A) with an attached hydroxyl group (substructure at node 43) was also associated with increased activity. This substructure resembles the reportedly privileged substructures quinoxaline (14) (Fig. 3A) and quinazoline (15) (Fig. 3A) (Horton et al., 2003). (The numbers of hydrogen atoms on quinoline and other aromatic substructures were not explicit leaving their preferred role as scaffolds or substituents ambiguous; however, enrichment in assay activity generally correlated with increasing number of explicit non-hydrogen substituents, P=0.0242; see Supplementary Fig. S2). These and many of the other discriminating substructures, e.g. naphthalene (node 13), are aromatic ring systems which are consistent with previous claims of general privilege for bicyclic aromatic substructures.
Fig. 3.
Validated discriminating substructures and related compounds. (A) Structures of quinoline, quinoxaline and quinazoline. Quinoline (13) (present at node 43) is associated with an increase in activity and similar to reported privileged substructures quinoxaline (14) and quinazoline (15). (B) Structure of vitamin K1, vitamin K2, naphthoquinone, 1,3-indandione and phenindione. Naphthoquinone (18) and 1,3-indandione (19), similar to vitamin K1 (16) and vitamin K2 (17), were identified as enriched in activity by the decision tree. Phenindione (20) which is an FDA-approved anti-coagulant containing 1,3-indandione is shown. (C) Structures of NSC636679, NSC634791 and NSC618757. NSC636679 (21), NSC634791 (22) and NSC618757 (23) contain the substructure at node 7 identified as enriched in the tree; these compounds inhibit cancer cell growth by inhibiting the ABCB1 (MDR1) membrane transport protein. (D) Structure of NIH. NIH is (24) a metal chelator containing the most discriminating substructure (node 1, root of the tree).
Interestingly, many of the discriminating substructures we identified have structural homology to naturally occurring molecules. For example, naphthoquinone (18) (substructure at node 25) (Fig. 3B) was identified as enriched in biological activity in the Chembridge library. This substructure comprises a significant portion of vitamins K1 (16) and K2 (17) (Fig. 3B). Vitamins K1 and K2 are essential nutrients involved in the regulation of at least 11 Gla-proteins involved in blood coagulation, bone metabolism and vascular biology. Interestingly, the non-hydrogen atom substitutions on substructure at node 25 correspond perfectly to the substituents present in vitamin K (Shearer, 2000). Another enriched substructure, 1,3-indandione (substructure at node 28) (19) is structurally similar to naphthoquinone (18) and is present in a variety of biologically active compounds including the FDA-approved anti-coagulant phenindione (20) and pesticides (Braselton et al., 1992) (Fig. 3B). 1,3-indandione has homology to a structural component of vitamins K1 and K2 and competes with vitamin K binding (Mount and Feldman, 1983).
In contrast to reportedly privileged substructures, some of the discriminating substructures associated with bioactivity are neither flat nor aromatic (Fig. 2 and Fig. S1; substructures at nodes 7 and 35). These substructures contain rings and double bonds that contribute to rigidity (a feature of many reported privileged substructures), but they also have sp3-hydridized atoms that make them richer in three-dimensional geometry. Notably, compounds in the Chembridge Diverse Set E library containing the substructure at node 7 are structurally homologous to three compounds present in the NCI library—NSC636679 (21), NSC634791 (22) and NSC618757 (23) (Fig. 3C). These compounds have been reported to inhibit cancer cell growth through inhibition of ABCB1 (MDR1), a membrane transport protein implicated in multi-drug resistance of cancers (Szakács et al., 2004). Each of the compounds contain the substructure at node 7 flanked by aromatic rings, forming a symmetric molecule. Examination of the ABCB1 structure (Seigneuret and Garnier-Suillerot, 2003) suggests that molecules may interact with ABCB1's two ATP-binding sites. This observation motivates the exploration of other molecular scaffolds with rich three-dimensional geometries and symmetries.
