Abstract
Over the past few years it has become evident that the intermediate filament proteins, the types A and B nuclear lamins, not only provide a structural framework for the nucleus, but are also essential for many aspects of normal nuclear function. Insights into lamin-related functions have been derived from studies of the remarkably large number of disease-causing mutations in the human lamin A gene. This review provides an up-to-date overview of the functions of nuclear lamins, emphasizing their roles in epigenetics, chromatin organization, DNA replication, transcription, and DNA repair. In addition, we discuss recent evidence supporting the importance of lamins in viral infections.
[Keywords: Lamins, chromatin, epigenetics, nuclear architecture, laminopathies, viruses]
In eukaryotic cells, chromatin is tightly packed in a highly organized fashion within a nucleus that is composed of two main compartments: the nucleoplasm and the nuclear envelope (NE). There are also subcompartments in the nucleus containing factors involved in essential nuclear functions such as DNA replication, transcription, and RNA splicing (Prasanth and Spector 2006; Spector 2006). The NE separates nuclear functions from cytoplasmic functions and at its inner surface it provides a docking site for chromatin. The major structural elements of the NE are the inner nuclear membrane (INM), the outer nuclear membrane (ONM), the nuclear pore complexes (NPCs), and the nuclear lamina. The lamina is comprised of a complex meshwork of proteins closely associated with the INM and attached to the periphery of NPCs and to chromatin (Fawcett 1966; Patrizi and Poger 1967; Aaronson and Blobel 1975). The main constituents of the lamina are the type V intermediate filament (IF) proteins, the nuclear lamins. Lamins are also found, in lower concentrations, distributed throughout the nucleoplasm. The organization of lamins at the nuclear periphery as well as within the nucleoplasm is influenced by numerous lamin-binding proteins (Dorner et al. 2007; Schirmer and Foisner 2007; Wagner and Krohne 2007).
This review focuses on the role of nuclear lamins in the organization and regulation of chromatin in the interphase nucleus—specifically, the involvement of lamins in essential processes such as transcription, DNA replication, DNA repair, and various epigenetic phenomena involved in the regulation of euchromatin–heterochromatin transitions. Emphasis is also placed on the remarkable array of disease-causing mutations in the human lamin A gene, from which many of the most recent insights into lamin functions have been derived. In addition, we also discuss emerging ideas regarding the roles of lamins in viral infections.
General properties of the nuclear lamins
Nuclear lamins were initially described as the major protein components of detergent-high salt resistant “lamina” fractions of rat liver and chicken erythrocyte nuclei (Aaronson and Blobel 1975; Gerace et al. 1978). Subsequently it was shown that they are members of the IF protein family (Aebi et al. 1986; Goldman et al. 1986; McKeon et al. 1986). Lamin genes are found in all metazoa examined to date, but are absent in plants and unicellular organisms (Meier 2001; Melcer et al. 2007). Lamins are divided into A and B types based on sequence homologies. In mammals, two major A-type lamins (lamin A and C) and two major B-type lamins (lamin B1 and B2) have been characterized, in addition to the minor isoforms lamin AΔ10 (Machiels et al. 1996) and germ cell-specific lamins C2 (Furukawa et al. 1994) and B3 (Furukawa and Hotta 1993). While lamins B1 and B2–B3 are encoded by different genes (LMNB1 and LMNB2, respectively), A-type lamins are derived from one gene (LMNA) by alternative splicing (Broers et al. 2006; Schumacher et al. 2006; Verstraeten et al. 2007).
Lamins as structural components of the nucleus
The lamins are composed of a long central α-helical rod domain, flanked by globular N-terminal (head) and C-terminal (tail) domains (Fig. 1A). Like most IF proteins, lamins self-assemble into higher-order structures whose basic subunit is a coiled-coil dimer formed by in parallel and in register interactions (Stuurman et al. 1998; Herrmann and Foisner 2003). Unlike cytoplasmic IFs, lamin dimers tend to form paracrystals in vitro rather than 10-nm filaments (Stuurman et al. 1998; Herrmann and Foisner 2003; Melcer et al. 2007). In addition to the central rod domain (Schirmer et al. 2001; Strelkov et al. 2004), it has been shown that the head and tail domains are also involved in lamin assembly (Sasse et al. 1998; Stuurman et al. 1998; Herrmann and Foisner 2003; Shumaker et al. 2005; Isobe et al. 2007). Although A- and B-type lamins appear to interact with each other in vitro (Georgatos et al. 1988; Ye and Worman 1995; Schirmer and Gerace 2004), little is known about the composition and structure of lamin polymers within the lamina, and even less is known about their nucleoplasmic structures. The structures that have been described within the lamina range from an orthogonal meshwork of 10- to 15-nm fibers seen in Xenopus oocyte germinal vesicles (Aebi et al. 1986; Stewart and Whytock 1988) to irregular filamentous meshworks reported in mammalian cells (Capco et al. 1982; de Graaf et al. 1991; Belmont et al. 1993). Within the lamina of interphase cells, both A- and B-type lamin structures are relatively stable, as revealed by their long recovery time in fluorescence recovery after photobleaching (FRAP) experiments (Broers et al. 1999; Moir et al. 2000c).
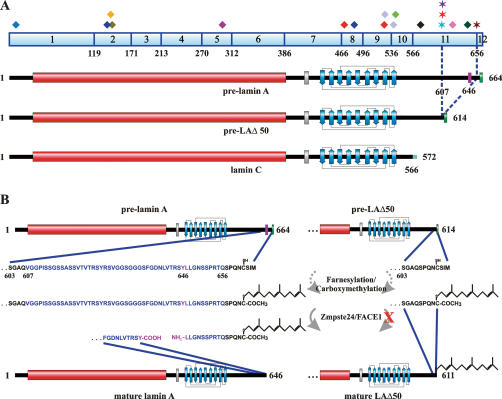
Figure 1.
