Abstract
Motivation: Chromatin immunoprecipitation (ChIP) experiments followed by array hybridization, or ChIP-chip, is a powerful approach for identifying transcription factor binding sites (TFBS) and has been widely used. Recently, massively parallel sequencing coupled with ChIP experiments (ChIP-seq) has been increasingly used as an alternative to ChIP-chip, offering cost-effective genome-wide coverage and resolution up to a single base pair. For many well-studied TFs, both ChIP-seq and ChIP-chip experiments have been applied and their data are publicly available. Previous analyses have revealed substantial technology-specific binding signals despite strong correlation between the two sets of results. Therefore, it is of interest to see whether the two data sources can be combined to enhance the detection of TFBS.
Results: In this work, hierarchical hidden Markov model (HHMM) is proposed for combining data from ChIP-seq and ChIP-chip. In HHMM, inference results from individual HMMs in ChIP-seq and ChIP-chip experiments are summarized by a higher level HMM. Simulation studies show the advantage of HHMM when data from both technologies co-exist. Analysis of two well-studied TFs, NRSF and CCCTC-binding factor (CTCF), also suggests that HHMM yields improved TFBS identification in comparison to analyses using individual data sources or a simple merger of the two.
Availability: Source code for the software ChIPmeta is freely available for download at http://www.umich.edu/∼hwchoi/HHMMsoftware.zip, implemented in C and supported on linux.
Contact: ghoshd@psu.edu; qin@umich.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Chromatin immunoprecipitation (ChIP) is a powerful method for isolating a transcription factor (TF) bound to DNA sequences in vivo (Orlando and Paro, 1993; Solomon et al., 1988). In ChIP experiments, cells are first treated with reagents such as formaldehyde inducing protein–DNA crosslinks, and DNA is isolated and fragmented afterwards. An antibody specific to a TF is added to precipitate the interacting pairs, and their crosslinks are reversed. The resulting DNA fragments are direct evidence for physical interactions between the TF and its target genes. These DNA segments can be simultaneously mapped to the genome with array-based hybridization, known as ChIP-chip (Iyer et al., 2001; Ren et al., 2000). ChIP-chip has been widely used for identifying TF binding sites (TFBS). Recently, ChIP experiment coupled with massively parallel sequencing (Bentley et al., 2008), or ChIP-seq, has been proposed as an alternative (Barski et al., 2007; Chen et al., 2008; Johnson et al., 2007; Mikkelsen et al., 2007; Robertson et al., 2007; Schones et al., 2008; Shivaswamy et al., 2008). ChIP-seq offers genome-wide coverage in a single base pair resolution at low cost (Park, 2008).
Although a number of previous studies have demonstrated the power of ChIP-seq, it has also been shown that different mapping strategies may identify mutually exclusive peak regions as candidate binding sites. For instance, Robertson et al. (2007) reported that the overlap between the ChIP-enriched regions identified by ChIP-seq and ChIP-chip is around 60% in signal transducer and activator of transcription protein 1 (STAT1) data. Euskirchen et al. (2007) found that ChIP-chip and ChIP-PET (Loh et al., 2006; Wei et al., 2006), a sequencing-based method, are frequently complementary to each other in identifying validated targets when the signal is not sufficiently strong. The evidence by Robertson et al. (2007) suggests that massively parallel sequencing may not work well for all DNA fragments uniformly. For example, the sequencing can be biased toward certain parts of the genome due to the complex chromatin structure of DNA molecules in their native form. Also, sequence reads may also have reduced sensitivity in the genomic regions where repeat sequences appear frequently. For those DNA fragments, other mapping methods not relying on direct sequencing, e.g. ChIP-chip, can be a valuable source to complement the weakness of the sequencing technology.
For many of the existing ChIP-seq data, ChIP-chip experiments have also been conducted and the data are publicly available. It is desirable to take advantage of existing ChIP-chip datasets to assist TFBS identification using ChIP-seq. While such a joint analysis has a promise, it is a challenging task to account for the heterogeneity of data from the ChIP-chip and ChIP-seq platforms. This is because the two technologies show vastly different behavior in terms of sensitivity and specificity. Specifically, the peaks identified by ChIP-seq are expected to form regions that are much sharper than those in ChIP-chip due to its superior resolution, whereas ChIP-chip tends to report broader regions with moderate significance including potential false positives. Hence, the signals from the two data sources have to be appropriately weighted in order to keep the overall false positive rates low in the joint analysis.
