Abstract
Background
Benign prostatic hyperplasia (BPH) is an age-related enlargement of the prostate, characterized by increased proliferation of stromal and epithelial cells. Despite its prevalence, the etiology of BPH is unknown.
Methods
The Brown Norway rat is a model for age-dependent, lobe-specific hyperplasia of the prostate. Histological analyses of the dorsal and lateral lobes from aged rats reveal focal areas characterized by increased numbers of luminal epithelial cells, whereas the ventral lobe is unaffected. This study examined differential gene expression by lobe and age in the Brown Norway rat prostate. The objective was to identify genes with different levels of expression in the prostate lobes from 4-month (young) and 24-month (old) animals, and to subsequently link changes in gene expression to mechanisms of prostate aging.
Results
The number of age-dependent differentially expressed genes was greatest in the dorsal compared to the ventral and lateral lobes. Minimal redundancy was observed among the differentially expressed genes in the three lobes. Age-related changes in the expression levels of fourteen candidate genes in the dorsal, lateral and ventral lobes were confirmed by quantitative RT-PCR. Genes that exhibited age-related differences in their expression were associated with proliferation, oxidative stress, and prostate cancer progression, including topoisomerase II alpha (Topo2a), aurora kinase B (Aurkb), stathmin 1 (Stmn1), and glutathione S-transferase pi. Immunohistochemistry for Topo2a, Aurkb, and Stmn1 confirmed age-related changes in protein localization in the lateral lobe of young and aged prostates.
Conclusion
These findings provide clues to the molecular events associated with aging in the prostate.
Keywords: prostate, hyperplasia, age, microarray, gene expression
Introduction
Benign prostatic hyperplasia (BPH) is an age-related benign overgrowth of the prostate in men characterized by increased numbers of cells, despite a decrease in circulating testosterone levels. BPH is a major public health concern as it occurs in ∼70% of men by the 7th decade of life, leading to lower urinary tract symptoms and clinical morbidity [1]. The precise etiology of BPH is unknown, however it is postulated that hyperplasia arises from age-associated alterations in epithelial-stromal interactions and the prostatic hormonal milieu [2].
The aging Brown Norway rat serves as a model for age-dependent, lobe-specific hyperplasia of the prostate [3, 4]. Histological analyses in aged rats reveal increased numbers of luminal secretory epithelial cells in the dorsal and lateral lobes of the prostate compared to young rats, whereas cell numbers in the ventral lobe appear unaffected by age. Serum testosterone levels significantly decline with increasing age in rats, similar to what occurs in aging men [5]. Castration of rats causes regression of the prostate characterized by massive apoptosis of epithelial cells in the ventral lobe, whereas apoptosis is not observed during regression of the dorsal and lateral lobes in which age-dependent hyperplasia has been identified [6]. Increased cell survival and expression of androgen receptor and cell cycle regulatory proteins have been reported by our laboratory in the lateral and dorsal lobes of aged Brown Norway rats and thereby are postulated to play important roles in the development of age-dependent and lobe-specific spontaneous epithelial hyperplasia [7-9].
Age-related differences in gene expression have been investigated in other rat models to elucidate the effects of aging on the prostate. Growth factor expression in the dorsal and ventral prostate lobes of aging Sprague-Dawley rats was reported previously [10]. Based upon RT-PCR and immunohistochemical analyses, the mRNA and protein levels of the growth factors, TGFα, TGFβ, EGF, and KGF and their receptors were shown to be invariant with age in these lobes during aging [10]. Microarray analyses of the ventral lobe of Noble rats revealed modest changes in gene expression with age [11]. Among the gene perturbations associated with age were those involved in protein turnover, secretion, cell survival, oxidative stress defenses and extracellular matrix remodeling [11]. The aging ACI/Seg rat is a well-characterized model of spontaneous prostate carcinoma that progresses from microscopic lesions to gross carcinoma [12]. To elucidate the molecular mechanisms underlying prostate cancer progression in this model, Reyes et al. [13] profiled the expression of genes in the dorsolateral lobes of aging rats. They reported age-related alterations in the genes for growth factors and the regulation of energy metabolism concurrent with the initiation and progression of prostate cancer.
It has been speculated that the susceptibility of the specific zones of the human prostate to BPH and prostate cancer may lie in the differential gene expression of the stromal and epithelial cells comprising the zones [14]. The question remains as to whether differences in gene expression between lobes contribute to the pathology observed with age in the dorsal and lateral lobes of the Brown-Norway rat prostate. The current study is an investigation of gene expression by lobe and age in the Brown Norway rat prostate using cDNA microarray analysis. Differentially expressed genes were associated with processes affected by aging including cell proliferation, immune response, oxidative stress, and protein modification. Fourteen genes were confirmed by quantitative RT-PCR to display altered levels of expression with respect to age and lobe. Large numbers of genes were differentially expressed in the dorsal and lateral lobes as a function of age, the same lobes which exhibit susceptibility to hyperplasia and cancer in rodent models [4, 15-19]. Our studies provide detailed gene expression profiles of all three prostate lobes of the Brown-Norway rat, and shed light on the molecular alterations that accompany aging.
Materials and Methods
Animals
Adult male Brown-Norway rats of 4 (young) and 24 (aged) months of age were purchased from Harlan (Indianapolis, IN) by special arrangement with the National Institute on Aging (Bethesda, MD). The animals were maintained in accordance with the NIH Guide for the Care and Use of Laboratory Animals, and animal protocols were approved by the Johns Hopkins University Animal Care and Use Committee. Individual lobes of the prostate were dissected as described previously [4] and were either fixed in paraformaldehyde for processing and embedding or snap frozen in liquid nitrogen for RNA isolation. As positive controls for epithelial cell proliferation in the immunohistochemistry studies, rats were castrated and two weeks later were implanted with testosterone-filled Silastic capsules (young rats, 0.5 cm capsule; old rats, 1.0 cm capsule) for 3 days as previously described [9].
RNA Isolation
Total RNA was isolated from individual prostate lobes of young (n=2) and old (n=2) animals using RNA STAT-60 reagent (Tel-Test, Friendswood, TX) according to the manufacturer’s instruction and further purified by the RNA mini-kit (Qiagen, Valencia, CA). The concentration of RNA was measured by reading the absorbance at 260 nm and the purity and integrity were verified by the 260/280 nm ratio > 1.8 and by denaturing agarose gel electrophoresis.
Microarray Hybridizations
The Affymetrix gene chip platform as described previously [20] was used to determine transcriptional changes in the prostate lobes with respect to age. Ten micrograms of total RNA from each of the samples was used to create the target for the microarray. The biotinylated cytosine and uridine triphosphate labeled cRNA was fragmented, hybridized to RAE230 2.0 arrays (Affymetrix, Santa Clara, CA), and stained in accordance with the manufacturer’s standard protocol. The arrays were stained and washed utilizing the Affymetrix GeneChip Fluidics Station 400 and scanned using a GeneArray Scanner 2500A (Agilent, Palo Alto, CA). The resulting data were viewed and preliminary assessment was made using GCOS software (Affymetrix). All reactions and microarray hybridization procedures were performed in the Laboratory for Biotechnology and Bioanalysis I (LBBI) at Washington State University.
Absolute and Statistical Analysis for Microarrays
Microarray output was examined visually for excessive background noise and physical anomalies. The default GCOS statistical values were used for all analyses. All probe sets on each array were scaled to a mean target signal intensity of 125, with the signal correlating to the amount of transcript in the sample. An absolute analysis using GCOS was performed to assess the relative abundance of the 45,000 represented transcripts based on signal and detection (present, absent, or marginal). Although 15,923 probe sets are represented on these arrays, there is a certain level of redundancy within and between each array, which results in a lower number of unique transcripts represented on the RAE230 2.0 chipset.