Some substructures (notably the substructures at nodes 10, 24 and 27) are ‘underprivileged’, i.e. associated with decreased biological activity. Among these substructures are long carbon chains and chains of other sp3-hybridized atoms which are highly flexible and therefore likely increase the entropic cost of protein binding, in contrast to the privileged ring systems, which have less flexibility and predictably smaller entropic barriers to binding drug pockets: substructures only containing sp3-hybridized carbons were enriched in significantly fewer assays than substructures only containing aromatic carbons (P=0.0006, see Supplementary Fig. S2) and substructures lacking rings were enriched in significantly fewer assays than substructures containing rings (P=0.0036, see Supplementary Fig. S2). Underprivileged substructures should not necessarily be excluded from chemical libraries, as they may provide binding specificity, an important property of successful drugs; in fact, certain substructures (notably those at nodes 37 and 40) become favorable in the presence of other substructures as depicted by the tree, but are not favored when considered individually (Table S1), demonstrating the ‘conditional’ privilege of certain substructures. Different aliphatic carbon chains distinguish the various K vitamins suggesting physiological significance of these substructures (Fig. 3B). It is also interesting that twice as many substructures were found to be enriched in activity than were found to be deficient.
The most discriminating substructure (node 1, the root node of the tree) represents the entire active portion (having sigma-orbital electron pairs) of the known metal chelator NIH (24) (Darnell and Richardson, 1999; Le and Richardson, 2004; Liang and Richardson, 2003) (Fig. 3D). This substructure is associated with a significant increase in biological activity. Metal chelators are reported to promote transcription non-specifically (Darnell and Richardson, 1999; Le and Richardson, 2004; Liang and Richardson, 2003) and inhibit other proteins affecting cell growth (Darnell and Richardson, 1999), so that this substructure is expected to correlate with bioactivity; many metal chelators were observed to confound our excluded gene reporter assays of the Chembridge Diverse Set E library (Randy King, HMS, personal communication).
To provide intuition about the compound sets corresponding to nodes in the tree, Supplementary Figure S3 shows representative structures for each leaf node of the tree shown in Figure 2 and Supplementary Fig. S1.
A separate tree generated using the same assay data with a fold-change threshold for activity yielded similar discriminating substructures (data not shown). Interestingly, the new tree included an additional three-dimensional substructure—a tricyclic ring system with a seven-membered ring resembling that of benzodiazepine (see Supplementary Fig. S4). The privileged status of this ring system was not evident in the original tree (perhaps due to the selection of two of its component rings in the original tree).
We explored an alternative definition of activity, requiring compounds to score in two or more assays. A tree trained using this definition contained many of the substructures present in the original tree and was similarly predictive of activity in independent assays despite the exclusion of 75% of compounds defined as active under the more permissive definition of activity (Fig. S9).
3.2 Substructures associated with general bioactivity
The significance of each substructure's enrichment or depletion for activity in the decision tree was also confirmed by a statistical test of association (α=0.01). Here, we tested the association of each substructure with activity in each individual assay. Because this test considered all compounds, significant associations indicate ‘unconditionally’ privileged (or underprivileged) substructures. An expanded set of 59 assays was examined (Table S1), including additional pure protein- or cell-extract-based assays and cellular toxicity assays, in addition to the 24 assays used to develop the decision tree. Substructures showing significant enrichment or depletion (α=0.05) in three or more assays were considered broadly enriched. Nearly all discriminating substructures selected for use in the decision tree were corroborated by significant enrichment or depletion (the enrichment of many tree nodes in bioactivity exceeded the frequency of actives in our training set, Table S2). For example, the Indole scaffold (substructure at node 16) was enriched in 18 individual assays—consistent with its role as enriching for activity in the decision tree. There were however, a number of exceptions, e.g. substructures at nodes 9, 14, 37 and 40. These substructures, selected as discriminating in the presence of other substructures (marked ‘Combo’ in Table S1), did not show enhanced activity in individual assays or tended to be depleted in activity. These substructures are conditionally privileged, i.e. are associated with heightened activity only in the presence of other substructures. Thus, the decision tree reveals ‘rules of privilege’ that associate biological activity with specific combinations of substructures.
Given that the decision tree selected only 43 substructures out of 4860 available, many other privileged substructures are likely present in the Chembridge Diverse Set E library. Some of these unselected substructures may not have been selected due to their similarity to substructures already selected. Examination of recent chemical literature (DeSimone et al., 2004; Horton et al., 2003) identified additional substructures previously reported to be ‘privileged’ in the Chembridge Diverse Set E library. We found that many of these are indeed broadly enriched (as defined above) (Fig. S5), including biphenyl (25), 1, 4-dihydropyridine (27), chromone (31), quinoxaline (33), indole (35) and benzimidazole (36).