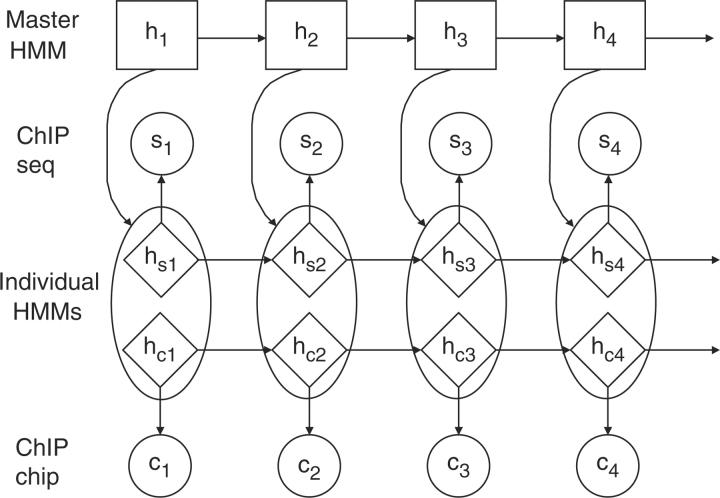
To this end, hierarchical hidden Markov model (HHMM), a collection of multiple individual-level HMMs governed by a population or master-level HMM, is developed in this work. HMMs have been frequently used to analyze ChIP-chip data in the literature (Du et al., 2006; Huber et al., 2006; Humburg et al., 2008; Ji and Wong, 2005; Li et al., 2005; Munch et al., 2006). The structure of the HHMM model is illustrated in Figure 1. In this method, individual-level HMMs function as de-noising filters that convert the raw data into inferred binary hidden states representing ChIP-enrichment and background noise, and the master-level HMM uses individual-level hidden states as a basis to infer the underlying true states. In this process, individual-level HMMs serve as a buffer to reduce the heterogeneity present in raw ChIP-chip and ChIP-seq data, and the master-level HMM summarizes their ChIP-enrichment status to produce the final probability score.
Fig. 1.
HHMM framework with the master process in the top layer and the multiple individual processes in the bottom layer. The hidden states in ChIP-seq and ChIP-chip data are considered as emission from the master process.
Development of HHMMs has been proposed previously in the literature (e.g. Bui et al. 2004; Fine et al. 1998). Recently, Shah et al. (2007) used this class of models for accurately detecting boundary points of copy number changes across multiple samples in genome-wide array-comparative genomic hybridization (aCGH) data. In their model, hidden states in the individual samples exchange mutual feedback with the hidden state in the master level. In contrast, for our problem, each data source is represented as an individual HMM, whose inferred hidden states are then modeled as the bivariate emission probabilities of the master-level HMM.
2 METHODS
2.1 Data
Data generated from ChIP-chip and ChIP-seq experiments are different. ChIP-chip data are fluorescent intensity levels from microarrays reflecting the amount of DNA fragments hybridized to the probes. Probes on tiling arrays are usually 36–50 nt long. Elevated intensity levels from multiple adjacent probes indicate ChIP-enrichment. In contrast, ChIP-seq data are sequencing reads that map to the reference genome. Reads piled up at a tight neighborhood indicate ChIP-enrichment.
Because a HMM framework was adopted, the data are first summarized into fragment counts in units of windows of fixed size (25 nt in this study and adjustable) along the genome. Dissecting chromosomes into windows of equal length has been used previously in the ChIP-seq literature (Mikkelsen et al., 2007). Since the start and end positions of ChIP-chip probes do not exactly overlap with these windows, ChIP-chip probe boundaries were redefined so as to match the ChIP-seq windows (later in the master-level HMM). With typical probes having length >25 nt, one ChIP-chip probe can be mapped to multiple windows. The impact of varying window sizes is further discussed in Section 3.
2.2 HHMM
Let the ChIP-seq count data and the ChIP-chip intensity data be denoted by S=(s1,…, sT) and C=(c1,…, cT), respectively, for a chromosome that has been divided into T windows. We assume that the number of windows is identical in the two data. It is assumed that each data source follows its own independent HMM. Their respective hidden states are denoted by hs=(hs1,…, hsT) and hc=(hc1,…, hcT). As shown in Figure 1, these hidden states are modeled as bivariate random variables in the emission of master HMM, whose hidden states are denoted by h=(h1,…, hT). Both the individual-level states (hs, hc) and the master-level states h consist of either ChIP enriched (denoted 1) or background (denoted 0) states. Note that the ultimate goal of HHMM is to infer the master-level hidden states h.
The model parameters are now specified in the individual level first. The three main components of HMM—initial state distribution, transition probability matrix and emission (Rabiner, 1989)—are defined. The initial state distribution π(hs1) and π(hc1) and the transition probabilities As and Ac can either be fixed or estimated from the data. Each matrix has two rows and two columns, with probability moving from one state in the row to another state in the column. In the latter case, one can assume that each row of As and Ac follows multinomial distribution and estimate the probabilities from the frequency of relevant moves in the inference of hs and hc, respectively. Parametric models are used to describe emission probabilities in the ChIP-enriched states and the background states.