The absolute analysis from GCOS was imported into GeneSpring 7.0 software (Silicon Genetics, Redwood City, CA). The age and lobe dependent data (n=2) was normalized within GeneSpring using the default/recommended normalization methods. These included setting of signal values below 0.01 to 0.01, total chip normalization to the 50th percentile, and normalization of each gene to the median. These normalizations allowed for the visualization of data based on relative abundance at any given time point rather than compared to a specific control value.
Data restrictions and analytical tools in GeneSpring were applied to isolate noteworthy and possibly important patterns of gene expression across age and lobes. Transcripts expressed differentially at a statistically significant level were determined using a P-value cutoff of 0.05 and using a Benjamini and Hochberg False discovery rate multiple testing correction. This was applied to all samples (two time points and three lobes) and considered all transcripts represented on the arrays. Subsequently, expression restrictions were applied to the transcripts expressed in a significant manner. These restrictions were designed so that the remaining transcripts met the following requirements in addition to being expressed in a significant manner: 1) each transcript must have a signal value of at least 100 in a minimum of 1 out of 2 age time points and in at least one replicate and 2) the range of the replicates must not exceed 1 (in the normalized scale). The resulting transcripts were screened using Excel (Microsoft, Redmond, WA) for redundant UniGene entries. Transcripts that passed these restrictions were considered for further analysis that included clustering and Venn diagrams.
Real Time RT-PCR
Total RNA was isolated from individual prostate lobes of 4-month-old (n=5) and 24-month-old (n=7) rats using the RNeasy Midi Kit (Qiagen), and treated with DNaseI (Ambion, Austin, TX). For each sample, 1 μg of RNA was then converted to cDNA using the iScript cDNA Synthesis kit (Bio-Rad) and real-time quantitative PCR was performed using SYBR Green Supermix (Qiagen) and the ABI Prism 7700 detection system (Applied Biosystems, Foster City, CA). Intron-spanning primers were designed for selected genes using the Primer 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3.cgi/) and synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences for these genes are shown in Table 1. Alternatively, primer pairs for the following genes were purchased from SuperArray Bioscience (Frederick, MD) and were based upon proprietary nucleotide sequences: aurora kinase B, stathmin 1, prostaglandin D2 synthase, lipoprotein lipase, colony stimulating factor 1 receptor, seminal vesicle protein 4, cystatin related protein 2, prostatic steroid binding protein C2, glutathione S-transferase mu 6, tropomyosin 1, and mesothelin. The real-time PCR reactions were performed in triplicate using the following protocol: denaturation for 90 sec at 95°C, followed by 40 cycles of 10 sec at 95°C, annealing for 20 sec at 55°C, elongation for 10 sec at 72°C, and a final elongation step for 1 min at 72°C. Melt curve analyses were performed after each run to ensure a single product. Standard curves were constructed for each gene using serial 10-fold dilutions of normal 4-month-old ventral prostate cDNA to serve as a template for absolute quantification of gene expression (Applied Biosystems User Bulletin 2). Expression of each target gene was normalized to the housekeeping gene GAPDH. Statistical differences between gene expression in young and aged animals were determined by Student’s t-test (p<0.05).
Table 1.
Primer sequences designed for quantitative RT-PCR analysis
Immunohistochemistry
For immunohistochemistry, prostate lobes from animals of 4 and 24 months of age were dissected and immersion fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight. Tissues were processed and embedded in paraffin blocks for sectioning. Tissue sections (5 μm) were mounted on standard glass slides, deparaffinized in xylene and rehydrated through an ethanol series. For antigen retrieval, slides were boiled in 10 mM citrate buffer (pH 6.0) for 8 min and allowed to cool to room temperature. Nonspecific binding was blocked for 20 min using 1% bovine serum albumin in PBS (PBA). Primary antibodies included anti-mouse ki-67 (clone MM1, NovaCastro Laboratories; 1:200 dilution), topoisomerase II alpha (Cat# 1769; Epigenomics Biotechnology, Lake Placid, NY; 1:200 dilution), aurora kinase B (ab2254; Abcam Laboratories, Inc, Cambridge, MA; 1:200 dilution) and stathmin-1 (ab47468; Abcam; 1:300 dilution). Antigen localization was visualized using the Vectastain Elite ABC Kit (Vector Laboratories, Burlingame, CA). Signal detection was performed using diaminobenzidine as the chromagen (Sigma, St. Louis, MO) for 2 to 5 min. Tissue sections were counterstained with hematoxylin, dehydrated, and mounted.
Results
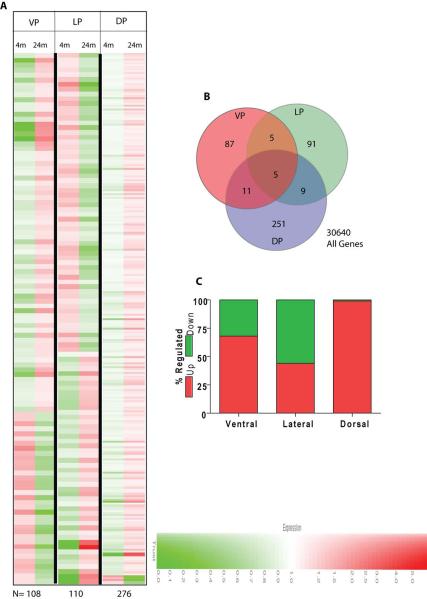
Analysis was performed to determine the age-dependent pattern of gene expression between individual lobes of the prostate (Fig. 1 and Tables 2, 3 and 4). Age dependent differential gene expression (DEG) was determined by comparing genes between 4 and 24 months of age in each lobe that satisfied the pre-determined constraints as indicated in materials and methods. The change in expression pattern with age was visualized through cluster analysis (Fig. 1A). Lobe-specific and age-dependent genes were then determined by partitioning the genes in Venn diagrams (Fig. 1B). As shown in the Venn diagram, minimal redundancy was observed among the genes that were differentially expressed in each lobe as a factor of age. The greatest number of DEGs was observed in the dorsal lobe (276 genes) as compared to the ventral (108 genes) and lateral (110 genes) lobes. Surprisingly, 99% of the DEGs in the dorsal lobe were up-regulated at 24 months as compared to 4 months of age (Fig. 1C). The lateral lobe had approximately equal numbers of genes that were up- and down-regulated between 4 and 24 months of age (Fig. 1C). These are significant observations because they suggest that differential gene expression in each lobe is distinct during aging.
Figure 1.
A: Cluster analysis of differentially expressed genes (DEG) between 4 and 24 months of age. The DEG clusters with ≥ 2-fold change and obeying the pre-defined limits as outlined in Materials and Methods are shown. The genes in red represent increase whereas green represents a decrease in expression. The intensity of the color correlates with fold change (color key shown at the bottom). The total number of DEG are indicated at the bottom (N). B: Lobe specific or common DEG. The gene list generated in A above for each lobe was used. C: The percentage of up- or down-regulated genes in each lobe. Of note is the observation that in the dorsal prostate lobe only 4 (∼1.5%) of genes were down-regulated.
Table 2.
