Abstract
Motivation: Development of high-throughput technology makes it possible to measure expressions of thousands of genes simultaneously. Genes have the inherent pathway structure, where pathways are composed of multiple genes with coordinated biological functions. It is of great interest to identify differential gene pathways that are associated with the variations of phenotypes.
Results: We propose the following approach for detecting differential gene pathways. First, we construct gene pathways using databases such as KEGG or GO. Second, for each pathway, we extract a small number of representative features, which are linear combinations of gene expressions and/or their transformations. Specifically, we propose using (i) principal components (PCs) of gene expression sets, (ii) PCs of expanded gene expression sets and (iii) expanded sets of PCs of gene expressions, as the representative features. Third, we identify differential gene pathways as those with representative features significantly associated with the variations of phenotypes, particularly disease clinical outcomes, in regression models. The false discovery rate approach is used to adjust for multiple comparisons. Analysis of three gene expression datasets suggests that (i) the proposed approach can effectively identify differential gene pathways; (ii) PCs that explain only a small amount of variations of gene expressions may bear significant associations between gene pathways and phenotypes; (iii) including second-order terms of gene expressions may lead to identification of new differential gene pathways; (iv) the proposed approach is relatively insensitive to additional noises; and (v) the proposed approach can identify gene pathways missed by alternative approaches.
Contact: shuangge.ma@yale.edu
Supplementary information:Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
In the past decade, we have witnessed a period of unparalleled development in the field of bioinformatics. Among the many newly encountered bioinformatics problems, identification of differential genomic markers or sets of markers has attracted extensive attention (Allison et al., 2006; Lesk, 2002; Wong, 2004). Identification of those differential markers or marker sets may reveal the genomic forces that drive variations of phenotypes.
Microarray technology makes it possible to measure expressions of thousands of genes simultaneously. In this article, we focus on microarray studies where clinical outcomes or phenotypes are measured along with expressions of thousands of genes. Genes or gene sets that have significant associations with the phenotypes/clinical outcomes in regression models will be referred to as ‘differentially expressed’. We note that the proposed methodologies can be extended to accommodate studies without clinical outcomes but with multiple experimental conditions.
With individual genes, there have been many publications investigating methodologies for identifying differential genes. We refer to McLachlan et al. (2004) and Knudsen (2006) for a comprehensive review. Identification of differential genes typically involves (i) computing a significance statistic for each individual gene. Shrinkage, penalization and thresholding methods have been proposed to remove extreme measurements due to randomness; and (ii) adjusting for multiple testing and identifying differential genes. Specifically, the false discovery rate (FDR) approaches have been proposed (Benjamini and Yekutieli, 2001).
Recent biomedical studies have suggested that variations of certain phenotypes, especially clinical outcomes of complex diseases such as cancer, are associated with differential expressions of multiple genes instead of a single gene. Such an observation has motivated researchers to define clusters of genes, instead of individual genes, as functional genomic units. These clusters are composed of multiple genes with coordinated biological functions, and have been referred to as ‘pathways’.
Detection of differential gene pathways or clusters has also been extensively investigated. Well-known examples include the gene set enrichment analysis (GSEA; Subramanian et al., 2005), the global test (Goeman et al., 2004) and the maxmean approach (Efron and Tibshirani, 2007), among others. We refer to Allison et al. (2006), Tintle et al. (2008), Nettleton et al. (2008), Ackermann and Strimmer (2009), Goeman and Buhlmann (2007) and Nam and Kim (2008) for comprehensive reviews of existing approaches. We note that the validity of different approaches is built on different assumptions of underlying data and model structures. With practical data, numerical studies in Tintle et al. (2008), Nettleton et al. (2008), Sneddon (2004) and references therein have suggested that different approaches may identify different sets of differential gene clusters, and there is no approach dominatingly better than the alternatives.
In this article, our goal is to identify gene pathways with significant associations with the clinical outcomes in regression models. Here, the gene pathways are constructed using existing pathological information of genes. Since a pathway may contain a large number of genes (more than the sample size), straightforward model fitting may lead to saturated models and improper significance. To tackle this problem, we propose first extracting a small number of representative features, which are linear combinations of gene expressions and/or their transformations, from each gene pathway. Regression models will be fit and significance will be defined with those representative features.
We propose using the principal components (PCs) of gene expressions and/or their transformations as the representative features of gene pathways. Detection of differential gene pathways using PC analysis (PCA)-based approaches has been investigated in Kong et al. (2006), Chen et al. (2008) and other articles. Compared with Kong et al. (2006), the proposed approach is applicable to not only data with binary outcomes, but also data with other type of outcomes. Compared with Chen et al. (2008), we investigate the possible contributions beyond the first PC. Such an aspect has been neglected in most previous studies. No supervised screening is conducted to avoid possible exclusion of important genes because of the screening. Inference is based on the permutation test to avoid possible overfitting. In addition, we consider the effects of higher order terms, which have been ignored in Kong et al. (2006) and Chen et al. (2008). Although higher order gene effects have been discussed in Jiang and Gentleman (2007) and other articles, they have not been seriously investigated with PCA-based approaches. In this article, we investigate whether the associations between the gene pathways and clinical outcomes can be attributed to higher order terms, particularly second-order terms, of gene expressions.