3.3 The ability of discriminating substructures to enrich for bioactivity
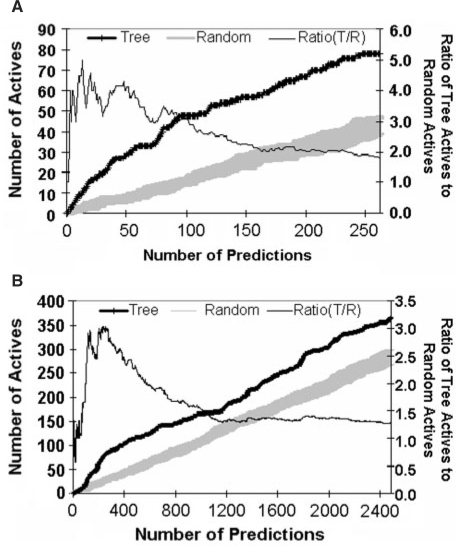
We wondered whether these substructures in combination could enrich for activity to an extent that would be practically useful. To examine this question, we trained a decision tree using a randomly selected 90% of Chembridge Diverse Set E library compounds, reserving the remaining 10% of compounds for testing. The resulting tree contained 39 discriminating substructures. Comparing these to the 43 discriminating substructures in the original tree, we found that 31 were identical, three differed only by a few explicit hydrogen atoms and the remaining five were structurally homologous. Each compound in the test set was mapped to a leaf node in the new tree based on substructure composition and assigned the corresponding activity score. Ranking compounds by activity score (Fig. 4A) revealed a substantial enrichment for active compounds; strikingly, there were 2–5 times more active compounds amongst the top 240-scoring compounds than among randomly chosen compounds. Similar results were obtained from a tree trained on only 50% of the data.
Fig. 4.
Enrichment of compounds for bioactivity based on substructure composition. (A) Recovery of active compounds in the 10% Chembridge Diverse Set E Test Set. A decision tree built using 90% of the Chembridge Diverse Set E library was used to rank the remaining 10% of compounds by rate of bioactivity expected given the substructure composition. The number of actives retrieved by the decision tree (wide black line) is 2–5 times greater than that retrieved by random selection indicated by the gray shaded area showing the mean ± 1 SD. (B) Recovery of compounds that inhibit cancer cell growth in the NCI Library. The original decision tree was used to rank NCI compounds according to the rate of activity against cancer cell growth expected given substructure composition. The number of actives retrieved by the decision tree rankings (wide black line) is 1.5–3 times greater than that retrieved by random selection indicated by the gray shaded area showing the mean ± 1 SD.
We wondered whether a decision tree trained on one set of assays to enrich for activity would be practically useful when applied to an independent set of assays. To this end, we labeled compounds in the Chembridge Diverse Set E library as active or inactive based on an assay measuring arginine methyltransferase inhibition (Cheng et al., 2004), which was not included in the original training set of 24 assays. Among the top-ranked 1200 compounds, the frequency of active compounds was 1.5–3 times greater than among randomly ranked compounds (Supplementary Fig. S6). Another assay not used in training, the extent of killing of Neu-overexpressing ‘oncogenic’ cells (Fantin et al., 2002) relative to wild-type cells, yielded a frequency of activity among top-ranked compounds that was 1.5–4 times higher than random compounds (Supplementary Fig. S7). Thus, the rules of privilege learned from one set of assays can be generally applied to substantially enrich for independent biological activities.
Because chemical libraries vary in their substructure composition, we wondered whether the rules of privilege learned from one chemical library would apply to independently constructed chemical libraries. To examine this question, we examined an assay of inhibition of histone acetylation in the Chembridge Microformat library. (This assay was among those applied to the Chembridge Diverse Set E library, which were used to train the original decision tree.) Each Microformat compound was mapped to a leaf node in the original decision tree (Fig. 2) and assigned the corresponding activity score. Amongst the top-ranked 450 compounds, the frequency of activity was 1.5–4 times higher than that of randomly chosen compounds (Supplementary Fig. S8). Repeating all of the above analyses after first removing compounds containing the substructure at node 1, the suspected metal chelators produced similar results. Thus, the rules of privilege determined from one chemical library allow substantial enrichment for activity within independent chemical libraries.
We wondered whether rules of privilege also had the power to enrich for bioactivity when both the chemical library and biological assays were independent of those used to train the decision tree (Fig. 2). Compounds of an NCI compound library were examined and identified as active if they scored in at least one assay measuring cancer cell line growth inhibition. Each NCI compound was mapped to a leaf node in the original decision tree (Fig. 2) and assigned the corresponding activity score. Amongst the top 1000 compounds, the frequency of activity was 1.5–3 times higher than that of randomly chosen compounds (Fig. 4B). This finding validates the concept of privileged substructure and shows that substructure properties learned from one dataset may be applied generally to multiple independent chemical libraries and bioactivities.