Structure of nuclear lamins. (A) Schematic drawing of a pre-lamin polypeptide chain. The central α-helical rod domain (red), the NLS (gray), the Ig-fold (blue; its simplified structure indicating the nine β-sheets is depicted), and the C-terminal −CAAX box (green) are shown. (B) Post-translational processing of pre-lamin A, B1, and B2: A farnesyl group is attached to the cystein residue of the −CAAX box by a farnesyltransferase; the last three residues (−AAX) are proteolytically cleaved off by an AAX endopeptidase; the carboxylic acid group (−COOH) of the C-terminal cysteine residue is methylated by a carboxyl methyltransferase. These steps lead to mature lamin B1 and B2. In the case of farnesylated/carboxymethylated pre-lamin A, an additional 15 C-terminal residues, including the farnesylated/carboxymethylated cysteine, are cleaved off by Zmpste24/FACE1. Inhibition of this step either due to mutations or deficiency in Zmpste24/FACE1 (indicated by the red X) or by protease inhibitors (PIs) can lead to severe phenotypes in mice and humans (for details, see the text). Note that inhibition of farnesylation by farnesyltransferase inhibitors (FTIs) results in a complete loss of processing. (C) Schematic drawings of the C-terminal tail domains of mature lamins A, C, B1, and B2. The following amino acid positions are indicated: start of the tail domain, first residue of the NLS, start and end of the Ig-fold, and C-terminal residue of the respective mature lamin. Note that lamins B1 and B2 are farnesylated and carboxymethylated, while lamins A and C are not.
Lamins contain a structural motif similar to a type s immunoglobulin fold (Ig-fold) within their C-terminal tail domain (Dhe-Paganon et al. 2002; Krimm et al. 2002) and a nuclear localization signal (NLS) between the central rod and the Ig-fold (Fig. 1A) that is required for the transport of lamins into the nucleus (Loewinger and McKeon 1988). Additionally, a −CAAX box is located at the C terminus of lamins B1 and B2 and lamin A, which leads to extensive post-translational processing of these proteins (Fig. 1; Young et al. 2005; Rusinol and Sinensky 2006). Initially expressed as pre-lamins, they undergo several modifications to become mature lamins (Fig. 1B). In the first step, a farnesyltransferase farnesylates the cysteine residue of the −CAAX box and subsequently the −AAX is removed by an endopeptidase, most likely Rce1 (Ras-converting enzyme 1) and/or Zmpste24 (Zinc metalloprotease related to Ste24p)/FACE1 (Rusinol and Sinensky 2006). In a third step the cysteine residue is carboxymethylated, a process catalyzed by the enzyme isoprenylcysteine carboxyl methyltransferase (Icmt). While the maturation of B-type lamins is terminated at this step resulting in their permanent farnesylation and carboxymethylation (Fig. 1C), lamin A is further processed by Zmpste24/FACE1 to form mature lamin A (Fig. 1B). This latter step results in the removal of an additional 15 amino acids from the C terminus of lamin A, including the farnesylated/carboxymethylated cysteine residue (Fig. 2B; Corrigan et al. 2005). It is noteworthy that the temporal sequence of these processing steps is critical; i.e., if farnesylation is blocked, the post-translational processing of lamins is inhibited (Fig. 1B; Rusinol and Sinensky 2006). The modifications of the −CAAX box are thought to play a role in targeting the lamins to the INM and also in establishing protein–protein interactions (Rusinol and Sinensky 2006). Interestingly, the inhibition of lamin farnesylation, either by replacing the cysteine residue of the −CAAX box or by farnesyltransferase inhibitors (FTIs) (Fig. 1B) does not inhibit their incorporation into the nuclear lamina (Sasseville and Raymond 1995; Broers et al. 1999; Pendas et al. 2002; Gruber et al. 2005). Lamin C, which lacks the 98 C-terminal amino acids present in pre-lamin A and contains a unique C terminus comprised of six amino acids lacking a −CAAX box (Figs. 1C, 2A; Fisher et al. 1986; McKeon et al. 1986), also becomes incorporated into the nuclear lamina (Fong et al. 2006b). It is noteworthy that the most variable region of the lamins, the C-terminal tail domain (Fig. 1C), contains the binding sites for most of the lamin-binding proteins (Goldman et al. 2002; Zastrow et al. 2004; Dorner et al. 2007; Schirmer and Foisner 2007; Wagner and Krohne 2007).
Figure 2.