In Section 2, we describe construction of gene pathways and their representative features via PCA. In Section 3, we describe the proposed PCA-based approach. In Section 4, we analyze three gene expression datasets to further investigate the proposed approach. Analysis of sensitivity to additional noises and comparisons with alternative approaches are conducted. Discussions of the proposed approach are provided in Section 5. The article concludes with Section 6. A heuristic discussion of the theoretical validity of the proposed approaches are provided in the Supplementary Material.
2 METHODS
2.1 Gene pathways
Genes have the inherent pathway structure, where pathways are composed of multiple genes with coordinated pathological functions. In recent years, more and more attention has been drawn towards pathway-based methods for analyzing gene expression data. ‘Pathway analysis is a promising tool to identify the mechanisms that underlie diseases, adaptive physiological compensatory responses, and new avenues for investigation’ (Curtis et al., 2005).
Although pathways can be viewed as an interactive dynamic network, in this study, we choose a simpler point of view and think of pathways as static gene clusters. Such a perspective has been adopted in studies, such as Wei and Li (2007), Pang and Zhao (2008) and Shi and Ma (2008). Of special note, (i) for a small number of genes, the present pathway information can be partial or even wrong. However, this does not prevent pathways from being a useful tool for gene expression data analysis; (ii) different gene pathways may have overlapping genes, since one gene may have multiple pathological functions; (iii) in our numerical study, we retrieve pathway information from KEGG. The pathway structure can be further refined, if more databases such as BioCarta or GO are utilized; and (iv) some genes may not be annotated. Pathway information for those genes is not available. Those genes will be excluded from analysis in this study. Such an approach has been adopted by Wei and Li (2007).
2.2 Principal component analysis
PCA is a dimension reduction method and has been extensively used in gene expression analysis (McLachlan et al., 2004; Sharov et al., 2005). With gene expression data, dimension reduction or variable selection are usually needed to extract a small number of representative features that can represent the effects of all genes. Variable selection methods can be used when there are a small number of strong signals. In contrast, dimension reduction, including PCA, may perform better when there exist a large number of weak signals.
Consider a feature set O composed of m variables {X1,…, Xm}. In the context of gene expression analysis, the Xis may denote gene expressions and/or their transformations. With the PCA, any linear combinations of the Xis can be rewritten as
where U1,…, Uk are the k PCs and k is the rank of O. Particularly, Uis have unit norms and are the linear combinations of Xj, j=1,…, m, with Ui being orthogonal to Uj for i≠j. Variation explained by Ui decreases as i increases.
When the Xis are the gene expressions, the PCs have been referred to as ‘super genes’, ‘latent variables’ and ‘latent causes’ among other terminologies. The rationale of using PCA in pathway-based gene expression analysis is that the effects of a pathway on the clinical outcome can be captured by a small number of ‘super genes’, and expressions of those super genes are linear combinations of expressions of the genes. The super genes may correspond to the linear combinations of genes that best explain the variations of gene expression. We refer to Johnson and Wichern (2001) for more discussions of PCA techniques, and McLachlan et al. (2004) for their applications in gene expression analysis.
2.3 Expanded gene expression set
For the set O={X1,…, Xm}, we define its second-order expanded set as EO=O∪{Xi Xj: i,j=1,…, m}. That is, the expanded feature set is composed of the original features and their second-order terms. In a similar manner, we can define expanded feature sets with even higher order terms. We focus on the second-order expanded set in this article, since it is relatively simple and has an affordable computational cost.
In bioinformatics studies, with the number of input features much larger than the sample sizes, attention has been mainly focused on the linear effects. In the context of gene expression data, some articles have argued heuristically that using linear effects of genes can be better than using non-linear effects, although non-linear effects can be more flexible (Zhang et al., 2006). In the context of genome wide association studies, publications such as Carrasquillo et al. (2002) have shown that including the second-order interactions may improve identification and classification. With gene expression data, although we expect the linear effects of genes to capture most of their associations with the clinical outcomes, there is no reason why higher order terms should have no detectable contributions. In the context of gene expression analysis, there are a few studies showing that transformations, which include the second-order terms as special cases, may improve identification of differential genes (Xiong, 2006). In addition, Jiang and Gentleman (2007) discusses the possibility of non-linear gene effects in pathway analysis.
In this article, we will focus on the second-order terms of gene expression, including quadratics and interactions. Other transformations of gene expressions are possible, but of less interest due to the lack of interpretability.
2.4 Construction of representative features
A key step of the proposed approach is to construct a small number of representative features for each gene pathway. The representative features are expected to capture most of the associations between genes within a pathway and the clinical outcome. Motivated by the successes of PCA with gene expression data and the possibility of non-linear gene effects, we consider the following ways of constructing representative features. For a pathway composed of m genes, denote X1,…, Xm as the gene expressions.
-
(R1)
Consider O={X1,…, Xm}, i.e. the set composed of the m gene expression measurements. We select the first c PCs of O as the representative features. Since it is not clear how many PCs should be used, we consider c=1,…, c*, i.e. c* different sets of representative features composed of the first 1,…, c* PCs. In our numerical study, we set c*=5.