Differentially expressed genes in the dorsal lobe of young and aged rats
| Gene name | Common name | GenBank no. | Fold change |
|---|---|---|---|
| Proteasomal genes, protein modification, proteases, inhibitors | |||
| Protein phosphatase 1B | Ppp1r1b | AA942959 | 3 |
| Proteasome subunit beta 9 (prosome macropain) | Psmb9 | AI599350 | 2.6 |
| Mitogen activated protein kinase 1 | Mapk1 | AI229025 | 2.6 |
| Ubiquitin conjugating enzyme E2N | Ube2n | BI294702 | 1.7 |
| Ariadne ubiquitin-conjugating E2 binding protein homolog 1 | AriH1 | BF555731 | 1.6 |
| Protease (prosome macropain) 28 subunit, beta | Psme2 | NM_01725 | 2.1 |
| Serine peptidase inhibitor, clade A, member 5 | Serpin a5 | NM_02295 | 3.3 |
| Protein kinase inhibitor beta | Pkib | AF413572 | 2.4 |
| Extracellular matrix, cell adhesion, cytoskeleton | |||
| Tropomyosin 3, gamma | Tpm3 | NM_057208 | 2.5 |
| Integrin beta 8 (predicted) | BG 668993 | 4.5 | |
| Coronin, actin binding protein 1A | Coro1a | NM_13043 | 2.4 |
| Laminin beta 2 | Lamb2 | NM_01297 | 2.4 |
| Collagen, type 1, alpha 1 | Col1a1 | Z78279 | -1.5 |
| Claudin 1 | Cldn1 | AI137640 | 2.5 |
| Secretory proteins | |||
| Cystatin related protein 2 (Crp-2) | P22k15 | BI284943 | 17 |
| Seminal Vesicle Secretion-6 | Svs6 | BI284578 | 5.9 |
| Spermine binding protein | Sbp | NM_01302 | 6.3 |
| Seminal vesicle protein 4 | Svp4 | M25590 | 3.7 |
| Prostatic steroid binding protein C2 | Psbp | BI279003 | 2.7 |
| Immune response, inflammation | |||
| Mast cell protease 9 | Mcpt9 | NM_01932 | 7.4 |
| T cell receptor beta chain | Tcrb | AW919577 | 2 |
| Allograft inflammatory factor 1 | Aif1 | NM_01719 | 3 |
| Hematopoietic cell signal transducer | Hcst | BF419129 | 3.8 |
| Oxidative stress, DNA damage | |||
| Glutathione S-transferase pi 1 | Gstp1 | X02904 | 2.5 |
| Hypoxia upregulated 1 | Hyou1 | BI282904 | 1.5 |
| Selenoprotein W , muscle 1 | Sepw1 | NM_013027 | 2.2 |
| XPA binding protein 2 | Xab2 | AF277899 | 3 |
| Transcription factors, DNA binding proteins | |||
| Kruppel-like factor 5 | Klf5 | BF561079 | 2.2 |
| Ring finger protein 1 | Ring1 | BI300772 | 2.6 |
| Transporters | |||
| Solute carrier family 4 (predicted) | Slc4a1ap | BI290779 | 2.8 |
| Metabolism | |||
| Isocitrate dehydrogenase 3 alpha | Idh3a | NM_05363 | 2.8 |
| Cytochrome P450, family 2,subfamily c, polypeptide 70 | Cyp2c70 | M58041 | 7.3 |
| Endocytic transport | |||
| Restin | Rsn | NM_03174 | 2.2 |
| Epsin 1 | Epn1 | NM_05713 | 2.8 |
| Unknown | |||
| Secretoglobin, family 1D , member 2 | Scgb1d2 | BI285057 | 3.1 |
| Thioredoxin domain 4 (predicted) | Txndc4 | AI144967 | 2.9 |
Differentially expressed genes in the dorsal lobe and their classification based on presumed function. Fold changes are expressed as ratios of 24-month:4-month values. Positive fold change indicates up-regulation, whereas negative values indicate down-regulation on a Log2 scale.
Table 3.
Differentially expressed genes in the lateral lobe of young and aged rats
| Gene name | Common name | GenBank no. | Fold change |
|---|---|---|---|
| Proteasomal genes, protein modification, proteases | |||
| Cathepsin S | Ctss | NM_017320 | 2 |
| Extracellular matrix, cell adhesion, cytoskeleton | |||
| Kinesin 5A | Kif5a | BF415701 | 2.8 |
| Secretory proteins | |||
| Prostaglandin D2 synthase | Ptgds | J04488 | 1.6 |
| Carbonic anhydrase 3 | Ca3 | AB030829 | 2.4 |
| Immune reaction, inflammation | |||
| Colony stim ulating factor 1 receptor | Csf1r | BI285793 | 2 |
| Mitosis, cell cycle | |||
| Aurora kinase B | Aurkb | NM_053749 | -2.3 |
| Topoisomerase II alpha | Topo2a | BM385445 | -2.5 |
| Cell division cycle 2 homolog A | Cdc2a | NM_019296 | -3 |
| Cyclin A2 | Ccna2 | AA998516 | -3.1 |
| Stathmin | Stmn1 | NM_017166 | -3.1 |
| Cyclin B1 | Ccnb1 | X64589 | -1.5 |
| similar to Mki67 protein | AI714002 | -3.2 | |
| Synuclein, gamma | Sncg | NM_03168 | 1.6 |
| DNA synthesis, repair | |||
| Ribonucleotide reductase M2 | Rrm2 | BG379338 | -2.6 |
| High mobility group box 2 | Hmgb2 | NM_017187 | -1.6 |
| Metabolism | |||
| Lipoprotein lipase | Lpl | NM_01259 | 2.5 |
| Unknown | |||
| TRAF4 associated factor 1 | BM388478 | -4.2 | |
Differentially expressed genes in thelateral lobeand their classification based on presumed function. Fold changes are expressed as ratios of 24-month:4-month values. Positive fold change indicates up-regulation, whereas negative values indicate down-regulation on a Log2 scale.
Table 4.
Differentially expressed genes in the ventral lobe of young and aged rats
| Gene name | Common name | GenBank no. | Fold change |
|---|---|---|---|
| Proteasomal genes, protein modification, proteases | |||
| Cathepsin S | Ctss | NM_017320 | 2.3 |
| Plasminogen activator, urokinase | Plau | X63434 | 2.6 |
| Extracellular matrix, cell adhesion, cytoskeleton | |||
| Tropomyosin 1 alpha | Tpm1 | BM383411 | 1.3 |
| Collagen, type 1, alpha 1 | Col1a1 | Z78279 | -1.6 |
| Collagen, type III, alpha 1 | Col3a1 | BI275716 | -1.6 |
| Mesothelin | Msln | NM_03165 | 2.5 |
| Secretory proteins | |||
| Kallikrein 6 | Klk6 | BI291725 | -2.9 |
| Mitosis, cell cycle | |||
| Synuclein, gamma | Sncg | NM_03168 | 2.4 |
| Oxidative stress | |||
| Glutathione S-transferase, mu type 6 | Gstmu6 | BF289090 | 6 |
| Glutathione S-transferase, mu type 3 | Gstmu3 | NM_031154 | 2.1 |
| Transcription factors, DNA binding proteins | |||
| NK2 transcription factor related, locus 5 | Nkx2.5 | NM_053651 | -2.2 |
| Apoptosis | |||
| Clusterin | Clu | AF314657 | 3.1 |
| Annexin 1 | Anxa1 | NM_01290 | 2.3 |
| Intracellular signaling | |||
| Cyclin D1 | Ccnd1 | X75207 | -1.8 |
Differentially expressed genes in theventral lobe and their classification based on presumed function. Fold changes are expressed as ratios of 24-month:4-month values. Positive fold change indicates up-regulation, whereas negative values indicate down-regulation on a Log2 scale.