Activity of large NCI compounds (molecular weight >500) was poorly predicted based on substructures trained on the Chembridge library. The contributions of privileged substructures that we identified in a low-molecular weight library are likely to be diluted in larger molecules; furthermore, the mechanism of action of compounds with large molecular weights is likely to be qualitatively different from that of smaller compounds. Although privileged substructures may well exist among higher molecular weight compounds, these may need to be learned from a similar analysis applied to diverse biological assays of high-molecular weight compounds.
4 DISCUSSION
Our results validate the concept of privileged substructures, showing that many privileged substructures remain even after accounting for their overall abundance in the screened library. Moreover, privileged substructures identified as enriched for bioactivity in one library were also enriched for bioactivity within independent chemical libraries and assays were not used to learn rules of privilege. We confirmed several previously reported privileged substructures. We also identified ‘underprivileged’ substructures depleted in biological activity, e.g. long chains of sp3-hybridized atoms. While previously reported privileged substructures have had flat aromatic ring systems, we identified privileged substructures with three-dimensional geometries and others may be found in the analysis of libraries containing more three-dimensional substructure. Furthermore, the observation that privileged scaffolds contain diverse substituents suggests the broad activity associated with privileged substructures is the result of favorable scaffold entropy, while activity against a given target is determined by entropic contributions in combination with complementarity of shape and charge resulting from enthalpic contributions of substituents. For many assays, prioritization of compounds based on substructure double the frequency of active compounds. Therefore, the use of ‘rules of privilege’ to design new chemical libraries with a preference for particular combinations of substructure could have important implications for pharmaceutical discovery.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Stuart Schreiber of Harvard University for his support. We thank John Tallarico, Nathaniel Gray, Gabriel Berriz and Lan Zhang for their advice. We thank John Sullivan, Caroline Shamu and Su Chiang for their informatics assistance. We also thank the Harvard Biophysics Program for supporting this research.
Funding: Keck Foundation (to F.P.R.); National Institutes of Health (grants R01 HG0017115, R01 HG003224 and U01 HL81341 to F.P.R.).
Conflict of Interest: none declared.
REFERENCES
- Andrews PR, Lloyd EJ. Molecular conformation and biological activity of central nervous system active drugs. Med. Res. Rev. 1982;2:355–393. doi: 10.1002/med.2610020404. [DOI] [PubMed] [Google Scholar]
- Ariens EJ, et al. The Receptors, a Comprehensive Treatise. New York: Plenum Press; 1979. [Google Scholar]
- Bemis GW, Murcko MA. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- Bondensgaard K, et al. Recognition of privileged structures by G-protein coupled receptors. J. Med. Chem. 2004;47:888–899. doi: 10.1021/jm0309452. [DOI] [PubMed] [Google Scholar]
- Boyce M, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Braselton WE, Jr, et al. Confirmation of indandione rodenticide toxicoses by mass spectrometry/mass spectrometry. J. Vet. Diagn. Invest. 1992;4:441–446. doi: 10.1177/104063879200400413. [DOI] [PubMed] [Google Scholar]
- Cheng D, et al. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- Cook D, Holder . Mining Graph Data. Hoboken, N.J: Wiley-Interscience; 2006. [Google Scholar]
- Csizmadia F. JChem: Java applets and modules supporting chemical database handling from web browsers. J. Chem. Inf. Comput. Sci. 2000;40:323–324. doi: 10.1021/ci9902696. [DOI] [PubMed] [Google Scholar]
- Darnell G, Richardson DR. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents III: the effect of the ligands on molecular targets involved in proliferation. Blood. 1999;94:781–792. [PubMed] [Google Scholar]
- DeSimone RW, et al. Privileged structures: applications in drug discovery. Comb. Chem. High Throughput Screen. 2004;7:473–494. doi: 10.2174/1386207043328544. [DOI] [PubMed] [Google Scholar]
