Abstract
Macroautophagy is a dynamic process involving the rearrangement of subcellular membranes to sequester cytoplasm and organelles for delivery to the lysosome or vacuole where the sequestered cargo is degraded and recycled. This process takes place in all eukaryotic cells. It is highly regulated through the action of various kinases, phosphatases, and guanosine triphosphatases (GTPases). The core protein machinery that is necessary to drive formation and consumption of intermediates in the macroautophagy pathway includes a ubiquitin-like protein conjugation system and a protein complex that directs membrane docking and fusion at the lysosome or vacuole. Macroautophagy plays an important role in developmental processes, human disease, and cellular response to nutrient deprivation.
Normal cell growth and development requires a well-controlled balance between protein synthesis and organelle biogenesis versus protein degradation and organelle turnover. The major pathways for degradation of cellular constituents are autophagy and cytosolic turnover by the proteasome. These degradative pathways are particularly important during development and under certain environmental stress conditions. For example, cellular death and resorption are critical during processes that involve extensive cellular remodeling such as insect metamorphosis, postpartum luteal cell regression, differentiation, and aging (1, 2), as well as in preventing various disease states including cardiomyopathy and some types of cancer. In some cases, degradation and turnover of the entire cell occurs as part of programmed cell death. At other times, turnover occurs on a subcellular scale; under starvation conditions, cells need to scavenge nonessential proteins and organelles and to recycle the components for reuse in the cytosol.
In eukaryotic cells, the lysosome or vacuole is a major degradative organelle. This compartment contains a range of hydrolases that are able to degrade essentially any subcellular constituent ( proteins, lipids, nucleic acids, and carbohydrates). In addition, regulated turnover of organelles is confined to the lysosome. Cytoplasmic components are degraded within the lysosome by microautophagy, chaperone-mediated autophagy, and macroautophagy (3–5). In mammalian cells, microautophagy has not been well characterized, and chaperone-mediated autophagy is a secondary response that temporally follows macroautophagy. In this review, we will limit our discussion to macroautophagy, the major inducible pathway for general turnover of cytoplasmic components. The process of macroautophagy is seen in all nucleate cell types that have been analyzed. The morphology of the process is essentially the same in yeast, plants, and animal cells. In addition, the recent identification of genes encoding components of the autophagic machinery in several organisms indicates that the autophagic process is highly conserved.
Morphology, Biochemistry, and Genetics
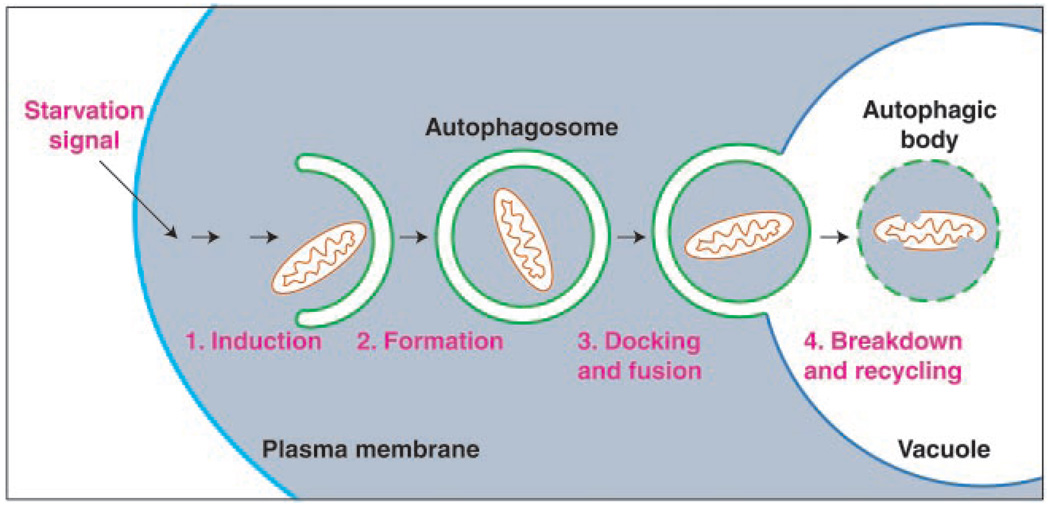
Macroautophagy is a dynamic process in which subcellular membranes undergo dramatic morphological changes [for reviews, see (5, 6)]. In this process, portions of cytoplasm are sequestered within double-membrane vesicles known as autophagic vacuoles in mammalian cells or autophagosomes in yeast (Fig. 1). For simplicity, we will use the term autophagosome. Macroautophagy has been extensively characterized morphologically in both yeast and mammalian systems. Genetic screens have been used in yeast to identify the genes that are involved in the autophagic process (Table 1). Screens for yeast mutants that were starvation-sensitive or defective in the degradation of specific cytosolic proteins (7, 8) produced apg and aut mutants, which overlap with mutants in the cytoplasm to vacuole targeting (Cvt) pathway, that were isolated because of defects in proteolytic processing of the resident vacuolar hydrolase aminopeptidase I (9–11). The overlap between the apg, aut, and cvt mutants was surprising, because the respective pathways, autophagy and Cvt, operate under distinct conditions. Autophagy is primarily used for degradation and is induced under starvation conditions. In contrast, the Cvt pathway is biosynthetic and operates under nutrient-rich conditions. However, biochemical and morphological studies show that in both pathways, the basic mechanism of cytoplasm-to-vacuole transport involves sequestration by a cytosolic double-membrane vesicle (12, 13). The major distinction between the two pathways appears to be the regulation of the size of the autophagosome. The process of macroautophagy can be broken down into at least four discrete steps: induction, formation of the autophagosome, autophagosome docking and fusion with the lysosome or vacuole, and autophagic body breakdown. The molecular details of these steps have begun to be elucidated.