With (R1), we assume that the PCs of the gene expressions can capture the associations between gene pathways and clinical outcomes. We will compare representative feature sets composed of different number of PCs. Such a comparison may partly answer the question of ‘how many PCs will be needed’, which has been ignored in many previous studies. To achieve such a comparison, we will focus on gene pathways with at least c* genes. Smaller pathways can be studied by investigating each gene separately, and thus the proposed approach is not needed.
-
(R2)
With O={X1,…, Xm}, we construct its second-order expanded set EO. We then select the first c PCs of EO as the representative features. Following the same rationale as with (R1), we will consider c=1,…, c*.
With (R2), we consider the sets composed of gene expressions and their second-order terms. We assume that the PCs of such sets can capture the associations between the pathways and clinical outcomes. Here, the PCs are linear combinations of gene expressions and their second-order terms.
-
(R3)
With O={X1,…, Xm}, we select its first d PCs. Denote P as the set composed of the d PCs, and EP as its second-order expanded set. Members of EP are selected as the representative features. We will consider d=1,…, d*. In our numerical study, we set d*=3.
With (R3), we first construct PCs of the gene expressions. The PCs and their second-order terms are selected as the representative features.
With (R1), it is assumed that linear effects of genes are sufficient. In contrast, with (R2) and (R3), it is assumed that higher order terms may have significant contributions beyond linear terms. The representative features defined in (R2) and (R3) are all linear combinations of gene expressions and their second-order terms. However, with c and d smaller than the full ranks of their corresponding sets, (R2) and (R3) are in general not equivalent. With the three different ways of defining the representative features, for a specific pathway, there are c*+c*+d* different sets of representative features.
3 IDENTIFICATION OF DIFFERENTIAL PATHWAYS
Consider gene expression data with clinical outcome Y. Identification of differential pathways consists of the following steps.
Construct gene pathways using information retrieved from public databases. Only genes with pathway information will be used in downstream analysis. In this study, the KEGG (http://www.genome.ad.jp/kegg/) is used to construct gene pathways. Specifically, since lymphoma and leukemia data are analyzed and the general scheme is ‘cancer-related’, for each gene, we search for its pathways using a list of manually picked keywords as suggested by http://www.sonycsl.co.jp/person/tetsuya/sub2.html.
- For each gene pathway:
- Construct the c*+c*+d* different sets of representative features.
- Fit a regression model with Y as the response and the representative features as the covariates. Compute a summary statistic T, which can measure the association between the outcome and covariates.
- Randomly permute the response Y, fit the same regression model, and compute the summary statistic.
- Repeat Step (c) for B times, and compute a permutation P-value for T. In our numerical study, we set B=50 000.
- The above procedure generates c*+c*+d* p-values for each specific pathway.
For each pathway, select l (which can be one or more) P-values based on the specific analysis of interest. Combine and analyze the l×M P-values from the M pathways using the FDR approach.
3.1 Statistical modeling
With gene expression data, we commonly encounter continuous, categorical and censored survival clinical outcomes. For each type of clinical outcome, there are multiple applicable models. With continuous outcomes, we propose using the linear regression model and the mean squared error as the summary statistic T. With categorical outcomes, the logistic model and the deviance are chosen as the default model and the summary statistic. With censored survival data, the Cox proportional hazards model and the statistic of the score test are chosen as the default model and the summary statistic. We note that when there are strong evidences of model misspecification, alternative models may need to be considered.
3.2 Controlling the FDR
Denote N as the total number of tests and P-values. Since multiple tests will be considered simultaneously, we use the following approach to control the FDR: (i) we set the expected FDR to q=0.2; (ii) we order the P-values across N tests p(1)≤p(2)≤…≤p(N); (iii) we let r be the largest i such that p(i)≤i/N×q/C(N); and (iv) pathways corresponding to p(1)…p(r) are defined as significantly differential.
With gene pathways constructed using pathological information, different pathways may share common genes. To account for the possible complicated correlations among P-values caused by overlapped pathways, we set C(N)=∑i=1N1/i as suggested in Benjamini and Yekutieli (2001).
4 DATA ANALYSIS
4.1 DLBCL data
DLBCL (diffuse large B-cell lymphoma) is a fast growing, aggressive form of non-Hodgkin's lymphoma (NHL). The DLBCL prognostic study was first reported in Rosenwald et al. (2002). This study retrospectively collected tumor-biopsy specimens and clinical data for 240 patients with untreated DLBCL. The median followup is 2.8 years, with 138 observed deaths. Lymphochip cDNA microarrays were used to measure expressions of 7399 genes. The raw data and detailed experiment protocol are available at http://llmpp.nih.gov/DLBCL/. We retrieve pathway information from KEGG as described in Section 3. A total of 1047 genes belong to 159 KEGG pathways, with sizes ranging from 1 to 127 and median size 7. Among the 159 gene pathways, 97 have sizes equal to or larger than 5 and will be studied in our downstream analysis.