Comparison of lamin A, lamin C, and LAΔ50 structures. (A) The 12 exons encoding lamin A and the residues 1–566 of lamin C (exons 1–10) are indicated along with their corresponding amino acid residues. In addition, the positions of 18 mutations associated with Hutchinson-Gilford progeria syndrome (HGPS) are shown (the number of incidents reported for each mutation is shown in parentheses) (http://www.umd.be:2000; http://www.dmd.nl): (blue diamond) 29 C > T (1); (dark-blue diamond) 412 G > A (1); (yellow diamond) 428 C > T (1); (green diamond) 433 G > A (1); (purple diamond) 899 G > A (1); (orange diamond) 1411 C > T + 1579 C > T (1); (dark-blue diamond) 1454 C > G (1); (light-blue diamond) 1583 C > T + 1619 C > T (1); (light-green diamond) 1626 G > C (4); (dark-purple diamond) 1733 A > T (1); (blue star) 1821 G > A (1); (red star) 1822 G > A (1); (purple star) 1824 C > T (37); (pink diamond) 1868 C > G (2); (dark-green diamond) 1930 C > T (1); (brown star) 1968 + 1 G > A (1). Mutations marked with a star lead to abnormal splicing of the last 150 nucleotides of exon 11, resulting in the expression of a mutant lamin A protein (pre-LAΔ50) lacking 50 amino acids from the C terminus of pre-lamin A (residues 607–656). These residues contain the second proteolytic cleavage site (magenta; residue 646) involved in pre-lamin A processing. For a description of the schematic drawing of pre-lamin A, pre-LAΔ50, and lamin C, see Figure 1, A and C. The six lamin C-specific residues (567–572) are indicated (light blue). Note that while most mutations affect both lamin A and C, some are specific for lamin A. (B) Differences in the post-translational processing between pre-lamin A and pre-LAΔ50. The amino acid sequences of pre-lamin A (residues 603–664) and pre-LAΔ50 (residues 603–614) are shown. Residues 607–656 in pre-lamin A, which are absent in pre-LAΔ50, are labeled blue, and the Zmpste24/FACE1 cleavage site (YL) within this region is labeled magenta. The first three steps, including farnesylation and carboxymethylation of the cystein residue of the −CAAX box (−CSIM), are identical for both proteins (for details, see Fig. 1B). In the last step, the 15 C-terminal residues—including the farnesylated/carboxymethylated cysteine residue—are cleaved off from pre-lamin A by Zmpste24/FACE1, resulting in mature lamin A (residues 1–646) and a 15-amino-acid-long farnesylated/carboxymethylated peptide. As the Zmpste24/FACE1 cleavage site is missing in the mutant protein, LAΔ50 remains farnesylated and carboxymethylated.
Both A- and B-type lamins are disassembled in a phosphorylation-dependent manner at the onset of mitosis (Fields and Thompson 1995) and upon dephosphorylation become incorporated into newly forming daughter cell nuclei during telophase/G1 (Fields and Thompson 1995; Thompson et al. 1997; Moir et al. 2000b). While B-type lamins relocalize exclusively to the periphery of reassembling daughter nuclei, A-type lamins accumulate in the nucleoplasm at the end of mitosis and in early G1 (Bridger et al. 1993; Moir et al. 2000c; Dechat et al. 2004). Although most prevalent in early G1, nucleoplasmic lamins A/C are present throughout interphase (Goldman et al. 1992; Broers et al. 1999; Moir et al. 2000c) and are thought to play a role in retinoblastoma-mediated cell proliferation (Johnson et al. 2004; Pekovic et al. 2007), initiation of DNA replication (Kennedy et al. 2000), and RNA splicing (Jagatheesan et al. 1999; Kumaran et al. 2002), although the latter function is controversial (Vecerova et al. 2004). Lamin A/C structures present in the nucleoplasm appear to be more dynamic and less tightly bound to the nucleoskeleton than those associated with the lamina, based on detergent extractability and FRAP analyses (Broers et al. 1999; Moir et al. 2000c; Muralikrishna et al. 2004). Lamin B is also present in the nucleoplasm but as a more stable structure compared with A-type lamins (Moir et al. 2000c), and intranuclear lamin B foci are associated with sites of mid–late-S-phase replication (Moir et al. 1994). In addition, there is ultrastructural evidence suggesting that lamins are part of a nucleoskeleton distributed in the nucleoplasm (Hozak et al. 1995; Barboro et al. 2002).
As IF structural proteins, it is now well established that the lamins provide shape and mechanical stability to the nucleus (Houben et al. 2007). There is also evidence supporting a role for the lamins in maintaining the mechanical properties of the entire cell. For example, lamin A/C-deficient cells display impaired mechanotransduction, decreased mechanical stiffness, and defective cell migration (Houben et al. 2007; Lee et al. 2007). These changes in the micromechanical properties of cells may ultimately be explained by alterations in the linkages between the nucleoskeleton with the cytoskeleton. Important elements in such linkages include the integral proteins of the INM, Sun1 and Sun2, and of the ONM, nesprin-1, nesprin-2, and nesprin-3α. Specifically, it is thought that the connection between the cytoplasm and nucleoplasm is mediated by nesprins interacting with Sun proteins in the luminal space between the INM and the ONM (Padmakumar et al. 2005; Crisp et al. 2006; Ketema et al. 2007). Sun1 in turn interacts with lamin A, but not with lamin C, B1, or B2 in the lamina region (Haque et al. 2006). While in the cytoplasm, nesprin-1 and nesprin-2 are thought to bind to actin (Warren et al. 2005) and nesprin-3α binds to the cytoskeletal IF-binding protein, plectin (Wilhelmsen et al. 2005; Ketema et al. 2007), which also binds to actin and possibly microtubules (Wiche 1998).
Nuclear lamin expression during development and differentiation
Studies of the early development of Xenopus have shown that oocytes contain mainly one B-type lamin, lamin B3 (XLB3) (Benavente et al. 1985; Stick and Hausen 1985) and minor amounts of XLB1 and XLB2 (Lourim et al. 1996). During development XLB3 persists only to the tail bud stage (stage 24) and reappears later in some adult cells (Benavente et al. 1985; Stick and Hausen 1985). While XLB1 and XLB2 are up-regulated during the midblastula transition and gastrulation (stages 8–12) (Benavente et al. 1985; Stick and Hausen 1985), XLA is not found in early tailbud embryos (stage 24), but is present in tadpoles (stage 45) (Wolin et al. 1987). Similar changes in lamin expression have been reported in early developing Drosophila (Riemer et al. 1995) and chicken embryos (Lehner et al. 1987). In mice, B-type lamins are expressed throughout early development, while lamins A/C are first detected in the trophoblast on embryonic day 9, in the embryoblast on day 10, and in myoblasts and other mesenchymal tissues by day 11 (Stewart and Burke 1987; Rober et al. 1989). These data suggest that lamin A/C expression is closely correlated with cell differentiation. This has been confirmed in several studies using cultured cells (for example, see Guilly et al. 1990; Rober et al. 1990; Mattia et al. 1992; Lin and Worman 1997). More recently it has been shown that lamins B1 and B2, but not lamins A/C, are expressed in undifferentiated mouse and human embryonic stem (ES) cells (Constantinescu et al. 2006). Upon differentiation, human ES cells appear to express lamins A/C prior to the down-regulation of the pluripotency marker Oct-3/4, but after the down-regulation of Tra-1-60, Tra-1-81, and the stage-specific embryonic antigen-4 (SSEA-4). The absence of lamins A/C in undifferentiated human ES cells has been suggested to be responsible for the high deformability of the nuclei in these cells (Pajerowski et al. 2007). Differential expression of A- and B-type lamins has also been shown in adult human tissues (Broers et al. 1997; Lin and Worman 1997; Tilli et al. 2003) and during neurogenesis in the adult rat brain (Takamori et al. 2007).