Fig. 1.
Schematic model of macroautophagy in yeast. A signal transduction event regulated by the Tor kinase leads to the following: (1) The induction of autophagy. (2) Membrane from an unknown source sequesters cytosol and/or organelles (a mitochondrion is depicted) resulting in the formation of a double-membrane vesicle (300 to 900 nm) termed an autophagosome. (3) On completion, the autophagosome docks with the lysosome or vacuole. Fusion of the autophagosome outer membrane with the vacuole releases the inner vesicle into the vacuole lumen. The inner vesicle is termed an autophagic body. (4) Breakdown within the vacuole allows recycling of the degraded autophagic body and its hydrolyzed cargo (amino acids, fatty acids, sugars, and nucleotides). The morphology of macroautophagy in mammalian cells is similar to that shown; however, in mammalian cells autophagy can be induced by environmental cues other than starvation.
Table 1.
Genes required for the autophagy and Cvt pathways.
| Gene | Mammalian homolog |
Characteristics | References |
|---|---|---|---|
| Induction of autophagy and formation of autophagosome Kinase signaling system | |||
| Tor2 | mTor | Rapamycin-sensitive protein kinase | (15) |
| APG1 | ULK1 | SER or THR protein kinase, in complex with Apg13, Apg17, Cvt9 |
(23, 44) |
| APG6 | BECN1 | 63-kD peripheral membrane protein, binds Apg14 |
(62, 71) |
| APG9 | Integral membrane protein | (72) | |
| APG13 | Associates with and activates Apg1 kinase | (20) | |
| APG14 | Peripheral membrane protein, interacts with Apg6 |
(71) | |
| APG17 | Interacts with Apg1 kinase, specific for autophagy |
(20) | |
| CVT9 | Interacts with Apg1 kinase, specific for Cvt pathway |
(20) | |
| VAC8 | Armadillo repeat protein, interacts with Apg13 |
(21, 73) | |
| APG protein conjugation system | |||
| APG5 | hAPG5 | 34-kD protein that forms conjugate with Apg12 |
(29) |
| APG7 | HsGSA7 | E1-like ubiquitin activating enzyme | (30, 31) |
| APG10 | Protein-conjugating enzyme | (32) | |
| APG12 | hAPG12 | Ubiquitin type modifier | (29) |
| APG16 | Coiled-coil protein that binds Apg5 | (33) | |
| Size regulation of the autophagosome | |||
| AUT2 | Cysteine protease processes Aut7 | (74) | |
| AUT7 | GATE-16 MAP LC3 |
Up-regulated by starvation, regulates autophagosome size |
(36, 37, 75–77) |
| Docking and fusion | |||
| VAM3 | Syntaxin 7 | Vacuolar tSNARE | (38, 78, 79) |
| VAM7 | SNAP-25 family member, forms part of SNARE complex |
(39) | |
| VPS genes | Class C Vps complex composed of Vps11, 16, 18, 33, 39, and 41 |
(41, 42) | |
| YPT7 | Rab7 | Rab GTPase | (80) |
| Breakdown | |||
| CVT17 | Lipase homolog | (13, 45) | |
| PRB1 | Vacuolar proteinase | (43) | |
| VMA genes | V-ATPase | Vacuolar-type ATPase acidifies lumen | (81, 82) |
Induction of macroautophagy
Autophagic degradation is both developmentally and nutritionally regulated. Nonspecific autophagy is inhibited under nutrient-rich conditions and is induced by starvation. In mammalian cells, phosphorylation of ribosomal protein S6 strongly correlates with inhibition of macroautophagy (14). The activity of p70S6 kinase is regulated by mTor kinase (15, 16). Inhibition of phosphorylation resulting from inactivation of mTor by treatment with rapamycin induces autophagy even under nutrient-rich conditions; however, the details of the regulatory mechanism are not understood. As in mammalian cells, autophagy can be induced in yeast by rapamycin-dependent inhibition of Tor2 (17). In yeast, Tor2 phosphorylates Tap42 causing it to interact with protein phosphatase 2A (PP2A), resulting in a decrease in PP2A activity (18). Inhibition of Tor2 results in activation of PP2A and the induction of autophagy. In mammalian cells, inhibition of PP2A by okadaic acid has a strong inhibitory effect on autophagy (19). Thus, changes in localization and/or activity of PP2A may be one mechanism that controls autophagy, although other phosphatases are also involved in its regulation (20).