In the following, we briefly describe the two individual-level HMMs proposed to model ChIP-chip and ChIP-seq data, respectively. More details of the two models can be found in the Supplementary Material. We will then explain in details about the master-level HMM which is the main focus of this study. Note that the individual-level HMMs proposed here can be replaced by alternative methods to infer ChIP-enrichment in the individual data sources.
2.2.1 HMM in ChIP-chip
In ChIP-chip data, we use uniform and normal distributions to model the hybridization intensities in the ChIP-enriched states and the background states, respectively. The uniform–normal mixture model has been previously used to model differential gene expression in microarray data analysis (Parmigiani et al., 2002). While the assumption of uniform distribution for signals in the ChIP-enriched states may be an over-simplification due to possible variation in TF binding affinity across the genome, this assumption alleviates the computational loading of HHMM when no prior knowledge is given as to the preferential binding site enrichment. A possible replacement for the distribution in the ChIP-enriched states is another normal distribution (Li et al., 2005).
In the uniform-normal model, Ct|hct=1 ∼ 𝒰θc1 (·) and Ct|hct=0 ∼ 𝒩θc0 (·), where 𝒰 and 𝒩 denote the uniform and normal distributions in the ChIP-enriched states and the background states, respectively. The uniform distribution parameters θc1 are fixed as the minimum and maximum of intensities {Ct}t=1T, and the mean and variance parameters of the normal distribution θc0=(μc, σc2) will be estimated. Bayesian inference for HMM was implemented with conjugate priors and efficient sampling algorithm was presented in (Scott, 2002) (See Supplementary Material Section 1.1 for details).
2.2.2 HMM in ChIP-seq
In ChIP-seq data, overdispersion and higher proportion of zero counts must be accounted for in the model. We assume single sample analysis for now. We use generalized Poisson (GP) distribution and zero-inflated Poisson (ZIP) distribution to model read counts in the ChIP-enriched states and the background states, respectively (Consul, 1989; Johnson et al., 1992). For inference purposes, a latent variable Zt is defined for sequence count st at location t such that Zt=0 if st=0 and st is generated from the point mass at zero, and Zt=1 otherwise (thus, it is always the case Zt=1 if st>0). As in the model for ChIP-chip data, Bayesian inference was implemented (See Supplementary Material Section 1.2 for details).
The same model can be extended to two sample analysis (ChIP-treated sample and untreated control sample). In the paired design, we used a bivariate GP/ZIP distribution to model the read counts between the two types of samples. A HMM is then designed to perform inference on the ChIP-enriched/background states (See the Supplementary Material Section 1.2 for details).
2.2.3 Master HMM
In the master level, the initial state distribution π(h1) and transition probabilities A are defined the same way as in the individual level. For the emission, the data (hs, hc) are modeled with two multinomial distributions, i.e. (hst, hct)|ht=1 ∼ ℳθ1(·) and (hst, hct)|ht=0 ∼ ℳθ0(·), where the distribution for the enriched state ℳ denotes multinomial distribution, and θ1=(p100, p101, p110, p111) and θ0=(p000, p001, p010, p011) are their parameters for the ChIP-enriched states and the background states, respectively. These parameters are given a conjugate Dirichlet prior with parameters (γ100, γ101, γ110, γ111) and (γ000, γ001, γ010, γ011), respectively.
Given the posterior probability pairs (qst, qct) at all positions t=1,…, T estimated in the individual-level HMMs, hidden states in the master level are inferred as follows. Had (hst, hct) been observed directly, the likelihood for the master-level HMM would be
From the inference of individual HMM, {(qst, qct)}t=1T are computed, but the actual hidden states {(hst, hct)}t=1T remain still unknown. Treating this as a missing data problem, the likelihood is integrated over all four possibilities of (hst, hct) based on the marginal weights (qst, qct), i.e.
where gt=(qst)hst (1−qst)(1−hst) (qct)hct (1−qct)(1−hct). This multiplicative factor gt weights the four possible cases of (hst, hct) based on the product of their corresponding marginal posterior probabilities in ChIP-seq and ChIP-chip at position t, as an approximate solution to the missing data problem.
With this likelihood, imputation and posterior sampling steps are iterated as in the ChIP-chip case: (i) Imputation: draw h(i+1) ∼ π(h|hs, hc, θ0, θ1, A) using the forward–backward algorithm, and (ii) Posterior sampling: draw θj(i+1) ∼ π(θj|hs, hc, h(i+1), A) for j=0, 1. With the multinomial likelihood and the Dirichlet prior, the posterior is again Dirichlet distribution, thus θj=(pj00, pj01, pj10, pj11) are drawn from 𝒟(γj00+Hj00, γj01+Hj01, γj10+Hj10, γj11+Hj11) where Hjkl=∑t 1{hst=k, hct=l, ht=j} for k, l=0, 1 and j=0, 1.