4.1.1 Pathway identification using linear gene effects
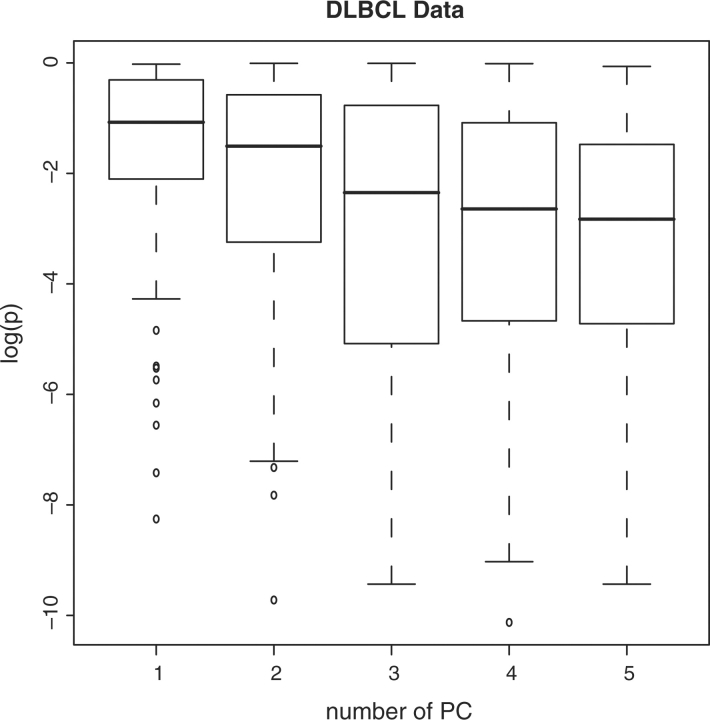
We first consider the representative features generated with (R1), and consider effects of different numbers of PCs. With c*=5, we show in Figure 1 the log P-values of the 97 pathways as a function of the number of PCs. It can be seen that, when the number of PCs increases, the P-values have an overall decreasing trend, which suggests that more pathways can be potentially identified as differential. The numbers of differential pathways identified using the first 1–5 PCs are 2, 11, 29, 29 and 29, respectively. Using the first three PCs can identify the most differential pathways, while keeping the number of representative features small. In the Supplementary Material, we provide detailed information on pathways identified with three PCs. Information on pathway names, pathway sizes and unadjusted P-values is available.
Fig. 1.
Analysis of DLBCL data with representative features (R1). Log P-values versus number of PCs.
We note that, although it is possible to get a P-value associated with T directly from the model fitting, we adopt the permutation approach to avoid any overly optimistic result.
Analysis of sensitivity to noises: as one reviewer pointed out, pathways, especially large pathways, may contain genes unrelated to the clinical outcomes. The proposed approach may potentially suffer from those noisy genes. To understand sensitivity to noises of the proposed approach, we consider the following analysis.
For a pathway with m genes, we add max(1, 10%×m) noisy genes, where expressions of the noisy genes are normally distributed with mean 0, and variance equal to the median variance of the original m genes. Thus, the ‘new’ pathway contains the original m genes as well as 10%m noises. In addition, we also consider 20% and 30% additional noises. With 10%, 20% and 30% additional noises, the proposed approach identifies 24, 28 and 24 pathways, respectively. The numbers of overlaps with pathways identified without noises are 24, 27 and 24, respectively, which suggests that the proposed analysis is relatively insensitive to noises.
Comparisons with alternative approaches: we analyze the DLBCL data with the following well-known alternative approaches and summarize the comparison results in Table 1: (i) the GSEA (Subramanian et al., 2005), which identifies two pathways as differential: the ribosome pathway, which has also been identified with the proposed approach, and the phenylpropanoid biosynthesis pathway missed by the proposed approach. Our preliminary investigation finds no association between the phenylpropanoid biosynthesis pathway and lymphoma progression; (ii) the maxmean approach (Efron and Tibshirani, 2007), which identifies no pathway as differential; and (c) the global test (Goeman et al., 2004), which identifies no pathway as differential.
Table 1.
Pathway identification using different approaches: number of pathways identified (number of overlap with the proposed approach)
| Approach | DLBCL | MCL | Leukemia |
|---|---|---|---|
| GSEA | 2 (1) | 69 (31) | 1 (1) |
| Maxmean | 0 (0) | 2 (2) | 2 (2) |
| Global test | 0 (0) | 68 (46) | 0 (0) |
| Kong's | NA | NA | 2 (2) |
| Univariate | 1 (1) | 77 (46) | 33 (19) |
| Proposed | 29 (29) | 50 (50) | 50 (50) |
NA: Kong's approach is only applicable to binary clinical outcomes.
4.1.2 Pathway identification using both linear and non-linear gene effects
We conduct the following analysis, which can account for contributions from non-linear gene effects.
Analysis I: in the first set of analysis of non-linear effects, we use the representative features generated with (R2) and different numbers of PCs. With c*=5, the numbers of differential pathways identified using the first 1–5 PCs are 0, 0, 1, 1 and 2, respectively. The Natural killer cell-mediated cytotoxicity pathway, which consists of 60 genes, is identified with # PC=5. This pathway is not identified using only linear effects. The significance of the natural killer cell-mediated cytotoxic pathway for NHL has been suggested in early biomedical studies (Mehta et al., 1989). Natural killer activity and antibody-dependent cellular cytotoxicity are two natural defense mechanisms that protect the host against various kinds of infections. The natural killer cells have a potential role in immune surveillance against virally infected cells and tumors, as well as in the regulation of normal stem-cell differentiation. The cells responsible for mediating the two activities are the large granular lymphocytes. Suppressions of natural killer and antibody-dependent cellular cytotoxicity have been observed in untreated NHL patients. In addition, natural killer cell-mediated cytotoxicity plays an important role in T-cell lymphomas (Neilan et al., 1983). Our analysis suggests that it is also related to B-cell lymphomas.