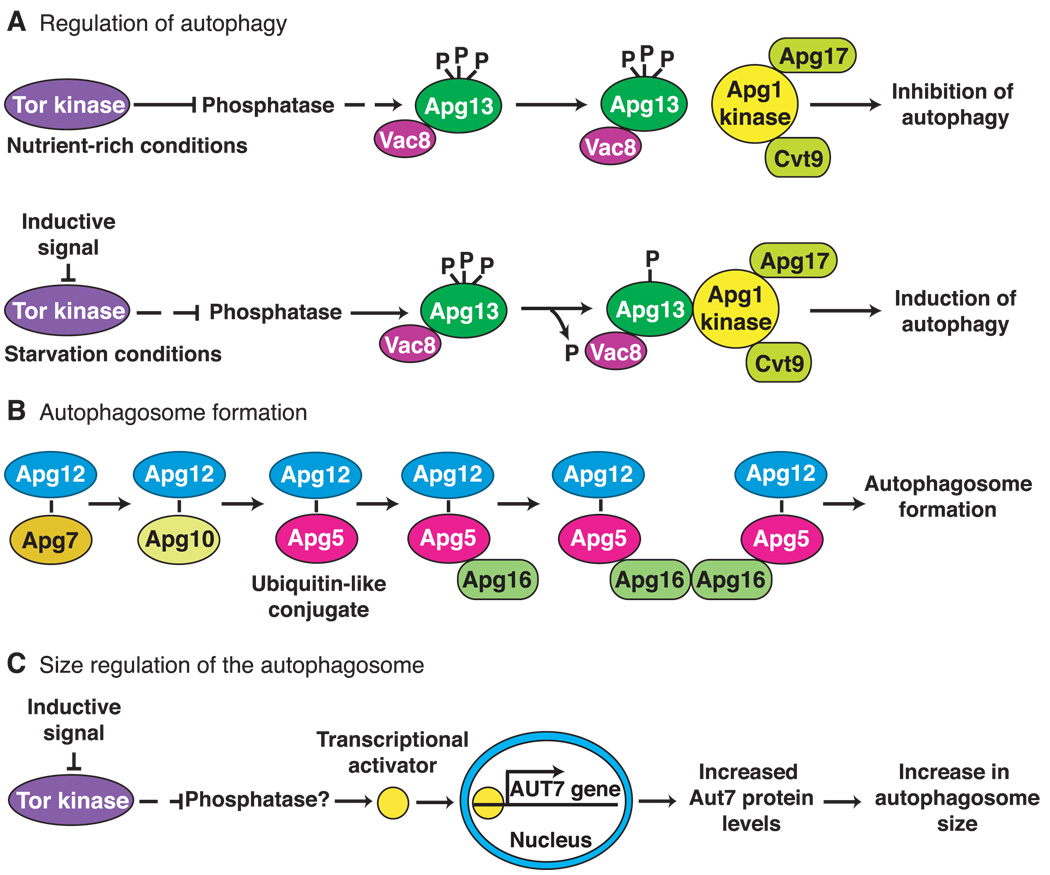
Although Tor2 regulates the expression of many genes, the majority of APG genes are constitutively expressed. In part, this is owing to the overlap between autophagy and the biosynthetic Cvt pathway. Accordingly, it appears that Tor2 may regulate the transition from the Cvt pathway to autophagy during the response to starvation. One of the downstream effectors of the yeast Tor2 kinase may be the Apg13 protein (Fig. 2). Apg13 is part of a dynamic protein complex, which includes the serine and threonine kinase Apg1 (20, 21). Under nutrient-rich conditions, Apg13 is highly phosphorylated and maintains a weak interaction with the Apg1 kinase. Upon inhibition of the Tor2 kinase, and presumably activation of a phosphatase, Apg13 is partially dephosphorylated and associates more tightly with Apg1, stimulating its kinase activity (Fig. 2). Thus, the Apg1 kinase appears to act downstream of Apg13; however, its target or targets have not been identified. The Apg1 kinase interacts with at least two other proteins, Apg17 and Cvt9. These two proteins have specific functions in either the autophagic or Cvt pathways, respectively (20). This finding fits well with the predicted role of the Apg13-Apg1 complex in controlling transition between biosynthetic and degradative pathways. The identification of APG1 homologs in Caenorhabditis elegans (22) and mice (23) suggests that the Apg1 kinase is required for autophagy in higher eukaryotes.