Prior was elicited to reflect the known technological difference between ChIP-seq versus ChIP-chip in terms of precision and sensitivity. Since ChIP-seq signals tend to be more sparse but more precise than ChIP-chip signals, elicitation of informative prior that elevates the confidence for ChIP-seq signals more than ChIP-chip signals was preferred for the master-level HMM. In fact, there are ways to conjecture the optimal posterior distribution in real data. For example, if one is aware of the false positive rates in ChIP-seq and ChIP-chip, then the posterior can be set so that the ratio p110/p101 is inversely proportional to the ratio of false positives. One can also learn this knowledge from preliminary motif search in TFBS identified in ChIP-seq and ChIP-chip and reflect the sensitivity ratios in p110/p101 and p111/p110. Through multiple simulations and real data analysis, it was found that the following prior works well: γ111=M/2, γ110=M/5 and γ101=M/10 in the ChIP-enriched windows, and γ0kl=1 in the background windows.
The elicited prior results in the posterior probability ratios 1<r01<r10<r11 where rkl=p1kl/p0kl. This requirement is important since the noise in the ChIP-chip data will substantially increase the number of windows with unique ChIP-chip signals H101 assigned to the ChIP-enriched state, and as a result the posterior probability of ChIP-enrichment for these windows will be overestimated unless informative prior is specified. Admission of ChIP-chip unique signals with higher frequency than ChIP-seq unique signals is likely to result in elevated false positive rates.
2.2.4 Regions with missing data
Due to the limitations in technology and the presence of repetitive regions, neither ChIP-seq nor ChIP-chip is able to completely survey all bases of the human genome. Regions that are inaccessible from both are marked and skipped. There are also regions on the genome that are uniquely accessible by either technology. When data from one source is missing, the inference of the hidden states at the upper level in HHMM will rely on the other data source alone. That is, using the marginal distribution (Bernoulli) of the joint distribution to model the observed (non-missing) data.
3 RESULTS
3.1 Simulation study
A simulation study was conducted in order to assess the performance of HHMM. The posterior probabilities were generated instead of the raw signals, as the focus of this simulation study is the assessment of master-level HMM, where the information from both data sources are combined.
First, the master-level hidden states h in a chromosome containing a 100 000 probes (25 Mb chromosome) were simulated from a stationary Markov chain with a transition probability matrix
Hidden state 1 denotes ChIP-enrichment. ChIP-enriched states have been accepted only when the probes formed a contiguous block, i.e. all ‘singletons’ in the ChIP-enriched state have been converted to the background state. This generates the baseline ‘truth’ where the true ChIP-enriched sites are 150 bp long on average (range from 75 bp to 1375 bp and IQR of 100–250 bp).
Given a value of hidden state ht=1 at each locus t, posterior probabilities P(hst=1|S) and P(hct=1|C) have been generated from Beta distributions with mean 0.9 and 0.8, respectively. In order to reflect higher resolution in ChIP-seq over ChIP-chip, data were generated so that each true ChIP-enriched region is almost exactly covered by a ChIP-seq peak region with ChIP-chip signals surrounding it. Negative signals (ht=0) have been placed as follows. Reflecting the actual false positive rates of <5% in ChIP-seq and 25% in ChIP-chip previously reported in analyses of real datasets, e.g. Robertson et al. (2007), these false positive signals were planted in blocks of 3–8 windows with probability 0.05 and 0.25 in the two datasets, respectively.
Datasets with four possible sampling behaviors (ps, pc) have been simulated. Sampling behavior here refers to the sensitivity of each data source producing signal within the true ChIP-enriched regions. Case I (ps=0.75, pc=0.9) and Case II (ps=0.6, pc=0.8) represent scenarios where ChIP-chip signals appear with a greater frequency (with a greater error rate) than ChIP-seq, which may represent the cases where the sequencing depth is low and hence a number of real ChIP-enriched regions are missed by ChIP-seq. Case III (ps=0.75, pc=0.75) and Case IV (ps=0.9, pc=0.9) represent scenarios where both data sources cover real binding site motif regions with a good sensitivity and some of the platform-specific regions host a good number of real motifs. Other scenarios of a varying range of combinations with the fixed ps/pc ratio have also been simulated, and the results were consistent.