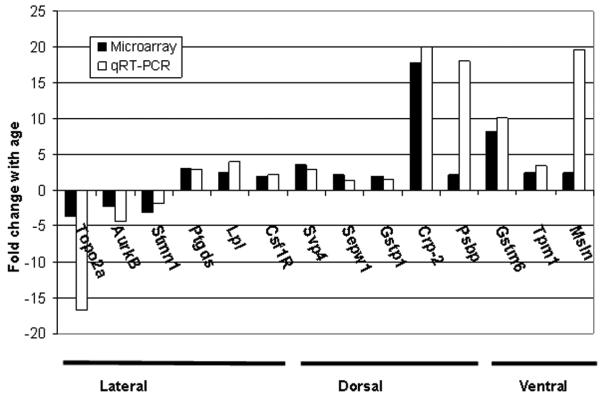
Using selection criteria where age-related gene expression values were increased or decreased by a minimum of 2-fold and the gene name was known (Tables 2-4), the differential expression of 14 genes (Table 5) was successfully verified in the different prostate lobes (Fig. 2) by quantitative RT-PCR analysis using rat-specific primers (Table 1). Specifically, 5 genes in the dorsal lobe were confirmed to have age-related increases in expression. These genes included seminal vesicle protein 4 (Svp4), selenoprotein w, muscle 1 (Sepw1), glutathione S-transferase pi 1 (Gstp1), cystatin related protein 2 (Crp2), and prostatic steroid binding protein C2 (Psbp). For the lateral lobe, 6 genes were confirmed to be differentially expressed with age. Topoisomerase II alpha (Topo2a), aurora kinase B (Aurkb), and stathmin 1 (Stmn1) were found to decrease significantly with age, whereas prostaglandin D2 synthase (Ptgds), lipoprotein lipase (Lpl), and colony stimulating factor 1 receptor (Csf1r) increased in the lateral lobe at 24 months of age (p<0.05). In the ventral lobe, 3 genes were confirmed to be up-regulated with age, including glutathione-S-transferase mu 6 (Gstm6), tropomyosin 1 (Tpm1) and mesothelin (Msln).
Table 5.
Genes from microarrays selected for verification by quantitative RT-PCR
| Gene | Gene name | 24m/4m | |
|---|---|---|---|
| LP | Topo2a | Topoisomerase II alpha | -3.7 |
| Aurkb | Aurora kinase B | -2.3 | |
| Stmn1 | Stathmin 1 | -3.1 | |
| Ptgds | Prostaglandin D2 synthase | 3.0 | |
| Lpl | Lipoprotein Lipase | 2.5 | |
| Csf1r | Colony stimulating factor 1 receptor | 2.0 | |
| DP | Svp4 | Seminal vesicle protein 4 | 3.5 |
| Sepw1 | Selenium protein W 1 | 2.2 | |
| Gstp1 | Glutathione S transferase pi 1 | 2.0 | |
| Crp-2 | Cystatin related protein 2 | 17.7 | |
| Psbp | Prostatic steroid binding protein C2 | 2.2 | |
| VP | Gstm6 | Glutathione S transferase mu 6 | 8.2 |
| Tpm1 | Tropomyosin 1 | 2.5 | |
| Msln | Mesothelin | 2.5 |
LP, lateral prostate lobe; DP, dorsal prostate lobe; VP, ventral prostate lobe. 24m/4m is the ratio of gene expression in each lobe from 24 month and 4 month old rats.
Figure 2.
Real Time RT-PCR analysis of genes by prostate lobe. Solid bars represent the fold difference in expression of 24 versus 4 months of age reported by cDNA microarray. Open bars represent fold difference in expression by qRT-PCR. Increases in expression with age are positive values, whereas decreases with age are negative. Quantitative differences in gene expression determined by qRT-PCR were statistically significant by Student’s t test (p<0.05).
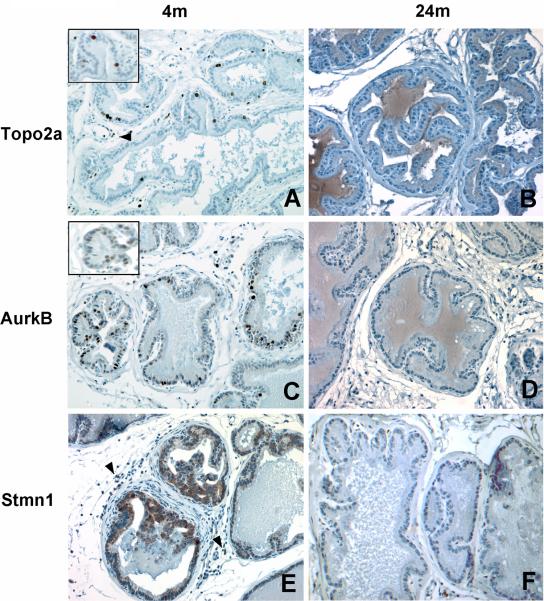
To determine whether age-related differences in protein expression correlated with those predicted by qRT-PCR of mRNA, we examined the expression of Topo2a, Aurkb, and Stmn1 proteins by immunohistochemistry in tissue sections from the lateral lobe. Topo2a and Stmn1 serve as substrates for Aurkb during mitosis [21, 22] and promote chromosome segregation during cell proliferation [23, 24]; activities that may have relevance to prostatic epithelial cell hyperplasia in the lateral lobe of aging rats. In the lateral lobe of 4 month old rats, Topo2a immunostaining was localized to nuclei of widely dispersed luminal epithelial cells as well as some basal epithelial cells and stromal cells (Fig. 3A). By contrast, in the lateral lobe of 24 month old animals, Topo2a immunostaining was weak or absent (Fig. 3B) except in focal areas of epithelial cell hyperplasia, as discussed below. Whereas Topo2a expression was also localized to nuclei of luminal epithelial cells in the ventral lobe of rats at 4 and 24 months of age, the overall frequency of expression in these cells was dramatically lower than that observed in the lateral lobe and no age-related difference was apparent (data not shown). In summary, the numbers of cells expressing Topo2a as detected by immunohistochemistry decreased with age in the lateral prostate; a finding consistent with the quantitative RT-PCR analysis of Topo2a mRNA transcript levels.
Figure 3.
Immunohistochemical staining of Topoisomerase 2 alpha, Aurora kinase B and Stathmin-1 in the lateral prostate lobe of young and aged rats. A, Strong nuclear staining of Topo2a at 4 months of age; B, Absence of Topo2a immunostaining at 24 months of age; C, Nuclear Aurkb immunostaining at 4 months of age; D, Absence of Aurkb immunostaining at 24 months of age; E, Cytoplasmic immunostaining of Stmn1 at 4 months of age; and F, Decreased immunostaining of Stmn1 at 24 months of age. Arrowheads indicate stromal cells with positive immunostaining. Tissues were counterstained with hematoxylin. Magnification in panels A-F, 400X; inset in A and C, 800 X.
Expression of Aurkb in the lateral lobe at 4 months of age was evident in selected luminal epithelial cells of the lateral prostate (Fig. 3C). Aurkb staining was predominantly nuclear and granular in appearance, and in a few luminal epithelial cells, staining was perinuclear. Occasional positively stained basal epithelial cells were also observed. By contrast, positively stained cells were rare in the lateral lobes of aged rats (Fig. 3D). Stmn1 displayed strong cytoplasmic staining in basal cells and occasional luminal epithelial cells in the lateral lobe of young rats (Fig. 3E), whereas a dramatic decrease in staining intensity of luminal epithelial cells was apparent in tissues from 24 month old rats (Fig.3F).
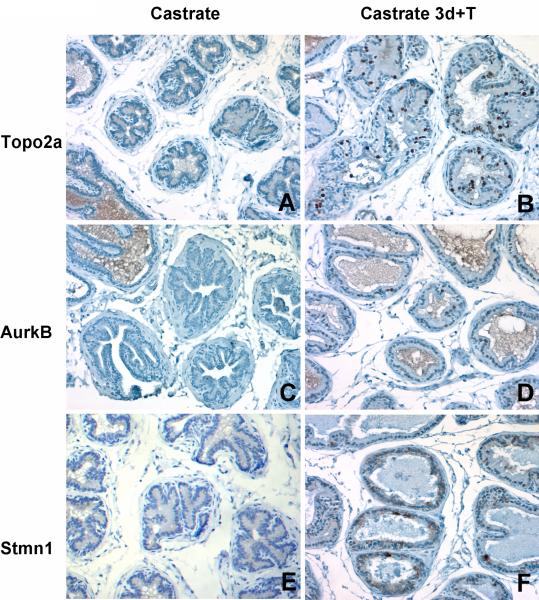
To confirm the specificity of Topo2a immunostaining in proliferating cells, we compared the immunohistochemical staining of Topo2a in prostate tissue sections from 4 month old animals that were castrated and subsequently administered testosterone for 3 days. Topo2a expression was not detected in the lateral lobe from castrated rats (Fig. 4A), whereas Topo2a immunostaining was localized to the nuclei of numerous epithelial cells in response to androgen treatment (Fig. 4B). Similarly, expression of Aurkb was detected in nuclei of many epithelial cells of the lateral lobe of castrated rats following testosterone replacement (Figs. 4C and 4D). Likewise, Stmn1 expression increased in response to androgen treatment of castrated rats, consistent with its role in cell proliferation (Figs. 4E and 4F). These findings indicate that the expression of Topo2a, Aurkb and Stmn1 proteins are coordinately increased in actively proliferating epithelial cells throughout the prostate of castrated rats following androgen replacement.