Fig. 2.
Molecular genetics of macroautophagy in yeast. (A) The Tor kinase exerts a negative regulatory effect on autophagy when cells are growing under nutrient-rich conditions. When starvation occurs, the Tor kinase is inactivated, and the negative regulation is relieved resulting in induction of autophagy. Most of the proteins required for autophagy are constitutively expressed and are used for biosynthetic import through the cytoplasm to vacuole targeting pathway under these conditions. The downstream effectors of Tor are likely to include phosphatases and kinases that modulate the phosphorylation state of Apg13. An inductive signal such as carbon or nitrogen starvation inactivates Tor and results in partial dephosphorylation of Apg13. This form of Apg13 associates more tightly with the Apg1 kinase and stimulates its activity. The function of Apg1 kinase is required for autophagosome formation. (B) The Apg7 (E1-like) and Apg10 proteins form thioester intermediates through a COOH-terminal glycine of Apg12. Apg12 is ultimately conjugated to Apg5 through an internal lysine residue in Apg5 in a process that is similar to ubiquitination. Apg16 binds the conjugated Apg5 protein noncovalently and dimerizes to form a complex that is required for formation and completion of the autophagosome. (C) Under nutrient-rich conditions, the Tor kinase negatively regulates the expression of the AUT7 gene resulting in basal levels of Aut7 synthesis. Under these conditions the Cvt pathway is operative and 150-nm Cvt vesicles are formed. Inhibition of Tor by transduction of an environmental signal or after treatment with rapamycin allows the activation of a presumed transcriptional activator protein that increases expression of AUT7. The resulting increase in Aut7 levels allows an expansion in the size of the autophagosome from 150 nm to a range of 300 to 900 nm.
Formation of the autophagosome
After induction of autophagy, a double-membrane vesicle begins to form in the cytosol, resulting in the sequestration of cytoplasmic components (Fig. 1). The origin of the sequestering membrane is not known, but for mammalian cells, it is generally thought to be the endoplasmic reticulum (24). However, an alternative compartment termed the phagophore may represent the donor membrane (25). Sequestration is highly regulated and is under the control of GTPases, phosphatidylinositol kinases, and various phosphatases. Increasing the levels of the class III phosphoinositide 3-kinase product phosphatidylinositol 3-phosphate in human colon cancer cells stimulates macroautophagy (26). In contrast, increases in class I phosphoinositide 3-kinase products, phosphatidylinositol 3,4-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, inhibit autophagy (26). In addition, the guanosine diphosphate (GDP)–bound form of the heterotrimeric G protein subunit Gαi3 is an activating factor for autophagy, whereas the GTP-bound form is inhibitory (27). Regulation by protein phosphatases and kinases is complex and not yet resolved (28).
Molecular genetic studies in yeast have identified some of the components required for autophagosome formation. The sequestration process involves a protein conjugation system (29) (Fig. 2). Apg12 is covalently attached to Apg5 through a COOH-terminal glycine and an internal lysine residue, respectively. This process requires the action of the Apg7 protein, which is homologous to the E1 family of ubiquitin-activating enzymes (30, 31), and Apg10, which functions as a protein-conjugating enzyme (32). A third protein, Apg16, binds to the conjugated Apg5 and dimerizes to link a pair of Apg12-Apg5 conjugates (33). This conjugation event is required for formation or completion of the sequestering vesicle (34). Human homologs of Apg5 and Apg12 have been identified and undergo a similar covalent linkage, indicating that this conjugation system is conserved in mammalian cells (35).
The level of most proteins involved in autophagy does not change during a shift from nutrient-rich to starvation conditions. In contrast, the AUT7 gene is highly up-regulated under starvation conditions or after rapamycin treatment (36, 37) (Fig. 2). Aut7 binds the forming autophagosome and becomes sequestered inside the vesicle along with the cytoplasmic cargo. The localization and upregulation of Aut7 suggests that it functions at least in part as a structural component during autophagosome formation.
Autophagosome docking and fusion
In mammalian cells, delivery to and fusion of the completed autophagosome with the lysosome (Fig. 1) depends on microtubules and maintenance of proper acidification. In yeast, components of the SNARE machinery, including the vacuolar syntaxin homolog Vam3p (38) and the SNAP-25 homolog Vam7p (39), are required for autophagosome docking and/or fusion with the vacuole (Table 1). In addition, the regulatory Rab GTPase, Ypt7 (30), and the class C Vps protein complex (13, 40–42) also function at this step to ensure the accurate and efficient fusion of the autophagosome with the vacuole.