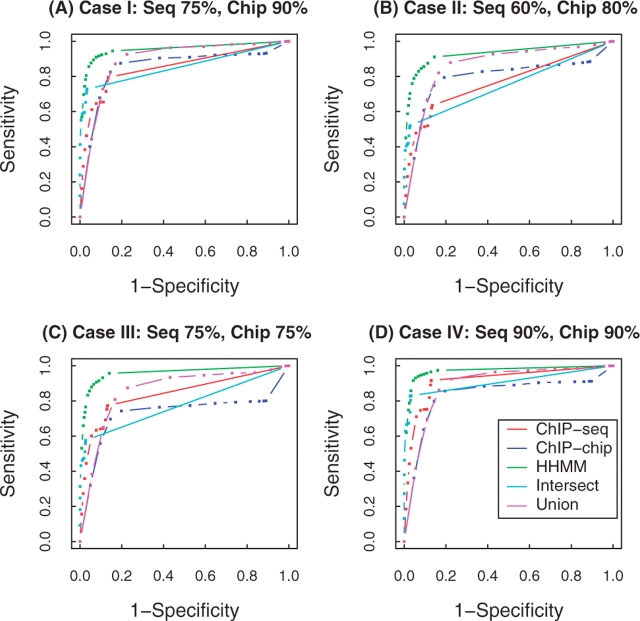
HHMM was compared with four other ways to identify ChIP-enriched regions with high probability: (i) ChIP-seq only: peak regions from ChIP-seq HMM; (ii) ChIP-chip only: peak regions from ChIP-chip HMM; (iii) Intersection: common peak regions in both sources; and (iv) Union: peak regions from either source. Figure 2 shows the receiver operating characteristic (ROC). In all examples, HHMM is the best performing method in terms of sensitivity followed by Union, outperforming both single-source analyses. More importantly, HHMM keeps the specificity higher than Union for nearly all decision points (square dots). The fact that the ROC curve bent to the right significantly for high specificity decision points in the Union indicates that blind picking of all signals would result in high false positive rates at a fixed specificity, mostly due to ChIP-chip data. HHMM removes most of the low-key negative signals, which can be seen in the upper left corner of the ROC curves.
Fig. 2.
Plots of the ROC in the four simulation datasets, comparing ChIP-seq only, ChIP-chip only, HHMM, Intersection and Union. Four different settings of ChIP-seq and ChIP-chip data were generated. Signal present in 75% and 90% (A); 60% and 80% (B);75% and 75% (C); and 90% and 90% (D) of the ChIP-enriched regions detected by ChIP-seq and ChIP-chip, respectively.
In sum, the results in Cases I through IV indicate that the area under the curve of the ROC is the highest in HHMM followed by Union and the number of positive calls is almost always highest in HHMM at fixed specificity. In all scenarios where either the more advanced mapping platform misses some of the true signals or both platforms complement the identification for each other, HHMM has the potential to collect the highest number of binding sites and, at the same time, to keep the false discovery rates lower than blind picking of all signals.
Meanwhile, another dataset was simulated with (ps, pc)=(0.8, 0.6), where the better performing platform ChIP-seq covers most of the signals picked up by ChIP-chip. Examination of ROC curve shows that HHMM, ChIP-seq only and Union methods perform equivalent to one another, indicating that there is no additional benefit earned by HHMM as expected. Also, this is consistent with the fact that the ROC improved the least in Case IV out of the four scenarios, where the number of overlapping signals in ChIP-seq and ChIP-chip is the largest among all.
3.2 Application to NRSF data
3.2.1 Data
HHMM was applied to a real dataset for the TF NRSF (Johnson et al., 2007). In the study, ChIP-seq was used to study genome-wide mapping of binding sites of NRSF, a neuron-restrictive silencer factor known for its negative regulation of many neuronal genes in non-neuronal cells (Schoenherr et al., 1996), were mapped to ∼2000 locations in the human genome using ChIP-seq. ChIP-seq data for the treated and control cell lines were available from the Illumina web site and an unpublished ChIP-chip data was also available in Gene Expression Omnibus (GSE7372). Since the array platform used in the ChIP-chip data (Nimblegen ENCODE tiling arrays) does not cover the whole genome, this section focuses on the 10 ENCODE regions each spanning 5 million bp, i.e. around 1% of the human genome.
High probability signals (0.9 and above) appeared in 26.5 and 272.9 kb in ChIP-seq and ChIP-chip data, respectively, indicating significant differences between the two platforms. Among these, 422 windows were overlapping, which accounts for 37% of ChIP-seq. The posterior probabilities were then combined into a single data as mentioned previously, and master-level HMM was fit.
3.2.2 Motif enrichment
For the peak regions identified by each method, we used two motif search engines to find TFBS. First, we used MatInspector (Cartharius et al., 2005) of Genomatix (http://www.genomatix.de) using the position weight matrix (PWM) reported in Schoenherr et al. (1996) (See Supplementary Material Section 2 for motif logo plot). Since the sequence alignment in MatInspector does not offer confidence assessment, we generated permuted sequences by randomly shuffling the nucleotides within each sequence in the peak regions and the motif search was reiterated, providing a reference for assessing the significance of the hits. The enrichment of TFBS motifs was tested by χ2 test in a contingency table setting. The rows of the 2 × 2 table indicate whether the motif search was done in the original sequence or in the permuted sequence, and the columns indicate whether the sequences contain motifs or not Frith et al. (2004). Second, we applied Clover to construct more realistic background sequences from a specific composition of real sequences (human chromosome 20). We used the same PWM used above for sequence alignment. Clover reports the motifs that are statistically significant matches compared with matches to background sequences at a certain significance threshold (e.g. P = 0.05).