Figure 4.
Immunohistochemical staining of Topo2a, Aurkb, and Stmn1 in the lateral lobe of 4 month old rats following castration-induced regression and testosterone replacement. The absence of Topo2a (A), Aurkb (C), and Stmn1 (E) immunostaining is seen after castration in contrast to the nuclear immunostaining of Topo2a (B) and Aurkb (D) in luminal epithelial cells and strong cytoplasmic immunostaining of Stmn1 (F) following androgen replacement for 3 days. Magnification, 400X.
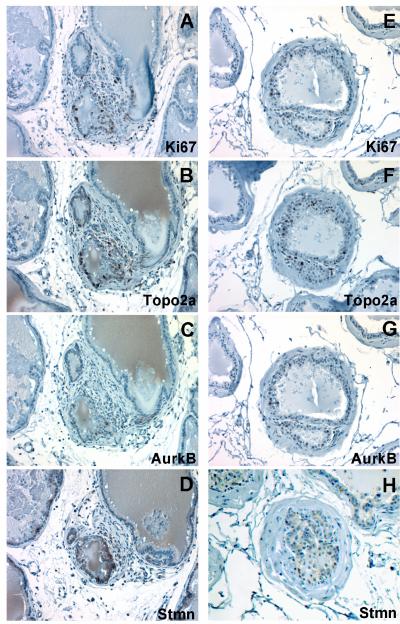
In the lateral and dorsal lobes of aging Brown Norway rats, actively dividing epithelial cells are found only within focal areas with histological appearance of hyperplasia [9]. With this in mind, we specifically looked for the expression of Topo2a, Aurkb and Stmn1 in epithelial cells of hyperplastic foci. Actively dividing epithelial cells were detected within these focal lesions in the lateral lobe of 24 month old rats using Ki67 as a biomarker for cell proliferation (Fig. 5A). In adjoining serial sections of the lateral lobe of aged rats, epithelia cells within the same foci showed preferential strong immunopositive staining for Topo2a (Fig. 5B). Similarly, the expression of Aurkb and Stmn1 was also detected in epithelial cells within hyperplastic foci (Fig. 5C and 5D, respectively). Similarly, epithelial cells within hyperplastic lesions in the dorsal lobe of aged rats were also preferentially positive for Topo2a, Aurkb, and Stmn1 (Fig. 5E-5H). These results reconcile our earlier observations of age-related reductions in Topo2a, Aurkb and Stmn1 mRNA transcript levels in samples of total RNA isolated from the heterogeneous populations of cells within the lateral lobe of aging rats that includes the increased numbers of total cells resulting from hyperplasia. Moreover, histological examination and immunohistochemical staining with Ki67 show that cellular hyperplasia is confined to foci which develop in the dorsal and lateral lobes over the time course of aging and these foci contain a subpopulation of cells that are actively proliferating at any given time point and are coincident with the positive immunostaining for Topo2a, Aurkb and Stmn1.
Figure 5.
Immunohistochemical detection and co-localization of Topo2a, Aurkb, and Stmn1 expression in focal regions of epithelial hyperplasia marked by Ki67 immunopositive cells in the lateral and dorsal lobes of 24 month old rats. Serial sections through a focal lesion in the lateral lobe shows immunostaining for Ki-67 (A), Topo2a (B), Aurkb (C), and Stmn1 (D). Similarly, serial sections through a focal lesion in the dorsal lobe shows immunostaining for Ki-67 (E), Topo2a (F), AurkB (G) and to a lesser extent, Stmn1 (H). Magnification, 400X.
Discussion
Our investigation of gene expression by lobe and age in the Brown Norway rat prostate revealed specific alterations in various cellular processes, including protein modification and degradation, extracellular matrix remodeling, immune response, oxidative stress and cell growth. Based upon microarray analyses, the dorsal lobe displayed the greatest number of differentially expressed genes related to age compared to the ventral and lateral lobes and interestingly, the vast majority (99%) of the differentially expressed genes in the dorsal lobe was up-regulated with age. By contrast, more than 50% of the 110 differentially expressed genes in the lateral lobe were down-regulated. These results may reflect the differences in histological changes observed in the lobes of aging rats. Quantitative RT-PCR analysis of select genes successfully confirmed changes in the expression of 14 genes as a function of age within specific lobes. Immunohistochemistry confirmed that changes in the lobe-specific expression of Topo2a, Aurkb, and Stmn1 genes correlated with changes in protein expression and cellular localization of these proteins in the prostate lobes. Specifically, these proteins were detected by immunohistochemistry in numerous epithelial cells in the young lateral lobe but there was a relative decrease in the total number of cells that express these proteins in the lateral lobe of old rats. However, the localized expression of these proteins in epithelial cells within focal, hyperplastic areas of the luminal epithelium that were also immunopositive for Ki67 in the lateral and dorsal lobes of aged rats suggested a role for Topo2a, Aurkb and Stmn1 in cell proliferation that contributes to the progression of age-dependent hyperplasia. Our observations represent a snapshot in time during the age-dependent development of hyperplasia in the dorsal and lateral lobes of aged rats.
Among the genes of known function and age-related magnitude of differential expression greater than 2-fold in the dorsal lobe were the 36 genes listed in Table 2, all except one of which was up-regulated with increasing age. These genes are associated with a broad range of cellular functions including protein degradation, protein modification, and extracellular matrix remodeling. Differential expression of genes that regulate these biological functions were reported previously in the aging rodent prostate [11, 13]. Alterations in the differentiated function of the prostate were noted by age-related increases in the expression of Psbp, Crp2, and Svp4 genes in the dorsal lobe revealed by microarray analysis and confirmed by quantitative RT-PCR. These genes encode major secretory proteins of the ventral prostate and seminal vesicles [25-27]. Only recently has attention been given to the quantitative analysis of prostatic secretory protein expression across all three lobes [28], and our findings provide further evidence that the expression of androgen-dependent secretory proteins is increased with age in the dorsal lobe of the rat prostate. Given that androgens regulate protein synthesis and secretion, the expression of these genes would have been expected to decline with the age-dependent reduction in serum androgen levels. However, we postulate that the observed increase in expression with age in the dorsal lobe may be the result of an increase in the number of secretory cells in the focal areas of epithelial hyperplasia and increased sensitivity to androgens with age. Microarray analysis also revealed age-related increases in the expression of genes associated with inflammation, such as mast cell protease and allograft inflammatory factor-1 in the dorsal lobe, findings similar to those observed in the aging ACI/Seg rat dorsolateral prostate [13]. These observations suggest an age-dependent, lobe-specific up-regulation of the immune response. Genes involved in oxidative stress protection were increased with age, including Gstp1 and Sepw1. Glutathione-S-transferases protect cells from oxidative stress and aberrant expression of the pi class of enzymes has been implicated in prostate carcinogenesis [29].