Analysis II: in the second set of analysis, we consider representative features generated with (R3), i.e. the expanded set generated with the first 1–3 PCs. The numbers of differential pathways identified are 3, 4 and 7, respectively. With three PCs of the gene expressions and their second-order terms, the cell-cycle pathway (81 genes) and the aminoacyl-tRNA biosynthesis pathway (11 genes) are identified as differential. Those two pathways are not identified using only linear effects. The KEGG cell-cycle pathway contains very important genes such as p53 and CDK1, which are associated with the progression of several cancers, including lymphomas. In addition, it has been suggested that cell-cycle regulators carry independent prognostic value in various subsets of lymphomas (Moller, 2003). It is very interesting that the cell-cycle pathway is identified as differential with non-linear effects. The implications of such a finding are worth further investigations. Aminoacyl-tRNA is tRNA to which its cognated amino acid is adhered. Its role is to deliver the amino acid to the ribosome where it will be incorporated into the polypeptide chain that is being produced. The specific linkage of the correct amino acid to each tRNA is accomplished by aminoacyl-tRNA synthetases. Although the aminoacyl-tRNA pathway has crucial biological functions in general, its connection with DLBCL progression is still not clear at this moment but is worth further biological investigations.
Loosely speaking, the feature sets used in the above analysis of non-linear effects contain the features used in the analysis of linear effects. However, with the second-order terms, the feature sets in the non-linear analysis are much larger than those in the linear analysis. Gene pathways contain noisy genes unrelated to the clinical outcomes. With second-order terms and higher dimensions, signals hidden in the features are further diluted, which makes it even harder for the PCA to pick up the associations with the clinical outcomes. This explains why fewer gene pathways are identified in the non-linear effects analysis.
As shown above, analysis using different sets of representative features may identify different sets of differential pathways. Considering that a pathway can be represented with different representative features, identification of differential pathways amounts to a ‘two-dimensional’ selection: for a specific pathway, selection of the representative features then can lead to the smallest P-value; and selection of differential pathways using those smallest P-values. Intuitively, this can be realized with a two-step procedure, one across different representative features for each pathway and one across multiple pathways, and two-step FDR control. In this article, we consider a one-step selection by pooling and analyzing P-values across multiple representative features for each pathway and across multiple pathways. Given that the total number of multiple comparisons to be accounted for remains the same with the two-step or one-step procedures, they should generate the same results. With the following analysis, we can determine not only which pathways are differential, but also which representative features reflect the differentiation. If only differential pathways are of interest, our theoretical investigation suggests that the following analysis may lead to slightly inflated false positive rates.
Analysis III: for each pathway, analysis using linear effects generates c* sets of representative features, and hence c* P-values. Non-linear analysis I also generates c* P-values for each pathway. In this set of analysis, for the M pathways, we consider all the (c*+c*)×M P-values together. Those P-values correspond to the significance measurements for the M pathways using (c*+c*) sets of representative features for each pathway. We use the FDR approach to identify differential pathways. Such an analysis has the advantage of revealing not only differential pathways, but also corresponding representative features that best represent effects of the pathways. We note that, with the FDR approach described in Section 3.2, an arbitrary covariance structure of the P-values is allowed. Given that each P-value is generated separately via permutations, and hence is consistent, identification of differential gene pathways in analysis III using FDR is valid. In this set of analysis, the natural killer cell mediated cytotoxicity pathway (60 genes) is identified as differential with representative features (R2) and #PC=5. This pathway is missed by using linear effects only.
Of note, one potential drawback is that by considering multiple representative features for each pathway and adjusting for more multiple comparisons, we may have less power to identify differential pathways.
Analysis IV: following a similar strategy as in analysis III, we consider the representative features generated using linear effects and using (R3). We combine and analyze the (c*+d*)×M P-values generated in Section 4.1.1 and analysis II. We identify the cell-cycle pathway (81 genes) and the aminoacyl-tRNA biosynthesis pathway (11 genes) as differential with representative features (R3) and #PC=3. These two pathways are not identified using linear effects only.
We note that, it is possible to follow analyses III and IV and consider all the (c*+c*+d*) sets of representative features for each pathway. However, we note that (R2) and (R3) are two different ways of accounting for non-linear effects. In addition, by considering more representative features, we may lose more power. Thus, analysis with all the (c*+c*+d*) sets of representative features is not pursued.
4.2 MCL data
Rosenwald et al. (2003) reported a study using microarray expression analysis in mantle cell lymphoma (MCL). Among 101 untreated patients with no history of lymphoma, 92 were classified as having MCL. Survival times of 64 patients were available and 28 patients were censored. The median survival time was 2.8 years (range 0.02–14.05 years). Lymphochip DNA microarrays were used to quantify mRNA expression in the lymphoma samples from the 92 patients. Gene expression data that contains expression values of 8810 cDNA elements is available at http://llmpp.nih.gov/MCL. Among the 8810 genes, 2011 belong to 176 known KEGG pathways. The pathways have sizes ranging from 1 to 259, with median size 14. Out of the 176 pathways, 134 have at least five genes.