Table 1 shows the result of the analysis using the probability threshold 0.9. The total number of motifs is the highest at 67 in the Union method, but the HHMM picks up 46 motifs while keeping the false positives less than half of the Union, indicating improved control of false positive rates, at the expense of fewer low ranking signals. This is congruent with the Clover analysis shown in the last two columns of Table 1, where the absolute number of motifs is the highest in the Union but the identification rate (per 1 kb) is the highest in HHMM (using the default P-value threshold 0.05).
Table 1.
Motif summary for the five methods in NRSF data
| Method | #Matcha (#Permuteb) | #Peaks | Coverage (kb) | ORc | χ2 | Match rated | #Clovere | Rate in Cloverf |
|---|---|---|---|---|---|---|---|---|
| HHMM | 46 (11) | 424 | 179.2 | 4.56 | 21.74 | 0.19 | 51 | 0.28 |
| Union | 67 (24) | 860 | 293.0 | 2.94 | 20.47 | 0.15 | 64 | 0.22 |
| ChIP-seq | 25 (4) | 61 | 26.5 | 9.89 | 18.09 | 0.79 | 37 | 1.39 |
| ChIP-chip | 52 (17) | 830 | 272.9 | 3.20 | 17.48 | 0.13 | 40 | 0.15 |
| Intersect | 10 (1) | 25 | 6.6 | 16.00 | 7.46 | 1.36 | 13 | 1.97 |
Regions containing at least one peak with probability 0.9 were selected and motifs with PWM of NRSF motif were searched.
a#Match is the number of peaks including a motif match in the original sequence.
b#Permute is the number of peaks including a motif match in the permuted sequence.
cOR stands for odds ratio.
dMatch rate is (#Match − #Permute)/1 kb.
e# Clover corresponds to the number of statistically significant motif hits in the selected peak regions at P-value threshold 0.05.
fRate in Clover refers to the # motif hits divided by the sequence coverage.
3.2.3 Comparison of individual HMMs
Since the performance of HHMM depends on the success of individual HMMs, we evaluated the individual-level HMMs in ChIP-seq and ChIP-chip data in comparison to cisGenome (Ji et al., 2008) in the NRSF data. CisGenome is an integrative system for detecting ChIP-enriched regions from ChIP-seq or ChIP-chip data. The same datasets used in the HHMM analysis were fed into CisGenome. All default arguments were used in CisGenome. The significance criterion was set at the posterior probability threshold 0.9 (default in HHMM) for HMMs in HHMM and FDR 0.1 for CisGenome (default in CisGenome).
The comparison in the ChIP-chip data reveals that a considerable overlap exists between the two methods. Our uniform–normal model and CisGenome identified 697 and 830 peaks, respectively, with 530 peaks (76%) identified in CisGenome in the peaks identified by our uniform–normal HMM. The peaks unique to each method are likely due to the distinct approach taken in TileMap (Ji and Wong, 2005) implemented in CisGenome. In the ChIP-seq data, GP-ZIP HMM identified 61 peaks with probability threshold 0.9 whereas CisGenome identified 37 peaks with FDR threshold 0.1. Thirty-five peaks from CisGenome (95%) have overlap with at least one region in the selected peak regions in our GP-ZIP HMM.
We also found that, for the ChIP-chip data, the peaks identified by CisGenome tend to be longer than the peaks identified by our uniform–normal HMM (589 bp versus 328 bp) on average, while, for the ChIP-seq data, peaks identified by CisGenome tend to be narrower than peaks identfied by our GP-ZIP HMM (136 bp versus 435 bp).
Despite the differences in the width of peak regions, both comparisons suggest that the individual-level HMMs in HHMM perform reasonably in concordance with other implementations such as CisGenome.
3.2.4 Impact of window size
We evaluated the impact of the window size in HHMM using 10 and 50 nt long windows in addition to the 25 nt window used above. In terms of sequence overlap, 25 and 50 nt windows gave consistent result (see Table 2). Peaks identified using 10 nt windows but missed using the other two larger windows from 3 to 8 windows (30 to 80 nt), indicating that they are short peak regions.
Table 2.
Impact of window sizes in HHMM analysis
| Window size (nt) | Peaks | Coverage (kb) | Overlap with |
||
|---|---|---|---|---|---|
| 10 nt | 25 nt | 50 nt | |||
| 10 | 1195 | 350.9 | – | – | – |
| 25 | 424 | 179.2 | 97% | – | – |
| 50 | 405 | 194.4 | 98% | 91% | – |
Finer window size helps to identify more motifs at the expense of extended computational loading.