Insights into the role of lamins during development come from mutations in the B-type Drosophila lamin Dm0 that cause lethality at different embryonic or late pupal stages, depending on the site of the mutation (Gruenbaum et al. 2003; Osouda et al. 2005). In addition, when the only Caenorhabditis elegans lamin, LMN-1, is knocked down, embryonic arrest occurs at the 100- to 300-cell stage (Liu et al. 2000). Further insights into the requirement for a specific lamin isoform during development have been obtained from knocking out either LMNA or LMNB1 in mice. The lamin A/C-deficient animals develop normally until birth, but have severe postnatal growth retardation and develop muscular dystrophy (Sullivan et al. 1999). The nuclei in LMNA-null mouse embryonic fibroblasts (MEFs) often display irregular shapes, a localized loss of B-type lamins, and the redistribution of emerin, a lamin A-associated integral protein of the INM (Clements et al. 2000), to the endoplasmic reticulum (ER) (Sullivan et al. 1999). In contrast, mice that are null for lamin A but express lamin C and the B-type lamins appear to be healthy. The nuclei in cells derived from these mice exhibit minimal shape changes and emerin localizes normally in the INM (Fong et al. 2006b). On the other hand, lamin B1-deficient mice die at birth and have defective lungs and bones (Vergnes et al. 2004). A high percentage of MEFs derived from these mice have misshapen nuclei and an abnormal distribution of lamin A/C. They are also polyploid and senesce prematurely. In humans, a recent case study described a lamin A/C deficiency in which the fetus survived early development, was born prematurely at 30 wk, and died immediately (Muchir et al. 2003; van Engelen et al. 2005). The patient was homozygous for a nonsense mutation in LMNA and displayed severe growth retardation, multiple bone fractures and poorly developed striated muscles. Cells derived from this patient had abnormally shaped nuclei with lobules lacking lamins B1, B2, and Nup153 (a NPC protein), and emerin was distributed to the ER (Muchir et al. 2003).
There is little information available on the regulation of lamin gene expression. Lamin A/C expression can be induced by retinoic acid in mouse embryonic carcinoma cells (Lebel et al. 1987) and there is a retinoic acid-responsive element in the LMNA promoter that is regulated by the transcription factors c-Jun and Sp1/Sp3 (Okumura et al. 2004). There are also other regulatory motifs in the LMNA promoter binding the transcription factors Sp1/3, c-Jun, and c-Fos, and the transcriptional coactivator CREB-binding protein (Muralikrishna and Parnaik 2001; Janaki Ramaiah and Parnaik 2006). Within the first intron of LMNA there are binding sites for the transcription factors, hepatocyte nuclear factor-3β and retinoic X receptor β (Arora et al. 2004). Recently, lamins A/C have been identified as p53 targets in mitomycin C-treated HCT116 colon carcinoma cells (Rahman-Roblick et al. 2007). Obviously more extensive studies are required in order to understand the regulation and function of the A- and B-type lamins during development and differentiation. Approaches to understanding the significance of the differential expression of lamins have involved silencing with RNAi in cultured cells. These studies have shown that down-regulation of either lamin B1 or B2 induces apoptosis in HeLa cells (Harborth et al. 2001). In contrast, HeLa cells in which lamins A/C have been silenced appeared to grow normally. Although this provides some useful information regarding the general requirements for A- and B-type lamins in cell growth, much more work is required to determine the specific functions of the A-type lamins in differentiation.
Nuclear lamins in disease and aging
Interest in nuclear lamins increased dramatically following the discovery that mutations in LMNA cause autosomal-dominant Emery-Dreifuss muscular dystrophy (AD-EDMD) (Bonne et al. 1999). Since this discovery, an enormous number of LMNA mutations, the vast majority of which are heterozygous, have been linked to many diseases including X-linked EDMD, familial partial lipodystrophy (FPLD), dilated cardiomyopathy, limb girdle muscular dystrophy 1B, congenital muscular dystrophy, and premature aging or progeroid syndromes. The latter include Hutchinson-Gilford progeria syndrome (HGPS), atypical Werner’s syndrome (WS), restricted dermopathy (RD), and mandibuloacral dysplasia (MAD) (Broers et al. 2006; Kudlow et al. 2007; Worman and Bonne 2007). Collectively, these diseases are known as the laminopathies. There are at least 11 distinct diseases (Worman and Bonne 2007) associated with >300 different mutations in LMNA producing ∼200 mutant lamin A/C proteins (for details, see http://www.umd.be:2000; http://www.dmd.nl; http://www.interfil.org). Remarkably, both the number of LMNA mutations and of their associated diseases continues to grow.