Autophagic body breakdown
Although the lysosome or vacuole is the terminal destination for endocytic and certain biosynthetic trafficking pathways, little is known about what happens to membranes after their delivery to this organelle. After fusion of the autophagosome with the vacuole, the outer membrane of the autophagosome is incorporated into the limiting membrane of the vacuole (Fig. 1). It is not known how this large patch of new membrane avoids degradation by the lipases and other hydrolases present within this organelle. The vacuole membrane itself contains a variety of glycoproteins that are presumed to play a role in protecting this membrane from degradation. Fusion causes the release of the single-membrane bound inner vesicle of the autophagosome, the autophagic body (Fig. 1), into the vacuole lumen. Because of the degradative nature of the lysosome or vacuole, the autophagic body is broken down. Efficient degradation is dependent on proteinase B, lumenal acidification, and the Cvt17 protein, a candidate lipase that may degrade the autophagic body (13, 43–45).
Specific Autophagic Pathways
Biosynthetic transport and organelle turnover
Autophagy is a ubiquitous process that occurs in plant, animal, and fungal cells. Although autophagy is generally considered to be nonselective, the protein components required for autophagic uptake are also used in specific transport events. For example, as noted above, autophagy in yeast overlaps with the biosynthetic Cvt pathway that transports aminopeptidase I (API) to the vacuole. API is synthesized as a precursor in the cytosol and rapidly oligomerizes into a dodecamer (46). The dodecameric precursor API assembles into a larger complex composed of multiple dodecamers, termed a Cvt complex. The Cvt complex is then enwrapped by a double-membrane that forms a cytosolic vesicle. This Cvt vesicle is smaller than an autophagosome and appears to exclude cytosol (12, 13). After fusion with the vacuole, the inner vesicle is degraded in the lumen, allowing release and maturation of precursor API.
In addition to the biosynthetic transport of precursor API, regulated autophagy functions in the selective sorting and turnover of certain cytoplasmic organelles. When methylotrophic yeasts or mammalian cells are grown on methanol or fatty acids, respectively, peroxisomes proliferate to allow maximal use of the available nutrients. Adaptation to preferred carbon sources such as glucose or ethanol result in the specific degradation of peroxisomes through an autophagic mechanism termed pexophagy (5). In some yeasts this degradation has been shown to occur by both micropexophagy, occurring at the vacuole surface, and macropexophagy, where the initial sequestration step takes place away from the vacuole (47). The proteins required for macroautophagy are also required for pexophagy (48, 49).
In general, maintenance of organelles is energetically costly. Organelles like the peroxisome are specifically degraded when they are no longer needed. In mitochondria, certain types of stress can lead to a loss of integrity of the inner and outer membranes resulting in depolarization, a process termed the “mitochondrial permeability transition” (50). It has been proposed that conversion of the mitochondrial permeability transition pore to an open state leads to mitochondrial degradation through macroautophagy and subsequently through apoptosis and necrosis (51). Macroautophagy then can protect cells from oxidative damage that could result from a loss of mitochondrial integrity without triggering apoptosis. However, lysosomal degradation of mitochondria can lead to clinical problems in patients lacking one or more lysosomal hydrolases. The lysosomal accumulation of subunit 9 of the mitochondrial ATPase due to inefficient degradation is associated with neuronal ceroid lipofuscinosis (Batten disease), a neurodegenerative disorder in children and young adults (52). In yeast, mitochondria have been detected within autophagosomes (43), providing additional evidence for mitochondrial degradation by macroautophagy.
Autophagy in development and disease
Macroautophagy plays a critical role in the homeostatic process of recycling proteins and organelles. However, autophagy has also been linked to developmental and pathological conditions. For example, intersegmental muscles are required in the larval and pupal stages of the moth life cycle and during emergence of the adult moth. These muscle cells are not needed in the adult and undergo cell death shortly after emergence. In this case, the characteristics of cell death are largely distinct from those associated with apoptosis and are proposed to occur through autophagy (53). Programmed cell death in developing neurons clearly involves apoptosis (54). However, cellular death in neurons may also involve autophagy (55, 56). Similarly, the loss of Rohon-Beard neurons from the spinal cord of Xenopus tadpoles requires the activity of lysosomal hydrolases and is thought to occur through autophagy-related cell death (57).