4.2.1 Pathway identification using linear gene effects
We first consider linear effects only and generate representative features with (R1). With FDR=0.2 and the number of PCs equal to 1… 5, the numbers of identified differential pathways are 36, 43, 40, 47 and 50, respectively. Using five PCs identifies the largest number of differential pathways. In the Supplementary Material, we provide detailed information on pathways identified with #PC=5.
Analysis of sensitivity to noises: we conduct the same sensitivity analysis as in Section 4.1.1. With 10%, 20% and 30% additional noises, the proposed approach identifies 55, 50 and 57 differential pathways. The numbers of overlaps with pathways identified without noises are 43, 48 and 44, respectively, which again suggests relative insensitivity to noises of the proposed approach.
Comparisons with alternative approaches: we consider comparisons with the following alternative approaches and show the results in Table 1: (i) the GSEA, which identifies 69 pathways as differential. Thirty-one pathways are identified by both the proposed approach and the GSEA; (ii) the maxmean approach, which identifies two pathways as differential, both of which are identified by the proposed approach; and (c) the global test, which identifies 68 differential pathways, 46 of which are also identified by the proposed approach.
4.2.2 Pathway identification using both linear and non-linear gene effects
We conduct the same analysis of non-linear gene effects as in Section 4.1.2.
Analysis I: with FDR=0.2, no gene pathway is identified as differential.
Analysis II: the numbers of differential pathways identified are 2, 3 and 5, respectively. The glycine, serine and threonine metabolism pathway, composed of 12 genes, is identified with # PC=3. This pathway is not identified using only linear effects. This pathway contains gene ALAS1, which has been identified as a lymphoma susceptibility gene in animal models (Shin et al., 2004). Genes GCAT and GLDC have been identified as susceptibility genes for cancers in general, and can be potentially linked with lymphoma progression.
Analysis III: no new gene pathway is identified beyond analysis using linear effects. This is caused by the large P-values obtained in analysis I.
Analysis IV: in this set of analysis, the Notch signaling pathway (40 genes), the metabolism of xenobiotics by cytochrome P450 pathway (30 genes), the galactose metabolism pathway (7 genes), and the glycine, serine and threonine metabolism pathway (12 genes), are identified as differential beyond analysis using linear effects. The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Notch signaling is dysregulated in many cancers, and faulty Notch signaling has been implicated in many diseases including T-cell acute lymphoblastic leukemia, cerebral autosomal dominant arteriopathy with sub-cortical infarcts and leukoencephalopathy, multiple sclerosis, tetralogy of fallot, alagille syndrome and myriad other disease states. The reactions in xenobiotics metabolism pathways are of particular interest in medicine as part of drug metabolism and as a factor contributing to multidrug resistance in infectious diseases and cancer chemotherapy. Induction of some P450s is a risk factor in several cancers since these enzymes can convert procarcinogens to carcinogens. P450 enzymes play a major role in drug interactions.
4.3 Leukemia data
The leukemia data contains gene expressions of two types of acute leukemia: acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) (Golub et al., 1999). Expression levels of 6817 genes were measured using Affymetrix oligonucleotide arrays. The data consists of 47 cases of ALL and 25 cases of AML, and is available at http://www.genome.wi.mit.edu/MPR. Among the 6817 genes, 1565 belong to 193 KEGG pathways. Pathway sizes range from 1 to 134, with median size 13. Out of the 193 pathways, 146 have sizes at least five.
4.3.1 Pathway identification using linear gene effects
With (R1) and number of PCs equal to 1… 5, 3, 9, 24, 42 and 50 pathways are identified as differential. Using five PCs identifies the most differential pathways. Detailed information on identified pathways is in the Supplementary Material.
Analysis of sensitivity to noises: with 10%, 20% and 30% additional noises, the proposed approach identifies 41, 37 and 41 differential pathways, respectively. There are 39, 36 and 39 pathways identified both with and without noises, respectively.
Comparisons with alternative approaches: we also analyze the Leukemia data using the following alternative approaches and show the results in Table 1: (i) The GSEA identifies one differential pathway, which is also identified with the proposed approach; (ii) the maxmean approach identifies two pathways, which are identified with the proposed approach; (iii) the global test, which identifies no pathway as differential; and (iv) the approach in Kong et al. (2006), which employs the PCA and Hotelling's test to define significance. With number of PCs equal to 1…5, the same two pathways are identified, both of which are identified with the proposed approach.
4.3.2 Pathway identification using both linear and non-linear effects
We now consider the non-linear effects.
Analysis I: with (R2) and number of PCs equal to 1… 5, 2, 0, 0, 0, 0 pathways are identified as differential. The tyrosine metabolism pathway is identified as differential beyond analysis using linear effects. Significance of the tyrosine metabolism in leukemia has been recognized for a long time (Ivanova and Kaverzneva, 1971).
Analysis II: in this set of analysis, the number of identified differential pathways are 0, 0 and 2, respectively, with no new pathway identified beyond analysis using linear effects only.
Analysis III: The tyrosine metabolism pathway is identified as differential beyond analysis using linear effects.
Analysis IV: no new gene pathway is identified beyond analysis using linear effects.