This inconsistent result with 10 nt window suggests that using microscopic windows may exaggerate short peaks, let alone increased computational loading. Based on the consistency in the analyses using 25 and 50 nt windows, windows of comparable size to sequence reads (20–30 nt) or array probes (36–50 nt) is deemed optimal for HHMM analysis.
3.3 Application to CTCF data
3.3.1 Data and model fit
For an example of genome-wide mapping, binding sites of CTCF were mapped using the data from Kim et al. (2007) and Barski et al. (2007). CTCF is a zinc finger protein that has a multivalent character as a TF (Dunn and Davie, 2003; Ohlsson et al., 2001) capable of participating in both repression and activation due to the combinatorial use of its 11 zinc fingers. CTCF zinc fingers can be selectively utilized based on the different needs of target genes, and thus the binding sites are likely to be more variable than other transcription factors. For instance, Kim et al. (2007) has reported 62 genes for which multiple CTCF binding sites were identified. We used the PWM reported in that study for motif search (See Supplementary Material Section 2 for the motif logo plot).
Individual HMM fits in this data showed that 419 457 windows in ChIP-seq and 3.4 million probes (7.1 million windows worth) in ChIP-chip had positive posterior probabilities, where 152 025 windows overlapped with each other (37% of ChIP-seq). Among these, 1.5 million windows had posterior probabilities 0.9 in at least one data source, and 1.2 million of these had 0.9 and above probability in the master level.
3.3.2 Motif enrichment
Table 3 presents the motif enrichment test results in MatInspector based on the data filtered at the probability threshold 0.9. For background sequences, we randomly shuffled the original sequences within each peak region as in the NRSF data. It is easy to see that HHMM and Union are the two methods that collect the highest number of TFBS motifs, but the number of hits in the permuted sequences shows almost a 3:4 ratio, indicating that the relative significance of motif search results is improved in HHMM.
Table 3.
Motif summary for the five methods in CTCF data
| Method | #Matcha (#Permuteb) | #Peaks | Coverage (Mb) | ORc | χ2 | Match rated |
|---|---|---|---|---|---|---|
| HHMM | 23 772 (4815) | 65 808 | 30.31 | 7.16 | 16 057.36 | 0.63 |
| Union | 26 788 (6200) | 83 325 | 40.08 | 5.89 | 16 018.71 | 0.51 |
| ChIP-seq | 16 771 (1836) | 25 372 | 9.33 | 25.00 | 18 926.85 | 1.60 |
| ChIP-chip | 16 599 (5134) | 69 246 | 33.83 | 3.94 | 7172.77 | 0.34 |
| Intersect | 6310 (719) | 9576 | 3.06 | 23.80 | 7023.18 | 1.83 |
Peak regions that contains a signal with probability 0.9 were selected and motifs with PWM of CTCF motif were searched.
a#Match is the number of peaks including a motif match in the original sequence.
b#Permute is the number of peaks including a motif match in the permuted sequence.
cOR stands for odds ratio.
dMatch rate is (#Match − #Permute)/1 kb.
Since the number of hits in a genome-wide data is extremely large, all χ2 tests reported extremely small P-values. However, the odds ratio of observing motifs in the selected regions was higher in HHMM (7.16) than in Union (5.89), and the match rate was also higher in HHMM (0.98/1 kb) than in Union (0.84/1 kb). This improvement is an obvious consequence of the fact that the regions picked by HHMM (30 Mb) is far narrower than those picked by Union (40 Mb) on average.
On the other hand, ChIP-seq data from Barski et al. (2007) seem to demonstrate the ultra-performance of ChIP-seq, where 62% of the motifs found in Union were identified, but the search regions are so specific that the number of hits in the permuted sequences is low (1836 in ChIP-seq, 6200 in Union) and therefore the odds ratio and the match rates are high. Nonetheless, it is the goal of HHMM to find a compromise between Union and ChIP-seq only analysis, in which extra 7000 motifs were saved by allowing some of the most significant ChIP-chip-specific regions at the expense of a reduced overall statistical significance of motif enrichment.
4 DISCUSSION
The availability of multiple experimental datasets profiling the activity of a specific TF is an important asset for delineating regulatory mechanisms. The proposed HHMM method not only identifies more binding sites with increased specificity, but also serves as an assessment of agreement and discrepancy between both technologies. It is noted that HHMM may not be optimal when the best performing experimental platform (ChIP-seq in this case) identifies most of the true ChIP-enriched regions, since additional information with a decreased precision will do nothing but dilute the signal with little contribution to finding extra binding site motifs (See the example of STAT1 data in the Supplementary Material Section 3). Nevertheless, it is difficult to expect that the new sequencing technology will always be able to provide perfect coverage of the genome in practice, and thus the previously deposited ChIP-chip datasets may be of significant value in improving TFBS identification in most cases.