There are many lines of evidence that connect macroautophagy to human disease. For example, elevated levels of autophagy are associated with neurodegenerative diseases such as Parkinson’s (58). The mechanism by which neuronal degeneration occurs is not well understood and clearly requires further study to allow the development of effective treatments. In contrast to the situation with Parkinson’s disease, lowered levels of autophagy are problematic in other diseases. A direct link to autophagy and heart disease was discovered by the finding that a deficiency of LAMP-2, a transmembrane protein in the late endosome and lysosome, is associated with the cardiomyopathic Danon’s disease (59), and LAMP-2–deficient mice are defective in the maturation of autophagosomes (60). Similarly, reduced levels of autophagy are seen in some forms of cancer (61). A key role for autophagy in controlling the unregulated cell growth that is associated with tumor development has been suggested through the study of a tumor suppressor gene, beclin 1 (62). Beclin 1 is the human homolog of the yeast autophagy gene APG6. Beclin 1 interacts with the antiapoptotic protein Bcl-2, which prevents the Bax-dependent release of mitochondrial cytochrome c. Decreased levels of the Beclin 1 protein have been correlated with the development or progression of breast tumors. In multicellular organisms, autophagy has also been linked with programmed cell death. Autophagy is specifically associated with type II (nonapoptotic) programmed cell death (53, 63). However, there is an indication that early stages of autophagy may play a role in type I (apoptotic) programmed cell death (64). Along these lines, a human apoptosis-specific protein was shown to be homologous to the APG5 gene product (65). Inhibition of autophagy may be therapeutic in certain neurodegenerative diseases, although its selective activation in specific cells could provide an avenue for cancer treatment.
An interesting connection between apoptosis and autophagy also has been seen in the coordinated regulation of Akt ( protein kinase B) and p70S6 kinase. The activity of p70S6 kinase is controlled through mTor but also through the action of the phosphoinositide-dependent protein kinase–1, PDK1 (66, 67). PDK1 is a multifunctional effector that regulates various kinases (68, 69). Phosphorylation of p70S6 kinase by mTor, or presumably PDK1, prevents autophagy. Class I phosphatidylinositol 3-kinase (PI 3-kinase) activity allows the membrane recruitment of Akt through its pleckstrin homology domain. Phosphorylation by PDK1 activates Akt and inhibits apoptosis (70). Thus, class I PI 3-kinase products may block both apoptosis and autophagy through PDK1. Along these lines, some of the same signals that trigger apoptosis may induce autophagy (55). Once it is activated, autophagy may be able to cause cell death even in the presence of apoptosis inhibitors. These findings support the idea that autophagy represents a second mechanism for programmed cell death. In fact, autophagy-related programmed cell death may have evolved before apoptosis (53).
Conclusions
The capacity for limited turnover of cytoplasmic contents by autophagy or for the programmed death of specific cells plays an important role in development. However, the capacity to degrade cells and cellular contents necessitates strict regulatory safeguards to prevent indiscriminate destruction of essential components. Accordingly, macroautophagy is regulated through the action of kinases (e.g., Tor and Apg1), phosphatases (e.g., PP2A), and GTPases (Gαi3 and Ypt7) that dictate the conditions under which it operates and the targets of the sequestration process.
Autophagy involves dynamic rearrangements of cellular membranes. This is not surprising as this process can result in the engulfment of entire organelles. Some aspects of autophagy are similar to those of other subcellular trafficking events, for example, the targeting, docking, and fusion of the autophagosome requires components such as Rabs and SNARES that are used in both the secretory and endocytic transport systems. However, other components of the autophagy system are specific (see Table 1). The origin of the sequestering membrane is not known, nor is it understood how this membrane can form an enwrapping vesicle in the cytosol. Formation of the autophagosome is topologically distinct from that of a transit vesicle that buds off a preexisting organelle and is likely to require a large number of gene products (Table 1), including those of a novel ubiquitinlike protein conjugation system (Fig. 2). The autophagic machinery is used for a range of processes that are carried out under different conditions and that display varying specificities.
The autophagic machinery is highly conserved in organisms as diverse as plants, animals, and yeast. Recent studies have demonstrated a role for autophagy in both promoting and preventing human diseases. In the last few years, tremendous advances have been made in identifying the molecular components required for macroautophagy through genetic studies in yeast. Combining these analyses with the pharmacological and biochemical information that is available from work in mammalian systems has begun to provide a detailed understanding of the mechanisms that underlie each stage of the autophagic process. Furthermore, workers in both plant and animal systems are exploiting the genomic data now available to identify homologs of the yeast APG genes for directed studies in these multicellular systems. Such studies should allow researchers to address tissue- and development-specific roles for autophagic degradation. Many questions still remain, including the following: What is the mechanism by which cells sense the starvation signal required to induce autophagy? What is the origin of the autophagosomal membrane? How does the autophagic machinery drive the deformation of membranes? How are specific substrates recognized as cargo for sequestration? However, with the progress that has been made in the last few years, it is likely that these and other important problems in the field of autophagy will be solved in the near future.