In stark contrast to LMNA, there are very few diseases associated with mutations in the LMNB1/B2 genes. These include autosomal-dominant leukodystrophy caused by a duplication of LMNB1 and acquired partial lipodystrophy caused by LMNB2 mutations (Hegele et al. 2006; Padiath et al. 2006). Mutations in several lamin-binding proteins have also been linked to diseases. For example, mutations in the lamin B receptor (LBR) cause Pelger-Huet anomaly and HEM/Greenberg skeletal dysplasia (Worman and Bonne 2007). Mutations in the lamina-associated polypeptide (LAP) 2α cause a form of dilated cardiomyopathy, and mutations in emerin cause a form of X-linked EDMD (Vlcek and Foisner 2007a; Worman and Bonne 2007). Interestingly, the LAP2α and emerin mutations are associated with diseases that can also be caused by LMNA mutations. Since both of these lamin-associated proteins specifically bind to lamin A (Zastrow et al. 2004), it is possible that these mutations inhibit the association of LAP2α or emerin with lamins A/C, causing at least some of the phenotypes observed in the respective diseases.
Because of its potential role as a model for human aging, the most intensively studied laminopathy to date has been the premature aging disease, HGPS. Investigations into the cellular and molecular basis of this rare disorder have provided numerous important insights into the structure and function of lamins A/C and into the mechanisms causing this devastating childhood disease. The most prevalent (65% of 57 HGPS cases analyzed) of the 18 LMNA mutations associated with HGPS (Fig. 2A) is the single nucleotide substitution, 1824 C > T (http://www.umd.be:2000; http://www.dmd.nl). This mutation activates a cryptic splice site resulting in the expression of a mutant lamin A lacking 50 amino acids within its C terminus (LAΔ50/progerin) (Fig. 2; De Sandre-Giovannoli et al. 2003; Eriksson et al. 2003). Besides the 1824 C > T mutation, three other HGPS mutations, 1822 G > A, 1821 G > A, and 1968 + 1 G > A, result in the same abnormal splicing event (Fig. 2A; Eriksson et al. 2003; Moulson et al. 2007). Since the deleted amino acids are just upstream of the −CAAX box and contain the second Zmpste24/FACE1 cleavage site, LAΔ50/progerin is permanently farnesylated and carboxymethylated (Fig. 2B; Glynn and Glover 2005; Dechat et al. 2007). This leads to an abnormal membrane association of the mutant protein throughout the cell cycle (Cao et al. 2007; Dechat et al. 2007). It has also been shown that LAΔ50/progerin is abnormally associated with the nuclear lamina (Goldman et al. 2004; Scaffidi and Misteli 2005; Dahl et al. 2006). These abnormalities contribute to several other phenotypes associated with HGPS, including lobulation of nuclei, thickening of the lamina, loss of peripheral heterochromatin, and alterations in specific heterochromatin marks (Figs. 3, 4; De Sandre-Giovannoli et al. 2003; Eriksson et al. 2003; Goldman et al. 2004; Scaffidi and Misteli 2005; Shumaker et al. 2006). In addition, expression of LAΔ50/progerin leads to mitotic defects comprising delays in cytokinesis and nuclear reassembly, abnormal chromosome segregation, and binucleation (Cao et al. 2007; Dechat et al. 2007). Many, but not all, of these phenotypes observed in cultured fibroblasts from HGPS patients become most obvious at late passage numbers. The latter finding is most likely attributable to the higher concentration of LAΔ50/progerin that has been reported in older cell cultures (Goldman et al. 2004; McClintock et al. 2006).
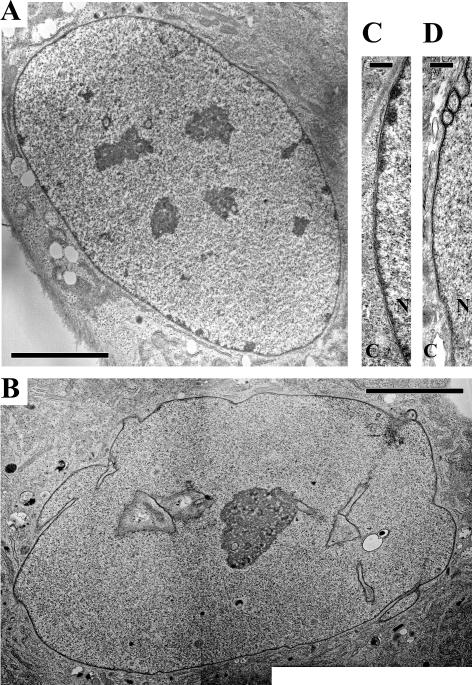
Figure 3.
Electron microscopic observation of normal and HGPS fibroblasts. Low-magnification views show peripheral heterochromatin and nucleoplasmic heterochromatic foci in the normal nucleus (A), which are both absent in the highly lobulated HGPS nucleus (B). A high-magnification view of the nuclear envelope of a HGPS nucleus (D) shows a loss of peripheral heterochromatin and a thickening of the nuclear lamina compared with a normal nucleus (C). (C) Cytoplasm; (N) nucleus. Bars: A,B, 5 μM; C,D, 300 nm.
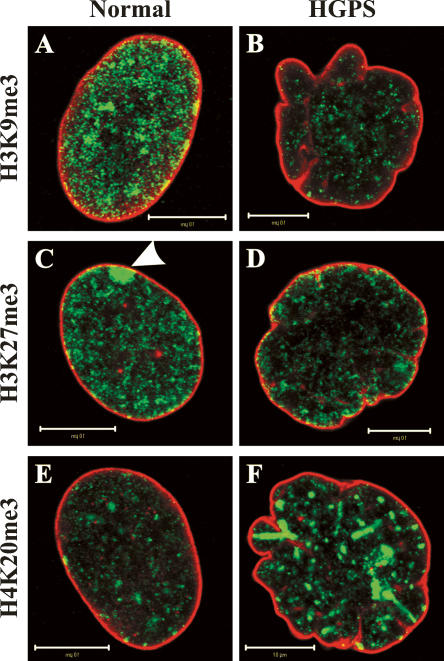
Figure 4.