- Dinnell K, et al. 2-Aryl indole NK1 receptor antagonists: optimisation of the 2-aryl ring and the indole nitrogen substituent. Bioorg. Med. Chem. Lett. 2001;11:1237–1240. doi: 10.1016/s0960-894x(01)00183-4. [DOI] [PubMed] [Google Scholar]
- Evans BE, et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 1988;31:2235–2246. doi: 10.1021/jm00120a002. [DOI] [PubMed] [Google Scholar]
- Fantin VR, et al. A novel mitochondriotoxic small molecule that selectively inhibits tumor cell growth. Cancer Cell. 2002;2:29–42. doi: 10.1016/s1535-6108(02)00082-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, et al. Exo1: a new chemical inhibitor of the exocytic pathway. Proc. Natl Acad. Sci. USA. 2003;100:6469–6474. doi: 10.1073/pnas.0631766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N, Goldszmidt M. Proceedings of the 12th Conference on Uncertainty in Artificial Intelligence. San Francisco, CA: Morgan Kaufmann; 1996. Learning Bayesian networks with local structure; pp. 252–262. [Google Scholar]
- Haggarty SJ, et al. Dissecting cellular processes using small molecules: identification of colchicine-like, taxol-like and other small molecules that perturb mitosis. Chem. Biol. 2000;7:275–286. doi: 10.1016/s1074-5521(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Haggarty SJ, et al. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl Acad. Sci. USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk PJ, et al. Privileged molecules for protein binding identified from NMR-based screening. J. Med. Chem. 2000;43:3443–3447. doi: 10.1021/jm000164q. [DOI] [PubMed] [Google Scholar]
- Heinelt U, et al. Solid-phase optimisation of achiral amidinobenzyl indoles as potent and selective factor Xa inhibitors. Bioorg. Med. Chem. Lett. 2001;11:227–230. doi: 10.1016/s0960-894x(00)00631-4. [DOI] [PubMed] [Google Scholar]
- Horton DA, et al. Exploring privileged structures: the combinatorial synthesis of cyclic peptides. J. Compt. Aided Mol. Des. 2002;16:415–431. doi: 10.1023/a:1020863921840. [DOI] [PubMed] [Google Scholar]
- Horton DA, et al. The combinatorial synthesis of bicyclic privileged structures or privileged substructures. Chem. Rev. 2003;103:893–930. doi: 10.1021/cr020033s. [DOI] [PubMed] [Google Scholar]
- Jacobson KA. Probing adenosine and P2 receptors: design of novel purines and nonpurines as selective ligands. Drug Dev. Res. 2001;52:178–186. doi: 10.1002/ddr.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, et al. Predicting gene function from patterns of annotation. Genome Res. 2003;13:896–904. doi: 10.1101/gr.440803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klabunde T, Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Klekota J, et al. Identifying biologically active compound classes using phenotypic screening data and sampling statistics. J. Chem. Inf. Model. 2005;45:1824–1836. doi: 10.1021/ci050087d. [DOI] [PubMed] [Google Scholar]
- Le NT, Richardson DR. Iron chelators with high antiproliferative activity up-regulate the expression of a growth inhibitory and metastasis suppressor gene: a link between iron metabolism and proliferation. Blood. 2004;104:2967–2975. doi: 10.1182/blood-2004-05-1866. [DOI] [PubMed] [Google Scholar]
- Lewell XQ, et al. RECAP–retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998;38:511–522. doi: 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]
- Liang SX, Richardson DR. The effect of potent iron chelators on the regulation of p53: examination of the expression, localization and DNA-binding activity of p53 and the transactivation of WAF1. Carcinogenesis. 2003;24:1601–1614. doi: 10.1093/carcin/bgg116. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Marsters JC, Jr, et al. Benzodiazepine peptidomimetic inhibitors of farnesyltransferase. Bioorg. Med. Chem. 1994;2:949–957. doi: 10.1016/s0968-0896(00)82044-1. [DOI] [PubMed] [Google Scholar]
- Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- McGaughey GB, et al. pi-Stacking interactions. Alive and well in proteins. J.∼Biol. Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- McGovern SL, et al. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J. Med. Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- Mount ME, Feldman BF. Mechanism of diphacinone rodenticide toxicosis in the dog and its therapeutic implications. Am. J. Vet. Res. 1983;44:2009–2017. [PubMed] [Google Scholar]
- Muller G. Medicinal chemistry of target family-directed masterkeys. Drug Discov. Today. 2003;8:681–691. doi: 10.1016/s1359-6446(03)02781-8. [DOI] [PubMed] [Google Scholar]