5 DISCUSSION
5.1 Using PCA in pathway identification
PCA has been considered for identification of differential pathways. In this study, we advance from Kong et al. (2006), Chen et al. (2008) and other PCA studies by considering possible contributions from PCs other than the first one, and/or contributions from non-linear effects. Analysis of three datasets suggests that, with the proposed approach, we are able to identify a reasonable number of differential pathways. For identified pathways, we can conclude that genes in those pathways are significantly associated with the clinical outcomes. In addition, such associations can be attributed to the linear combinations of genes and/or their transformations that explain relatively larger amount of variations. One possible drawback of the proposed approach is that PCs are used, which are linear combinations of all genes in pathways. Thus, biological interpretation of the identification results can be less lucid: we are able to conclude significance of pathways; however, conclusions on individual genes within pathways can only be based on their loadings in the PCs and are difficult to draw.
5.2 How many PCs will be needed?
In several previous studies, ad hoc arguments have suggested that the first one or two PCs may satisfactorily capture properties of gene expressions. Our numerical studies in Section 4 suggest that more PCs may be needed for identification of differential pathways. In this study, we consider at most five PCs. If more PCs are considered, further restrictions on the size of the pathways will be needed. Of note, in other contexts of gene expression data analysis such as clustering, it has been suggested that PCs beyond the first one or two are needed (Yeung and Ruzzo, 2001).
It is not our intention to suggest that five PCs will be sufficient for all practical data analysis. Rather, we intend to raise the awareness of the extra information brought by PCs beyond the first one or two. In practical data analysis, we suggest that researchers explore different numbers of PCs, and select the proper number based on, for example, biological implications and predictive power of the set of identified differential pathways.
5.3 Comparisons with alternative approaches
Beyond comparisons conducted in Section 4, we have also considered a univariate approach suggested by one reviewer. We first compute the statistic T for each gene. Then within each pathway, the most significant T is selected as the statistic for the significance of the pathway. P-values are then obtained using permutation and the FDR is used for pathway identification. With this univariate approach, we identify 1, 77 and 33 differential pathways for the DLBCL, MCL and Leukemia data, respectively. The numbers of overlaps with pathways identified with the proposed approach using linear effects of genes are 1, 46 and 19, respectively. We note that, although univariate approaches may identify meaningful pathways for certain data, they are not the common practice of pathway analysis.
In Section 4, we conduct comparisons with alternatives including the GSEA, maxmean, global test and the approach in Kong et al. (2006). We are aware that there exist other approaches for identification of differential pathways. However, since they are less extensively adopted, we do not pursue comparisons with them. From Table 1, it is clear that the proposed approach can identify pathways significantly different from using existing approaches, and can provide a valuable alternative. Similarities of identified pathway sets using different approaches vary across different datasets. Such discrepancies have been noted in studies such as Sneddon (2004) and Tintle et al. (2008). We conducted comparisons using real datasets, instead of simulated data, since simulated gene expressions can be considerably different from those observed in practice. Analysis of real data has satisfactorily demonstrated the main properties of the proposed approach. Thus, we defer simulations, which can provide additional insights beyond real data analysis, to future studies.
With the DLBCL and Leukemia data, alternative approaches identify very small number of pathways. This is partly caused by the relatively conservative FDR control. If we ignore the possible correlations among P-values, then alternative approaches can identify many more pathways and generate results more similar to those using the proposed approach.
We note that it is possible to modify alternative approaches to make them more comparable with the proposed one. For example, as pointed out by one reviewer, instead of using gene expressions, it is possible to use the PCs in the global test. However, such modifications have not been seriously investigated in the literature and will not be further pursued here.
5.4 Non-linear effects in pathway analysis
In this study, we focus on gene pathways. The pathway sizes ranges from less than 10 to a few hundreds, which is considerably smaller than the total number of genes in a typical microarray study. The relatively smaller sizes of the pathways make it possible to consider second-order terms. In particular, most commercial software have efficient functions to compute singular value decomposition and hence the PCs. Thus, the computational cost with the proposed analysis of non-linear effects is quite affordable.
Our numerical studies suggest that using second-order terms may introduce a small number of differential pathways that are missed by using the first-order terms only. These pathways can have important pathological implications and suggest important new directions for further biomedical research.
We propose two possible ways of introducing the second-order terms. We expect their relative performance to be dependent on the underlying data and models. In practice, we suggest that researchers carefully consider both possibilities. We note that there are many other ways of introducing non-linear effects. The proposed approach is simply one of many possibilities. More refined comparison of different ways of defining non-linearity is beyond the scope of this article.
6 CONCLUSIONS
In this study, we propose identifying differential gene pathways by assessing significance of their representative features, which are defined as PCs and/or their transformations. Our numerical studies suggest that (i) the proposed approach can effectively identify differential gene pathways; (ii) PCs that explain a small proportion of the variations may bear significant associations with the clinical outcome; and (iii) non-linear effects need to be considered for identifying a small number of key pathways.
In this study, the representative features are selected using an unsupervised method. In recent studies such as Chen et al. (2008), it has been suggested that supervised selection methods may out perform unsupervised methods. The unsupervised method is adopted in this article since it is computationally easy and is still the common practice. Performance of the proposed approach may need to be further investigated and validated using independent studies. Specifically, the sets of identified differential pathways need to be confirmed with independent studies, which are not available at this moment.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the associate editor and three referees for very insightful comments, which have led to significant improvement of this article.