Alterations in histone methylation patterns in HGPS fibroblasts. Normal and HGPS fibroblasts from female donors were double-labeled with antibodies against lamins A/C (red) and against trimethylation of either Lys 9 in histone H3 (H3K9me3; A,B), Lys 27 in histone H3 (H3K27me3; C,D), or Lys 20 in histone H4 (H4K20me3; E,F) (all green). Note the decrease of H3K9me3 (B) and H3K27me3 (D) and the increase of H4K20me3 (F) in the lobulated HGPS nuclei compared with normal nuclei (A,C,E). The decrease in H3K27me3 is best observed at the inactive X chromosome, which is normally enriched in this histone modification (see arrowhead in C). Bars, 10 μM.
Evidence that LAΔ50/progerin may contribute directly to vascular disease comes from the finding that it appears to accumulate in vascular smooth muscle and endothelial cell nuclei in skin biopsies from an HGPS patient (McClintock et al. 2006). In addition, a progressive loss of vascular smooth muscle cells was found in an HGPS mouse model (Varga et al. 2006). In an effort to mimic HGPS in mice, additional models have been developed that display progeroid phenotypes to varying degrees (Stewart et al. 2007). Two of these animal models have provided new insights into disease mechanisms (Yang et al. 2006, 2008). In mice expressing lamin C and LAΔ50/progerin only (LMNAHG/LCO), the steady-state level of the mutant protein appears to be lower compared with mice also expressing lamin A (LMNAHG/+). While the latter animals display severe progeria-like disease phenotypes, the elimination of wild-type lamin A leads to improvements in the overall health of the animals. Mutations in Zmpste24/FACE1, leading to the accumulation of farnesylated and carboxymethylated pre-lamin A (Fig. 1B), have also been linked to HGPS, RD, and MAD (Young et al. 2005; Worman and Bonne 2007), and mice deficient in Zmpste24 resemble progeroid phenotypes (Stewart et al. 2007). Because permanent farnesylation of either LAΔ50/progerin or farnesylated pre-lamin A is thought to be responsible for the pathophysiological changes seen in these diseases, several attempts have been undertaken to rescue the cellular phenotypes by inhibiting farnesylation with FTIs (Glynn and Glover 2005; Toth et al. 2005; Cao et al. 2007; Dechat et al. 2007). In addition, specific silencing of LAΔ50/progerin has been used in attempts to ”rescue cells” expressing this mutant protein (Huang et al. 2005; Scaffidi and Misteli 2005). Although these approaches appeared to restore nuclear shape and some heterochromatin marks to a normal appearance in cells derived from HGPS patients, it is not clear if there is a complete reversal of disease phenotypes. When LMNAHG/+ mice or ZMPSTE24−/− mice were treated with an FTI, they showed an improvement in their overall health compared with untreated littermates (Fong et al. 2006a; Yang et al. 2006). Since there are no other possible treatments in sight, FTIs are now being tested in HGPS patients (for details, see http://www.progeriaresearch.org). However, the impact of these drugs on the processing and functions of wild-type lamins (including lamins B1 and B2 and lamin A expressed from the unaffected LMNA allele) must also be taken into consideration (Fig. 1B). Recent support for this comes from the finding that the distribution of B-type lamins is altered in the G1 phase of the cell cycle in cells treated with FTIs (Dechat et al. 2007).
Of great interest is the possibility that the altered lamins expressed in progeria patients may be involved not only in premature aging, but also in normal aging, as LAΔ50/progerin has been found in a small number of cells in cultures obtained from healthy individuals, especially in those from older donors (Scaffidi and Misteli 2006; Cao et al. 2007). Recently, it has also been shown that LAΔ50/progerin is expressed at low levels in skin biopsies from healthy individuals ranging in age from newborn to 97 yr and that the mutant protein seems to accumulate with age (McClintock et al. 2007). In addition, alterations in nuclear shape and structure, similar to those found in premature aging, have been described in dermal fibroblasts from old individuals (Scaffidi and Misteli 2006), in normal human fibroblasts with increasing passage number (Cao et al. 2007), and in aged C. elegans (Haithcock et al. 2005).
The role of nuclear lamins in chromatin organization and related functions
Heterochromatin is highly organized and in mammalian cells mainly associated with nucleoli and with regions of the lamina (Fig. 3A,C). Although these lamina regions are thought to provide a transcriptionally repressive environment, other regions of the lamina, which are not associated with heterochromatin (Fig. 3A,C), could conceivably be involved in gene expression. In support of this, it has previously been shown that a genetic locus, which has been targeted to the nuclear periphery by lamin B1, maintains its ability to be transcriptionally induced (Kumaran and Spector 2008). Beside heterochromatin, entire chromosomes are also organized into well-defined regions or territories within mammalian nuclei (Cremer et al. 2006). These territories are arranged in a nonrandom fashion, with gene-rich chromosomes, like human chromosome 19, residing preferentially deep inside the nucleus and gene-poor chromosomes, like human chromosome 18, more likely to be positioned in the region of the lamina (Croft et al. 1999). This is not only true for whole chromosome territories, but also for smaller domains of chromosomes (Kupper et al. 2007). Positions within these territories appear to be related to the control of gene expression and other nuclear activities (Fraser and Bickmore 2007; Lanctot et al. 2007). Recent models of nuclear architecture depict lamins and their associated proteins as determinants of chromosome positioning throughout the nucleus (Goldman et al. 2002; Dorner et al. 2007; Vlcek and Foisner 2007b). Thus the lamins would be either directly or indirectly involved in anchoring chromatin to the nuclear lamina and would also act as a nucleoplasmic scaffold for organizing chromatin elsewhere in the nucleus. In addition, lamins appear to play a role in several nuclear functions that require remodeling of chromatin, including DNA replication, DNA repair, and transcription.