For the lateral prostate, genes related to secretion and inflammation, including Ptgds and Csf1r, were up-regulated with age. Csf1, the ligand of Csf1r, controls macrophage production, differentiation and function. Elevated expression of Csf1r, a tyrosine kinase, is observed in urogenital sinus epithelial buds during mouse prostate development and differentiation [30]. Over-expression of Csf1r in human prostate tumors and cancer cell lines suggests that Csf1/Csf1r signaling may play a role in aberrant growth of prostate epithelial cells [30]. Lpl, a gene involved in lipid metabolism and frequently deleted in prostate cancer [31, 32], also increased with age in the lateral lobe. The microarray analysis also revealed significant decreases in the expression of cell cycle regulatory molecules and chromosomal binding proteins with age.
Interestingly, components of the mitotic regulatory network, including Topo2a, Aurkb and Stmn1 were down-regulated with age in the lateral lobe. Topo2a functions in DNA replication, transcription and chromosome condensation and up-regulation of its expression has been reported in tumors, including prostate cancer [33-35]. Topo2a serves as a substrate for Aurkb [20], a member of the Aurora family of serine-threonine kinases that associate with the centromeric regions of chromosomes during cell division. Specifically, Aurkb coordinates the scaffold assembly of multiple proteins at the centromere as cells prepare to divide [36]. This activity requires interaction with the chromosomal passenger proteins, INCENP and survivin [37, 38]. If Aurkb activity is blocked, mitosis is disrupted by improper chromosomal alignment, segregation, and cytokinesis [39, 40]. Aurkb is overexpressed in several human cancers, and its expression correlates with the malignant phenotype of human prostate cancer cell lines [41]. In cells that overexpress Aurkb, aneuploidy and chromosome destabilization is observed [42, 43]. Although we observed an overall decline in Topo2a and Aurkb mRNA levels by microarray analyses of total RNA isolated from the enlarged, hyperplastic lateral lobes of aged rats, Topo2a and Aurkb protein expression was localized to a subset of cells within focal areas of epithelial hyperplasia, thus suggesting that chromosomal instability may accompany the replication of cells within focal lesions of the aging prostate.
A third component of the mitotic regulatory network is Stmn1, a protein capable of sequestering tubulin and thereby facilitating the destabilization of microtubules [23]. Stmn1 is phosphorylated during mitosis and it has recently been identified as a substrate of Aurkb [22]. Although we do not know the phosphorylation status of Stmn1 in the lateral lobe, upregulation and differential phosphorylation of human Stmn1 have been observed in prostate cancer specimens and cell lines [44]. Interestingly, Topo2a, Aurkb, and Stmn1 are co-expressed in proliferating germ cells of the testis [24, 33, 41, 45]. Given their key roles in proper cell division, Stmn1, Aurkb, and Topo2a serve as robust markers of cell proliferation in the Brown Norway rat lateral prostate and if overexpressed could lead to the accumulation of chromosomal abnormalities.
The ventral lobe had the fewest number of differentially expressed genes related to age. Clusterin was among the genes that were up-regulated with increasing age in the ventral lobe, consistent with previous reports for the ventral prostate in aging Noble and Wistar rats [11, 46]. Clusterin was initially reported to be an androgen-repressed gene and its expression increased in the ventral lobe following castration [47-49]. Studies in our laboratory have confirmed the up-regulation of clusterin mRNA and protein levels in the ventral lobe of aged rats [50]. We also observed increased expression of tropomyosin 1 alpha (Tpm1) and mesothelin (Msln), genes associated with the cytoskeleton and cell adhesion, in the ventral lobe of aged rats. Tpm1 belongs to a large family of actin regulatory proteins involved in cytoskeleton remodeling. Their association with actin is modulated by phosphorylation, and in the case of Tpm1, this occurs through the ERK signaling pathway [51, 52]. Diminished expression of Tpm1 has been reported in prostate cancer, and this may result in decreased cell stability and altered morphology [53]. Less is known about the precise function of Msln, a cell surface molecule that is overexpressed in human cancers such as mesothelioma [54]. Msln displays limited expression in normal tissues and its expression in prostate tumors is very low [55]. We also found increased levels of glutathione S-transferase mu type 6 (Gstmu6) mRNA in the ventral lobe of aged rats. The catalytic properties of class mu Gst enzymes are still unknown, however their role to protect cell membranes from lipid peroxidation products has been suggested [56]. Further characterization of Gstmu isoenzyme expression in the prostate is needed, in light of recent findings that polymorphisms in Gstmu1 and Gstmu3 may increase prostate cancer risk [57, 58].
In conclusion, the current study addresses changes in gene expression in the dorsal, lateral, and ventral lobes of the aging Brown Norway rat prostate in an ongoing effort to understand the molecular mechanisms that accompany aging and the naturally-occurring spontaneous development of focal epithelial hyperplasia. Our findings provide evidence for differential expression of genes involved in an array of functions, including protein modification, cell cycle regulation, and oxidative damage protection in an age-dependent and lobe-specific manner. As a gene array study in whole tissue specimens, it is important to note that the expression of these genes is, in some cases, not limited to the epithelium. We can not rule out the possibility that altered expression of genes in the stroma may also play a role in the development of focal epithelial hyperplasia in the dorsal and lateral prostate. It is of significant interest to understand whether changes in the expression of epithelial- and stromal-specific genes can be detected within hyperplastic foci through the use of laser capture microdissection.
Our findings highlight the need to further characterize genetic changes in the aging prostate, and to analyze the role that the protein products of these genes play in the benign (proliferative) and malignant disease processes. The challenge remains to distinguish between genes that contribute to hyperplasia and those whose expression is intrinsic to aging in the prostate. With the widespread use of cDNA microarray analyses, it has become increasingly important to identify specific components of pathways that are perturbed during the development of hyperplasia and disease progression. The identification of these key pathways will provide additional molecular targets for therapeutic applications in benign prostate disease.
Acknowledgements
The authors (JC) wish to thank Ms. Michelle Schmidt in the Laboratory for Biotechnology and Bioanalysis at Washington State University for coordinating the initial analysis of microarray data and Ms. Anne Jedlick in the Johns Hopkins Malaria Institute Gene Array Core Facility for assistance in real-time quantitative PCR.
This work was supported by National Institutes of Health Grant R01-AG020999 (to T.R.B.) C.R.B. was supported by a post-doctoral fellowship from National Institutes of Health Grant T32-CA09110.