Funding: CA120988 National Cancer Institute (to S.M.) CA075142 (to M.R.K.).
Conflict of Interest: none declared.
REFERENCES
- Ackermann M, Strimmer K. A general modular framework for gene set enrichment analysis. BMC Bioinformatics. 2009;10:47. doi: 10.1186/1471-2105-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, et al. Microarray data analysis: from disarray to consolidation and consensus. Nat. Rev. Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29:1165–1188. [Google Scholar]
- Carrasquillo MM, et al. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat. Genet. 2002;32:237–244. doi: 10.1038/ng998. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. Supervised principal component analysis for gene set enrichment of microarray data with continuous or survival outcomes. Bioinformatics. 2008;24:2474–2481. doi: 10.1093/bioinformatics/btn458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis RK, et al. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–435. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. On testing the significance of sets of genes. Ann. Appl. Stat. 2007;1:107–129. [Google Scholar]
- Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- Goeman JJ, et al. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Golub TR, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Ivanova VD, Kaverzneva MM. Tyrosine metabolism in leukemia. Probl. Gematol. I Pereliv. Krovi. 1971;16:14–20. [PubMed] [Google Scholar]
- Jiang Z, Gentleman R. Extensions to gene set enrichment. Bioinformatics. 2007;23:306–313. doi: 10.1093/bioinformatics/btl599. [DOI] [PubMed] [Google Scholar]
- Johnson RA, Wichern DW. Applied Multivariate Statistical Analysis. Upper Saddle River, NJ: Prentice Hall; 2001. [Google Scholar]
- Knudsen S. Cancer Diagnostics with DNA Microarrays. Hoboken, NJ: Wiley; 2006. [Google Scholar]
- Kong SW, et al. A multivariate approach for integrating genome-wide expression data and biological knowledge. Bioinformatics. 2006;22:2373–2380. doi: 10.1093/bioinformatics/btl401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesk AM. Introduction to Bioinformatics. USA: Oxford University Press; 2002. [Google Scholar]
- McLachlan GJ, et al. Analyzing Microarray Gene Expression Data. Wiley-Interscience. 2004 [Google Scholar]
- Mehta BA, et al. In vitro modulation of natural killer cell activity in non-Hodgkin's lymphoma patients after therapy. Cancer Immunol. Immunother. 1989;28:148–152. doi: 10.1007/BF00199116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller MB. Molecular control of the cell cycle in cancer: biological and clinical aspects. Dan. Med.l Bull. 2003;50:118–138. [PubMed] [Google Scholar]
- Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief. Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- Neilan BA, et al. Natural cell-mediated cytotoxicity in cutaneous T-cell lymphomas. J. Invest. Dermatol. 1983;81:176–178. doi: 10.1111/1523-1747.ep12543616. [DOI] [PubMed] [Google Scholar]
- Nettleton D, et al. Identification of differentially expressed gene categories in microarray studies using nonparametric multivariate analysis. Bioinformatics. 2008;24:192–201. doi: 10.1093/bioinformatics/btm583. [DOI] [PubMed] [Google Scholar]
- Pang H, Zhao H. Building pathway clusters from random forests classification using class votes. BMC Bioinformatics. 2008;9:87. doi: 10.1186/1471-2105-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. NEJM. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- Sharov AA, et al. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- Shi M, Ma S. Identifying subset of genes that have influential impacts on cancer progression: a new approach to analyze cancer microarray data. Funct. Integr. Genomics. 2008;8:361–373. doi: 10.1007/s10142-008-0084-9. [DOI] [PubMed] [Google Scholar]
- Shin MS, et al. High-throughput retroviral tagging for identification of genes involved in initiation and progression of mouse splenic marginal zone lymphomas. Cancer Res. 2004;64:4419–4427. doi: 10.1158/0008-5472.CAN-03-3885. [DOI] [PubMed] [Google Scholar]
- Sneddon M. Pathway analysis. SoCalBSI 2004. 2004 Available at http://instructional1.calstatela.edu/jmomand2/2004/presentations/index.html. [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintle NL, et al. Gene set analyses for interpreting microarray experiments on prokaryotic organisms. BMC Bioinformatics. 2008;9:469. doi: 10.1186/1471-2105-9-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li H. Nonparametric pathway-based regression models for analysis of genomic data. Biostatistics. 2007;8:265–284. doi: 10.1093/biostatistics/kxl007. [DOI] [PubMed] [Google Scholar]
- Wong S. The Practical Bioinformatician. World Scientific Publishing Company. 2004 [Google Scholar]
- Xiong H. Non-linear tests for identifying differentially expressed genes or genetic networks. Bioinformatics. 2006;22:919–923. doi: 10.1093/bioinformatics/btl034. [DOI] [PubMed] [Google Scholar]
- Yeung KY, Ruzzo WL. Principal component analysis for clustering gene expression data. Bioinformatics. 2001;17:763–774. doi: 10.1093/bioinformatics/17.9.763. [DOI] [PubMed] [Google Scholar]
- Zhang H, et al. Gene selection using support vector machines with non-convex penalty. Bioinformatics. 2006;22:88–95. doi: 10.1093/bioinformatics/bti736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.