Interactions between nuclear lamins and chromatin
Early biochemical evidence supporting lamin/chromatin interactions came from the biochemical analysis of HeLa nuclear lamina preparations (Bouvier et al. 1985) and from UV cross-linking experiments using calf thymus or Ehrlich ascites tumor cell nuclear “matrices” (Boulikas 1986; Galcheva-Gargova and Dessev 1987). Furthermore, lamins A/C bind to mitotic chromosomes (Burke 1990; Glass and Gerace 1990), and polynucleosomal particles in vitro (Yuan et al. 1991). Furthermore, A-type lamins have been identified as conserved peripheral proteins of human metaphase chromosomes (Takata et al. 2007). The interaction between the lamins and chromatin appears to involve their non-α-helical C-terminal tail domain and the N- and C-terminal tail domains of core histones (Hoger et al. 1991; Yuan et al. 1991; Schmidt and Krohne 1995; Taniura et al. 1995; Goldberg et al. 1999; Mattout et al. 2007). Interestingly, this interaction may also involve the NLS of the lamins (Mattout et al. 2007). There are also reports that lamins can bind directly to DNA (Shoeman and Traub 1990; Baricheva et al. 1996; Rzepecki et al. 1998; Stierle et al. 2003) and that this may involve specific sequences in the matrix attachment/scaffold-associated regions (MARs/SARs) (Luderus et al. 1994; Zhao et al. 1996). This possibility is further supported by the finding that lamin Dm0 interacts with transcriptionally inactive DNA sequences in Drosophila (Pickersgill et al. 2006).
There is additional evidence that lamins interact with chromatin and DNA through lamin-binding proteins (Mattout-Drubezki and Gruenbaum 2003; Dorner et al. 2007; Schirmer and Foisner 2007). One example for such a cross-bridging protein is LBR, an integral protein of the INM, which interacts with lamin B, DNA, and the human heterochromatin protein 1 (HP1) (Worman et al. 1990; Ye and Worman 1994, 1996). Other proteins that interact with both lamins and chromatin are LAP2α and LAP2β (Foisner and Gerace 1993; Furukawa et al. 1997; Dechat et al. 1998, 2000). While LAP2α binds specifically to lamins A/C (Dechat et al. 2000), LAP2β interacts exclusively with B-type lamins (Foisner and Gerace 1993). Another proposed linker between the nuclear lamins and chromatin is the DNA-bridging protein, barrier-to-autointegration factor (BAF) (Segura-Totten and Wilson 2004; Margalit et al. 2007), which has been reported to bind to dsDNA (Zheng et al. 2000), to histones (Montes de Oca et al. 2005), to lamin-binding proteins containing a LEM (LAP2s, emerin, and MAN1) domain (Shumaker et al. 2001), and to A-type lamins (Holaska et al. 2003). The number of interactions between lamins, lamin-binding proteins, and DNA/chromatin is growing at an extremely rapid pace. However, the functional significance and further biochemical verification of these complex interactions remains to be determined.
The role of lamins in the organization of interphase chromatin and epigenetics
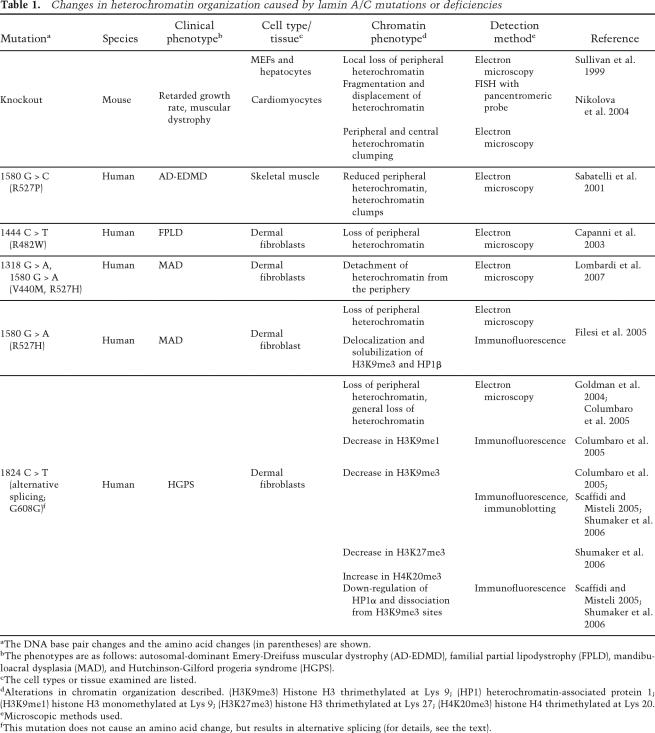
The importance of lamins in chromatin organization is most evident in cells derived from patients suffering from various laminopathies and in the LMNA-null mouse (Table 1). In fibroblasts and cardiomyocytes derived from LMNA-null mice there is a dissociation of heterochromatin from some regions of the nuclear periphery (Sullivan et al. 1999; Nikolova et al. 2004). Similar changes in peripheral heterochromatin or a more general loss of heterochromatin have been reported in cells from patients with LMNA mutations causing AD-EDMD (Sabatelli et al. 2001), FPLD (Capanni et al. 2003), MAD (Filesi et al. 2005; Lombardi et al. 2007), and HGPS (Fig. 3B,D; Goldman et al. 2004; Columbaro et al. 2005). In cells from HGPS patients, it has been shown that these abnormalities in chromatin organization are related to epigenetic changes, specifically to changes of specific histone methylations known to define heterochromatin. These histone methylations include the trimethylation of histone H3 at Lys 9 (H3K9me3) and of histone H4 at Lys 20 (H4K20me3), which define constitutive heterochromatin, and H3K27me3, which defines facultative heterochromatin (Martin and Zhang 2005; Sarma and Reinberg 2005). With respect to these epigenetic marks, a down-regulation of H3K9me3 and H3K27me3 and an up-regulation of H4K20me3 have been observed in HGPS fibroblasts (Fig. 4; Columbaro et al. 2005; Scaffidi and Misteli 2005; Shumaker et al. 2006). Changes in H3K9me3 patterns have also been observed in MAD fibroblasts (Filesi et al. 2005) and in cells derived from old healthy individuals expressing LAΔ50/progerin (Scaffidi and Misteli 2006). In addition to the changes in histone methylation, HP1α, which is normally associated with H3K9me3, was found to be down-regulated and partially dissociated from H3K9me3 sites in HGPS fibroblasts (Scaffidi and Misteli 2005; Shumaker et al. 2006). Taken together, these findings clearly demonstrate that the expression of mutant lamins leads to global changes in the epigenetic organization of chromatin, which undoubtedly contributes to the phenotypes observed in different laminopathies, including defects in DNA repair and alterations in gene expression (see below).