Although our method is designed for combining ChIP-chip and ChIP-seq data, the HHMM framework is rather general and can be applied to other scenarios where information collected from multiple sources may be integrated. The opportunities for this type of joint analyses frequently arise in biomedical research. With the rapid development of new technologies, there are often multiple assays co-existing, measuring the same or closely related quantities of interest. Also for measuring protein–DNA binding, a series of assays have been developed, e.g. ChIP-PCR, ChIP-chip, ChIP-PET and ChIP-seq. Since these assays often have different sensitivity and specificity, straightforward combinations such as Union and Intersection do not work well. HHMM, on the other hand, is built under a coherent probability framework that is able to handle heterogeneity in sensitivity and specificity from the individual data sources, and therefore allows for easy incorporation of data from multiple experimental platforms. Because technologies are constantly changing, the two-stage HHMM estimation framework allows straightforward incorporation of data from new platforms.
Supplementary Material
ACKNOWLEDGEMENTS
Authors are grateful to the three reviewers for their helpful comments that significantly improved the article.
Funding: National Institute of General Medical Sciences (grant R01GM72007 to D.G. in part); the Huck Institute for Life Sciences at Penn State University (to D.G. in part); Career development award from University of Michigan Prostate SPORE (to Z.S.Q.'s research, in part).
Conflict of Interest: none declared.
REFERENCES
- Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bentley D, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui H, et al. Proceedings of AAAI. San Jose, CA: 2004. Hierarchical hidden Markov models with general state hierarchy. [Google Scholar]
- Cartharius K, et al. Matinspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Consul P. Generalized Poisson Distributions. New York: Marcel Dekker; 1989. [Google Scholar]
- Dunn K, Davie J. The many roles of the transcriptional regulator CTCF. Biochem. Cell Biol. 2003;81:161–167. doi: 10.1139/o03-052. [DOI] [PubMed] [Google Scholar]
- Du J, et al. A supervised hidden Markov model framework for efficiently segmenting tiling array data in transcriptional and chIP-chip experiments: systematically incorporating validated biological knowledge. Bioinformatics. 2006;22:3016–3024. doi: 10.1093/bioinformatics/btl515. [DOI] [PubMed] [Google Scholar]
- Euskirchen G, et al. Mapping of transcription factor binding regions in mammalian cells by chip: Comparison of array- and sequencing-based technologies. Genome Res. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine S, et al. The hierarchical hidden Markov model: analysis and applications. Mach. Learn. 1998;32:41–62. [Google Scholar]
- Huber W, et al. Transcript mapping with high-density oligonucleotide tiling arrays. Bioinformatics. 2006;22:1963–1970. doi: 10.1093/bioinformatics/btl289. [DOI] [PubMed] [Google Scholar]
- Humburg P, et al. Parameter estimation for robust HMM analysis of chIP-chip data. BMC Bioinformatics. 2008;9:343. doi: 10.1186/1471-2105-9-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Ji H, Wong W. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Ji H, et al. An integrated software system for analyzing chip-chip and chip-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Johnson N, et al. Univariate Discrete Distributions. New York: John Wiley & Sons; 1992. [Google Scholar]
- Kim T, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, et al. A hidden Markov model for analyzing chIP-chip experiments on genome tiling arrays and its application to p53 binding sequences. Bioinformatics. 2005;21:i274–i282. doi: 10.1093/bioinformatics/bti1046. [DOI] [PubMed] [Google Scholar]
- Loh Y, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Mikkelsen T, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch K, et al. A hidden Markov model approach for determining expression from genomic tiling microarrays. BMC Bioinformatics. 2006;7:239. doi: 10.1186/1471-2105-7-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R, et al. CTCF is uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- Park P. Epigenetics meets next-generation sequencing. Epigenetics. 2008;3:318–321. doi: 10.4161/epi.3.6.7249. [DOI] [PubMed] [Google Scholar]
- Parmigiani G, et al. A statistical framework for expression-based molecular classification in cancer. J. R. Stat. Soc. B. 2002;64:717–736. [Google Scholar]
- Rabiner L. A tutorial on hidden Markov models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- Ren B, et al. Genome-wide location and function of DNA-associated proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Schoenherr C, et al. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl Acad. Sci. USA. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S. Bayesian methods for hidden Markov models: recursive computing in the 21st century. J. Am. Stat. Assoc. 2002;97:337–351. [Google Scholar]
- Shah S, et al. Modeling recurrent DNA copy number alterations in array CGH data. Bioinformatics. 2007;23:i450–i458. doi: 10.1093/bioinformatics/btm221. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, et al. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon M, et al. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone h4 is retained on a highly transcribed gene. Cell. 1988;53:937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- Wei C, et al. A global map of p53 transcription factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.