- Nieland TJ, et al. Discovery of chemical inhibitors of the selective transfer of lipids mediated by the HDL receptor SR-BI. Proc. Natl Acad. Sci. USA. 2002;99:15422–15427. doi: 10.1073/pnas.222421399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijssen S, Kok JN. Systems, Man and Cybernetics, 2004 IEEE International Conference. Vol. 5. Den Haag, Netherlands: IEEE20Press; 2004. Frequent graph mining and its application to molecular databases; pp. 4571–4577. [Google Scholar]
- Nilsson JW, et al. Solid-phase synthesis of libraries generated from a 4-phenyl-2-carboxy-piperazine scaffold. J. Comb. Chem. 2001;3:546–553. doi: 10.1021/cc010013o. [DOI] [PubMed] [Google Scholar]
- Patchett AA, Nargund RP. Privileged structures – an update. Annu. Rep. Med. Chem. 2000;35:289–298. [Google Scholar]
- Patchett AA, et al. Design and biological activities of L-163,191 (MK-0677): a potent, orally active growth hormone secretagogue. Proc. Natl Acad. Sci. USA. 1995;92:7001–7005. doi: 10.1073/pnas.92.15.7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripka WC, et al. Protein beta-turn mimetics I. Design, synthesis, and evaluation in model cyclic peptides. Tetrahedron. 1993;49:3593–3608. [Google Scholar]
- Rosenkranz HS, Klopman G. Evaluating the ability of CASE, an artificial intelligence structure-activity relational system, to predict structural alerts for genotoxicity. Mutagenesis. 1990;5:525–527. doi: 10.1093/mutage/5.6.525. [DOI] [PubMed] [Google Scholar]
- Rusinko A, III, et al. Analysis of a large structure/biological activity data set using recursive partitioning. J. Chem. Inf. Comput. Sci. 1999;39:1017–1026. doi: 10.1021/ci9903049. [DOI] [PubMed] [Google Scholar]
- Rusinko A, III, et al. Optimization of focused chemical libraries using recursive partitioning. Comb. Chem. High Throughput Screen. 2002;5:125–133. doi: 10.2174/1386207024607383. [DOI] [PubMed] [Google Scholar]
- Sangameswaran L, et al. Purification of a benzodiazepine from bovine brain and detection of benzodiazepine-like immunoreactivity in human brain. Proc. Natl Acad. Sci. USA. 1986;83:9236–9240. doi: 10.1073/pnas.83.23.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret M, Garnier-Suillerot A. A structural model for the open conformation of the mdr1 P-glycoprotein based on the MsbA crystal structure. J. Biol. Chem. 2003;278:30115–30124. doi: 10.1074/jbc.M302443200. [DOI] [PubMed] [Google Scholar]
- Shearer MJ. Role of vitamin K and Gla proteins in the pathophysiology of osteoporosis and vascular calcification. Curr. Opin. Clin. Nutr. Metab. Care. 2000;3:433–438. doi: 10.1097/00075197-200011000-00004. [DOI] [PubMed] [Google Scholar]
- Sheridan RP. Finding multiactivity substructures by mining databases of drug-like compounds. J. Chem. Inf. Comput. Sci. 2003;43:1037–1050. doi: 10.1021/ci030004y. [DOI] [PubMed] [Google Scholar]
- Szakács G, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6:129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- van Rhee AM. Use of recursion forests in the sequential screening process: consensus selection by multiple recursion trees. J. Chem. Inf. Comput. Sci. 2003;43:941–948. doi: 10.1021/ci034023j. [DOI] [PubMed] [Google Scholar]
- Wagener M, van Geerestein VJ. Potential drugs and nondrugs: prediction and identification of important structural features. J. Chem. Inf. Comput. Sci. 2000;40:280–292. doi: 10.1021/ci990266t. [DOI] [PubMed] [Google Scholar]
- Willoughby CA, et al. Combinatorial synthesis of 3-(amidoalkyl) and 3-(aminoalkyl)-2-arylindole derivatives: discovery of potent ligands for a variety of G-protein coupled receptors. Bioorg. Med. Chem. Lett. 2002;12:93–96. doi: 10.1016/s0960-894x(01)00665-5. [DOI] [PubMed] [Google Scholar]
- Yarrow JC, et al. Phenotypic screening of small molecule libraries by high throughput cell imaging. Comb. Chem. High Throughput Screen. 2003;6:279–286. doi: 10.2174/138620703106298527. [DOI] [PubMed] [Google Scholar]
- Yarrow JC, et al. Screening for cell migration inhibitors via automated microscopy reveals a Rho-kinase inhibitor. Chem. Biol. 2005;12:385–395. doi: 10.1016/j.chembiol.2005.01.015. [DOI] [PubMed] [Google Scholar]
- Young SS, Hawkins DM. Analysis of a 2(9) full factorial chemical library. J. Med. Chem. 1995;38:2784–2788. doi: 10.1021/jm00014a030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.