References and Notes
- 1.Mortimore GE, Miotto G, Venerando R, Kadowaki M. Subcell. Biochem. 1996;27:93. doi: 10.1007/978-1-4615-5833-0_4. [DOI] [PubMed] [Google Scholar]
- 2.Vittorini S, et al. J. Gerontol. A Biol. Sci. Med. Sci. 1999;54:B318. doi: 10.1093/gerona/54.8.b318. [DOI] [PubMed] [Google Scholar]
- 3.Dunn WA., Jr Trends Cell Biol. 1994;4:139. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 4.Cuervo AM, Dice JF. J. Mol. Med. 1998;76:6. doi: 10.1007/s001090050185. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Klionsky DJ. Annu. Rev. Biochem. 2000;69:303. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ, Ohsumi Y. Annu. Rev. Cell. Dev. Biol. 1999;15:1. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Thumm M, et al. FEBS Lett. 1994;349:275. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 8.Tsukada M, Ohsumi Y. FEBS Lett. 1993;333:169. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 9.Harding TM, Morano KA, Scott SV, Klionsky DJ. J. Cell Biol. 1995;131:591. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. J. Biol. Chem. 1996;271:17621. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 11.Scott SV, et al. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12304. [Google Scholar]
- 12.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. J. Cell Biol. 1997;139:1687. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott SV, Baba M, Ohsumi Y, Klionsky DJ. J. Cell Biol. 1997;138:37. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. J. Biol. Chem. 1995;270:2320. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- 15.Thomas G, Hall MN. Curr. Opin. Cell Biol. 1997;9:782. doi: 10.1016/s0955-0674(97)80078-6. [DOI] [PubMed] [Google Scholar]
- 16.Brown EJ. Nature. 1995;377:441. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 17.Noda T, Ohsumi Y. J. Biol. Chem. 1998;273:3963. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 18.Beck T, Hall MN. Nature. 1999;402:689. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 19.Holen I, Gordon PB, Seglen PO. Biochem. J. 1992;284:633. doi: 10.1042/bj2840633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamada Y, et al. J. Cell Biol. 2000;150:1507. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scott S, et al. J. Biol. Chem. 2000;275:25840. doi: 10.1074/jbc.M002813200. [DOI] [PubMed] [Google Scholar]
- 22.Ogura K, et al. Genes Dev. 1994;8:2389. doi: 10.1101/gad.8.20.2389. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, et al. Biochem. Biophys. Res. Commun. 1998;246:222. doi: 10.1006/bbrc.1998.8546. [DOI] [PubMed] [Google Scholar]
- 24.Dunn WA., Jr J. Cell Biol. 1990;110:1923. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strømhaug PE, Berg TO, Fengsrud M, Seglen PO. Biochem. J. 1998;335:217. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. J. Biol. Chem. 2000;275:992. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 27.Ogier-Denis E, Houri J-J, Bauvy C, Codogno P. J. Biol. Chem. 1996;271:28593. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- 28.Codogno P, Ogier-Denis E, Houri J-J. Cell. Signal. 1997;9:125. doi: 10.1016/s0898-6568(96)00130-1. [DOI] [PubMed] [Google Scholar]
- 29.Mizushima N, et al. Nature. 1998;395:395. [Google Scholar]
- 30.Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Mol. Biol. Cell. 1999;10:1337. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanida I, et al. Mol. Biol. Cell. 1999;10:1367. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shintani T, et al. EMBO J. 1999;18:5234. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima N, Noda T, Ohsumi Y. EMBO J. 1999;18:3888. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George MD, et al. Mol. Biol. Cell. 2000;11:969. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. J. Biol. Chem. 1998;273:33889. doi: 10.1074/jbc.273.51.33889. [DOI] [PubMed] [Google Scholar]
- 36.Huang WP, Scott SV, Kim J, Klionsky DJ. J. Biol. Chem. 2000;275:5845. doi: 10.1074/jbc.275.8.5845. [DOI] [PubMed] [Google Scholar]
- 37.Kirisako T, et al. J. Cell Biol. 1999;147:435. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darsow T, Rieder SE, Emr SD. J. Cell Biol. 1997;138:517. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato TK, Darsow T, Emr SD. Mol. Cell. Biol. 1998;18:5308. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rieder SE, Emr SD. Mol. Biol. Cell. 1997;8:2307. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato TK, Rehling P, Peterson MR, Emr SD. Mol. Cell. 2000;6:661. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 42.Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9402. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. J. Cell Biol. 1992;119:301. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Gene. 1997;192:245. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 45.Teter SA, Klionsky DJ. unpublished data. [Google Scholar]