References
- 1.Walsh PC. Campbell’s Urology. WB Saunders; Philadelphia: 1986. Benign prostatic hyperplasia; pp. 1248–1265. chap 27. [Google Scholar]
- 2.Oesterling JE. Benign prostatic hyperplasia: a review of its histogenesis and natural history. Prostate Suppl. 1996;6:67–73. [PubMed] [Google Scholar]
- 3.Banerjee PP, Banerjee S, Dorsey R, Zirkin BR, Brown TR. Age- and lobe-specific responses of the brown Norway rat prostate to androgen. Biol Reprod. 1994;51:675–684. doi: 10.1095/biolreprod51.4.675. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee PP, Banerjee S, Lai JM, Strandberg JD, Zirkin BR, Brown TR. Age-dependent and lobe-specific spontaneous hyperplasia in the Brown Norway rat prostate. Biol Reprod. 1998;59:1163–1170. doi: 10.1095/biolreprod59.5.1163. [DOI] [PubMed] [Google Scholar]
- 5.Zirkin BR, Chen H. Regulation of Leydig cell steroidogenic function during aging. Biol Reprod. 2000;63:977–981. doi: 10.1095/biolreprod63.4.977. [DOI] [PubMed] [Google Scholar]
- 6.Banerjee S, Banerjee PP, Brown TR. Castration-induced apoptotic cell death in the Brown Norway rat prostate decreases as a function of age. Endocrinology. 2000;141:821–832. doi: 10.1210/endo.141.2.7339. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee PP, Banerjee S, Brown TR. Increased androgen receptor expression correlates with development of age-dependent, lobe-specific spontaneous hyperplasia of the Brown Norway rat prostate. Endocrinology. 2001;142:4066–4075. doi: 10.1210/endo.142.9.8376. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee PP, Banerjee S, Brown TR. Bcl-2 protein expression correlates with cell survival and androgen independence in rat prostatic lobes. Endocrinology. 2002;143:825–832. doi: 10.1210/endo.143.5.8763. [DOI] [PubMed] [Google Scholar]
- 9.Yan J, Brown TR. Cell proliferation and expression of cell cycle regulatory proteins that control the G1/S transition are age dependent and lobe specific in the Brown Norway rat model of prostatic hyperplasia. Endocrinology. 2008;149:193–207. doi: 10.1210/en.2007-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Patra A, Yeh CC, Tanaka Y, Oh BR, Dahiya R. Effects of aging on growth factor gene and protein expression in the dorsal and ventral lobes of rat prostate. Biochem Biophys Res Commun. 2002;292:482–491. doi: 10.1006/bbrc.2002.6660. [DOI] [PubMed] [Google Scholar]
- 11.Lau KM, Tam NN, Thompson C, Cheng RY, Leung YK, Ho SM. Age-associated changes in histology and gene-expression profile in the rat ventral prostate. Lab Invest. 2003;83:743–757. doi: 10.1097/01.lab.0000069519.06988.24. [DOI] [PubMed] [Google Scholar]
- 12.Isaacs JT. The aging ACI/Seg versus Copenhagen male rat as a model system for the study of prostatic carcinogenesis. Cancer Res. 1984;44:5785–5796. [PubMed] [Google Scholar]
- 13.Reyes I, Reyes N, Iatropoulos M, Mittelman A, Geliebter J. Aging-associated changes in gene expression in the ACI rat prostate: Implications for carcinogenesis. Prostate. 2005;63:169–186. doi: 10.1002/pros.20164. [DOI] [PubMed] [Google Scholar]
- 14.van der Heul-Nieuwenhuijsen L, Hendriksen PJ, van der Kwast TH, Jenster G. Gene expression profiling of the human prostate zones. BJU Int. 2006;98:886–897. doi: 10.1111/j.1464-410X.2006.06427.x. [DOI] [PubMed] [Google Scholar]
- 15.Pollard M. Spontaneous prostate adenocarcinomas in aged germfree Wistar rats. J Natl Cancer Inst. 1973;51:1235–1241. doi: 10.1093/jnci/51.4.1235. [DOI] [PubMed] [Google Scholar]
- 16.Leav I, McNeal JE, Ziar J, Alroy J. The localization of transforming growth factor alpha and epidermal growth factor receptor in stromal and epithelial compartments of developing human prostate and hyperplastic, dysplastic, and carcinomatous lesions. Hum Pathol. 1998;29:668–675. doi: 10.1016/s0046-8177(98)90274-x. [DOI] [PubMed] [Google Scholar]
- 17.Hoover DM, Bendele AM, Hoffman WP, Foxworthy PS, Eacho PI. Effects of chronic treatment with the leukotriene D4 antagonist compound LY171883 on Fischer 344 rats and rhesus monkeys. Fund Appl Toxicol. 1990;14:123–130. doi: 10.1016/0272-0590(90)90238-f. [DOI] [PubMed] [Google Scholar]
- 18.Noble RL. Prostate carcinoma of the Nb rat in relation to hormones. Int Rev Exp Pathol. 1982;23:113–159. [PubMed] [Google Scholar]
- 19.Danielpour D, Kadomatsu K, Anzano MA, Smith JM, Sporn MB. Development and characterization of nontumorigenic and tumorigenic epithelial cell lines from rat dorsal-lateral prostate. Cancer Res. 1994;54:3413–3421. [PubMed] [Google Scholar]
- 20.Asirvatham AJ, Schmidt M, Gao B, Chaudhary J. Androgens regulate the immune/inflammatory response and cell survival pathways in rat ventral prostate epithelial cells. Endocrinology. 2006;147:257–271. doi: 10.1210/en.2005-0942. [DOI] [PubMed] [Google Scholar]
- 21.Morrison C, Henzing AJ, Jensen ON, Osheroff N, Dodson H, Kandels-Lewis SE, Adams RR, Earnshaw WC. Proteomic analysis of human metaphase chromosomes reveals topoisomerase II alpha as an Aurora B substrate. Nucl Acids Res. 2002;30:5318–5327. doi: 10.1093/nar/gkf665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadea BB, Ruderman JV. Aurora B is required for mitotic chromatin-induced phosphorylation of Op18/Stathmin. Proc Natl Acad Sci U S A. 2006;103:4493–4498. doi: 10.1073/pnas.0600702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curmi PA, Gavet O, Charbaut E, Ozon S, Lachkar-Colmerauer S, Manceau V, Siavoshian S, Maucuer A, Sobel A. Stathmin and its phosphoprotein family: general properties, biochemical and functional interaction with tubulin. Cell Struct Funct. 1999;24:345–357. doi: 10.1247/csf.24.345. [DOI] [PubMed] [Google Scholar]
- 24.Bakshi RP, Galande S, Bali P, Dighe R, Muniyappa K. Developmental and hormonal regulation of type II DNA topoisomerase in rat testis. J Mol Endocrinol. 2001;26:193–206. doi: 10.1677/jme.0.0260193. [DOI] [PubMed] [Google Scholar]
- 25.Metafora S, Peluso G, Persico P, Ravagnan G, Esposito C, Porta R. Immunosuppressive and anti-inflammatory properties of a major protein secreted from the epithelium of the rat seminal vesicles. Biochem Pharmacol. 1989;38:121–131. doi: 10.1016/0006-2952(89)90158-5. [DOI] [PubMed] [Google Scholar]
- 26.Vercaeren I, Vanaken H, Van Dorpe J, Verhoeven G, Heyns W. Expression of cystatin-related protein and of the C3-component of prostatic-binding protein during postnatal development in the rat ventral prostate and lacrimal gland. Cell Tissue Res. 1998;292:115–128. doi: 10.1007/s004410051041. [DOI] [PubMed] [Google Scholar]
- 27.Winderickx J, Hemschoote K, De Clercq N, Van Dijck P, Peeters B, Rombauts W, Verhoeven G, Heyns W. Tissue-specific expression and androgen regulation of different genes encoding rat prostatic 22-kilodalton glycoproteins homologous to human and rat cystatin. Mol Endocrinol. 1990;4:657–667. doi: 10.1210/mend-4-4-657. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki T, Fujimoto N, Kitamura S, Ohta S. Quantitative determination of lobe specificity of mRNA expression of androgen-dependent genes in the rat prostate gland. Endocr J. 2007;54:123–132. doi: 10.1507/endocrj.k06-142. [DOI] [PubMed] [Google Scholar]
- 29.Cookson MS, Reuter VE, Linkov I, Fair WR. Glutathione S-transferase PI (GST-pi) class expression by immunohistochemistry in benign and malignant prostate tissue. J Urol. 1997;157:673–676. [PubMed] [Google Scholar]
- 30.Ide H, Seligson DB, Memarzadeh S, Xin L, Horvath S, Dubey P, Flick MB, Kacinski BM, Palotie A, Witte ON. Expression of colony-stimulating factor 1 receptor during prostate development and prostate cancer progression. Proc Natl Acad Sci U S A. 2002;99:14404–14409. doi: 10.1073/pnas.222537099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacGrogan D, Levy A, Bostwick D, Wagner M, Wells D, Bookstein R. Loss of chromosome arm 8p loci in prostate cancer: mapping by quantitative allelic imbalance. Genes Chromosomes Cancer. 1994;10:151–159. doi: 10.1002/gcc.2870100302. [DOI] [PubMed] [Google Scholar]
- 32.Dhanasekaran SM, Dash A, Yu J, Maine IP, Laxman B, Tomlins SA, Creighton CJ, Menon A, Rubin MA, Chinnaiyan AM. Molecular profiling of human prostate tissues: insights into gene expression patterns of prostate development during puberty. FASEB J. 2005;19:243–245. doi: 10.1096/fj.04-2415fje. [DOI] [PubMed] [Google Scholar]
- 33.Cobb J, Reddy RK, Park C, Handel MA. Analysis of expression and function of topoisomerase I and II during meiosis in male mice. Mol Reprod Dev. 1997;46:489–498. doi: 10.1002/(SICI)1098-2795(199704)46:4<489::AID-MRD7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 34.Willman JH, Holden JA. Immunohistochemical staining for DNA topoisomerase II-alpha in benign, premalignant, and malignant lesions of the prostate. Prostate. 2000;42:280–286. doi: 10.1002/(sici)1097-0045(20000301)42:4<280::aid-pros5>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 35.Hughes C, Murphy A, Martin C, Fox E, Ring M, Sheils O, Loftus B, O’Leary J. Topoisomerase II-alpha expression increases with increasing Gleason score and with hormone insensitivity in prostate carcinoma. J Clin Pathol. 2006;59:721–724. doi: 10.1136/jcp.2005.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayama H, Brinkley WR, Sen S. The Aurora kinases: role in cell transformation and tumorigenesis. Cancer Metastasis Rev. 2003;22:451–464. doi: 10.1023/a:1023789416385. [DOI] [PubMed] [Google Scholar]
- 37.Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Delacour-Larose M, Thi MN, Dimitrov S, Molla A. Role of survivin phosphorylation by aurora B in mitosis. Cell Cycle. 2007;6:1878–1885. doi: 10.4161/cc.6.15.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 40.Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chieffi P, Cozzolino L, Kisslinger A, Libertini S, Staibano S, Mansueto G, De Rosa G, Villacci A, Vitale M, Linardopoulos S, Portella G, Tramontano D. Aurora B expression directly correlates with prostate cancer malignancy and influence prostate cell proliferation. Prostate. 2006;66:326–333. doi: 10.1002/pros.20345. [DOI] [PubMed] [Google Scholar]
- 42.Ota T, Suto S, Katayama H, Han ZB, Suzuki F, Maeda M, Tanino M, Terada Y, Tatsuka M. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 2002;62:5168–5177. [PubMed] [Google Scholar]
- 43.Tatsuka M, Katayama H, Ota T, Tanaka T, Odashima S, Suzuki F, Terada Y. Multinuclearity and increased ploidy caused by overexpression of the aurora- and Ipl1-like midbody-associated protein mitotic kinase in human cancer cells. Cancer Res. 1998;58:4811–4816. [PubMed] [Google Scholar]
- 44.Ghosh R, Gu G, Tillman E, Yuan J, Wang Y, Fazli L, Rennie PS, Kasper S. Increased expression and differential phosphorylation of stathmin may promote prostate cancer progression. Prostate. 2007;67:1038–1052. doi: 10.1002/pros.20601. [DOI] [PubMed] [Google Scholar]
- 45.Guillaume E, Evrard B, Com E, Moertz E, Jegou B, Pineau C. Proteome analysis of rat spermatogonia: reinvestigation of stathmin spatio-temporal expression within the testis. Mol Reprod Dev. 2001;60:439–445. doi: 10.1002/mrd.1108. [DOI] [PubMed] [Google Scholar]
- 46.Bettuzzi S, Strocchi P, Marinelli M, Astancolle S, Davalli P, Corti A. Gene relaxation and aging: changes in the abundance of rat ventral prostate SGP-2 (clusterin) and ornithine decarboxylase mRNAs. FEBS Lett. 1994;348:255–258. doi: 10.1016/0014-5793(94)00609-1. [DOI] [PubMed] [Google Scholar]
- 47.Marinelli M, Quaglino D, Bettuzzi S, Strocchi P, Davalli P, Corti A. Increased levels of clusterin mRNA in the ventral prostate of the aging rat are associated to increases in cuboidal (atrophic) cell population and not to changes in apoptotic activity. Biochem Cell Biol. 1994;72:515–521. doi: 10.1139/o94-069. [DOI] [PubMed] [Google Scholar]
- 48.Rosenberg ME, Silkensen J. Clusterin: physiologic and pathophysiologic considerations. Int J Biochem Cell Biol. 1995;27:633–645. doi: 10.1016/1357-2725(95)00027-m. [DOI] [PubMed] [Google Scholar]
- 49.Sensibar JA, Griswold MD, Sylvester SR, Buttyan R, Bardin CW, Cheng CY, Dudek S, Lee C. Prostatic ductal system in rats: regional variation in localization of an androgen-repressed gene product, sulfated glycoprotein-2. Endocrinology. 1991;128:2091–2102. doi: 10.1210/endo-128-4-2091. [DOI] [PubMed] [Google Scholar]
- 50.Omwancha J, Anway MD, Brown TR. Differential age-associated regulation of clusterin expression in prostate lobes of Brown Norway rats. Prostate. doi: 10.1002/pros.20866. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sano K, Maeda K, Oda T, Maeda Y. The effect of single residue substitutions of serine-283 on the strength of head-to-tail interaction and actin binding properties of rabbit skeletal muscle alpha-tropomyosin. J Biochem. 2000;127:1095–1102. doi: 10.1093/oxfordjournals.jbchem.a022703. [DOI] [PubMed] [Google Scholar]
- 52.Houle F, Rousseau S, Morrice N, Luc M, Mongrain S, Turner CE, Tanaka S, Moreau P, Huot J. Extracellular signal-regulated kinase mediates phosphorylation of tropomyosin-1 to promote cytoskeleton remodeling in response to oxidative stress: impact on membrane blebbing. Mol Biol Cell. 2003;14:1418–1432. doi: 10.1091/mbc.E02-04-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alaiya AA, Oppermann M, Langridge J, Roblick U, Egevad L, Brindstedt S, Hellstrom M, Linder S, Bergman T, Jornvall H, Auer G. Identification of proteins in human prostate tumor material by two-dimensional gel electrophoresis and mass spectrometry. Cell Mol Life Sci. 2001;58:307–311. doi: 10.1007/PL00000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hassan R, Bera T, Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res. 2004;10:3937–3942. doi: 10.1158/1078-0432.CCR-03-0801. [DOI] [PubMed] [Google Scholar]
- 55.Frierson HF, Jr, Moskaluk CA, Powell SM, Zhang H, Cerilli LA, Stoler MH, Cathro H, ampton GM. Large-scale molecular and tissue microarray analysis of mesothelin expression in common human carcinomas. Hum Pathol. 2003;34:605–609. doi: 10.1016/s0046-8177(03)00177-1. [DOI] [PubMed] [Google Scholar]
- 56.Rao AV, Shaha C. Multiple glutathione S-transferase isoforms are present on male germ cell plasma membrane. FEBS Lett. 2001;507:174–180. doi: 10.1016/s0014-5793(01)02958-1. [DOI] [PubMed] [Google Scholar]
- 57.Agalliu I, Langeberg WJ, Lampe JW, Salinas CA, Stanford JL. Glutathione S-transferase M1, T1, and P1 polymorphisms and prostate cancer risk in middle-aged men. Prostate. 2006;66:146–156. doi: 10.1002/pros.20305. [DOI] [PubMed] [Google Scholar]
- 58.Medeiros R, Vasconcelos A, Costa S, Pinto D, Ferreira P, Lobo F, Morais A, Oliveira J, Lopes C. Metabolic susceptibility genes and prostate cancer risk in a southern European population: the role of glutathione S-transferases GSTM1, GSTM3, and GSTT1 genetic polymorphisms. Prostate. 2004;58:414–420. doi: 10.1002/pros.10348. [DOI] [PubMed] [Google Scholar]