- 46.Kim J, Scott SV, Oda M, Klionsky DJ. J. Cell Biol. 1997;137:609. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuttle DL, Dunn WA., Jr J. Cell Sci. 1995;108:25. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 48.Yuan W, Stromhaug PE, Dunn WA., Jr Mol. Biol. Cell. 1999;10:1353. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutchins MU, Veenhuis M, Klionsky DJ. J. Cell Sci. 1999;112:4079. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 50.Crompton M. Biochem. J. 1999;341:233. [PMC free article] [PubMed] [Google Scholar]
- 51.Lemasters JJ, et al. Biochim. Biophys. Acta. 1998;1366:177. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- 52.Dawson G, Cho S. J. Neurosci. Res. 2000;60:133. doi: 10.1002/(SICI)1097-4547(20000415)60:2<133::AID-JNR1>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz LM, Smith SW, Jones ME, Osborne BA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:980. doi: 10.1073/pnas.90.3.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dudek H, et al. Science. 1997;275:661. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 55.Xue L, Fletcher GC, Tolkovsky AM. Mol. Cell. Neurosci. 1999;14:180. doi: 10.1006/mcne.1999.0780. [DOI] [PubMed] [Google Scholar]
- 56.Isahara K, et al. Neuroscience. 1999;91:233. doi: 10.1016/s0306-4522(98)00566-1. [DOI] [PubMed] [Google Scholar]
- 57.Lamborghini JE. J. Comp. Neurol. 1987;264:47. doi: 10.1002/cne.902640105. [DOI] [PubMed] [Google Scholar]
- 58.Anglade P, et al. Histol. Histopathol. 1997;12:25. [PubMed] [Google Scholar]
- 59.Nishino I, et al. Nature. 2000;406:906. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka Y, et al. Nature. 2000;406:902. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 61.Schulte-Hermann R, et al. Toxicol. Pathol. 1997;25:89. doi: 10.1177/019262339702500117. [DOI] [PubMed] [Google Scholar]
- 62.Liang XH, et al. Nature. 1999;402:672. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 63.Bursch W, et al. J. Cell Sci. 2000;113:1189. doi: 10.1242/jcs.113.7.1189. [DOI] [PubMed] [Google Scholar]
- 64.Jia L, et al. Br. J. Haematol. 1997;98:673. doi: 10.1046/j.1365-2141.1997.2623081.x. [DOI] [PubMed] [Google Scholar]
- 65.Hammond EM, et al. FEBS Lett. 1998;425:391. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- 66.Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. Curr. Biol. 1998;8:69. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 67.Pullen N, et al. Science. 1998;279:707. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 68.Vanhaesebroeck B, Alessi DR. Biochem. J. 2000;346:561. [PMC free article] [PubMed] [Google Scholar]
- 69.Coffer PJ, Jin J, Woodgett JR. Biochem. J. 1998;335:1. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hemmings BA. Science. 1997;275:628. doi: 10.1126/science.275.5300.628. [DOI] [PubMed] [Google Scholar]
- 71.Kametaka S, Okano T, Ohsumi M, Ohsumi Y. J. Biol. Chem. 1998;273:22284. doi: 10.1074/jbc.273.35.22284. [DOI] [PubMed] [Google Scholar]
- 72.Noda T, et al. J. Cell Biol. 2000;148:465. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang YX, Catlett NL, Weisman LS. J. Cell Biol. 1998;140:1063. doi: 10.1083/jcb.140.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kirisako T, et al. J. Cell Biol. 2000;151:263. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. EMBO J. 2000;19:1494. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lang T, et al. EMBO J. 1998;17:3597. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kabeya Y, et al. EMBO J. 2000;19:5720. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wada Y, Nakamura N, Ohsumi Y, Hirata A. J. Cell Sci. 1997;110:1299. doi: 10.1242/jcs.110.11.1299. [DOI] [PubMed] [Google Scholar]
- 79.Mullock BM, et al. Mol. Biol. Cell. 2000;11:3137. doi: 10.1091/mbc.11.9.3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haas A, Scheglmann D, Lazar T, Gallwitz D, Wickner W. EMBO J. 1995;14:5258. doi: 10.1002/j.1460-2075.1995.tb00210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Graham LA, Powell B, Stevens TH. J. Exp. Biol. 2000;203:61. doi: 10.1242/jeb.203.1.61. [DOI] [PubMed] [Google Scholar]
- 82.Nakamura N, Matsuura A, Wada Y, Ohsumi Y. J. Biochem. 1997;121:338. doi: 10.1093/oxfordjournals.jbchem.a021592. [DOI] [PubMed] [Google Scholar]
- 83.The authors would like to thank J. Kim, T. Sato, A. Wurmser, and S. Scott for helpful comments. D.J.K. is supported by Public Health Service grant GM53396 from NIH and S.D.E. is supported by NIH grant CA58689 and is an Investigator with the Howard Hughes Medical Institute.