Table 1.
Changes in heterochromatin organization caused by lamin A/C mutations or deficiencies
aThe DNA base pair changes and the amino acid changes (in parentheses) are shown.
bThe phenotypes are as follows: autosomal-dominant Emery-Dreifuss muscular dystrophy (AD-EDMD), familial partial lipodystrophy (FPLD), mandibuloacral dysplasia (MAD), and Hutchinson-Gilford progeria syndrome (HGPS).
cThe cell types or tissue examined are listed.
dAlterations in chromatin organization described. (H3K9me3) Histone H3 thrimethylated at Lys 9; (HP1) heterochromatin-associated protein 1; (H3K9me1) histone H3 monomethylated at Lys 9; (H3K27me3) histone H3 thrimethylated at Lys 27; (H4K20me3) histone H4 thrimethylated at Lys 20.
eMicroscopic methods used.
fThis mutation does not cause an amino acid change, but results in alternative splicing (for details, see the text).
The positioning of chromosome territories may also involve lamins. For example, there is some evidence that chromosome territories 13 and 18, normally positioned at the nuclear periphery, are displaced into the nuclear interior in the cells from some types of laminopathy patients (Meaburn et al. 2007). Furthermore, MEFs deficient in either lamin B1 or the lamin B1 processing enzyme Rce1 display global changes in gene expression and a movement of the gene-poor chromosome 18 away from the lamina region (Malhas et al. 2007). The latter was accompanied by the up-regulation of a gene cluster on that chromosome. Further evidence supporting a role for lamins in chromosome positioning comes from the finding that the normal localization at the nuclear periphery of a genetic locus on human chromosome 4 associated with facioscapulohumeral muscular dystrophy (FSHD) is altered in human lamin A/C-null cells (Masny et al. 2004).
Studies of lamin degradation during apoptosis also support a role for lamins in chromatin organization. Following the induction of apoptosis, cell death and DNA condensation are greatly delayed in cells expressing mutants of lamins A or B that render them uncleavable by caspases (Rao et al. 1996). In addition, inhibition of caspase-6-mediated lamin A/C cleavage blocks chromatin condensation and apoptotic body formation (Ruchaud et al. 2002). Furthermore, expression of caspase-8 in mesenchymal stem cells alters lamin organization, causing deformation of the NE. These changes are accompanied by telomere aggregation and the translocation of centromeres to more peripheral locations (Raz et al. 2006).
Lamins in the epigenetic regulation of chromatin: consequences and potential mechanisms
Although little is known about the detailed mechanisms underlying the specific interactions between lamins and the organization of chromatin, some insights have been revealed in studies of the human laminopathies. For example, the decrease of the heterochromatin mark H3K9me3 in pericentric regions observed in HGPS cells carrying the 1824C > T mutation is accompanied by an up-regulation of pericentric chromosome 9 sat III transcripts (Shumaker et al. 2006). This suggests that there is a link between changes in histone methylation caused by LAΔ50/progerin and transcriptional activation of normally heterochromatic regions. Furthermore, the expression of LAΔ50/progerin also results in the loss of H3K27me3 on the inactive X chromosome (Xi), which leads to some decondensation of the Xi (Shumaker et al. 2006). These changes frequently occur prior to alterations in nuclear shape and do not result in a dissociation of XIST RNA from the Xi. However, it remains unknown whether there is transcriptional activation of the Xi in HGPS. The reduction in H3K27me3 is probably the direct result of the significant decrease in the expression of the histone methyltransferase (HMT), enhancer of zeste homolog (EZH2), that has been reported in HGPS fibroblasts (Shumaker et al. 2006). Changes in histone methylation caused by the expression of mutant lamins could also account for telomere shortening reported in cells from HGPS and WS patients (Hofer et al. 2005), and in normal cells overexpressing HGPS lamin A mutants (Huang et al. 2008). Telomeres are normally enriched in the heterochromatic marks H3K9me3 and H4K20me3 (Blasco 2007). Furthermore, a decrease in H4K20me3 has been related to telomere elongation (Benetti et al. 2007). Therefore the increase in H4K20me3 and decrease in H3K9me3 (Shumaker et al. 2006) may be the cause of telomere shortening observed in HGPS.
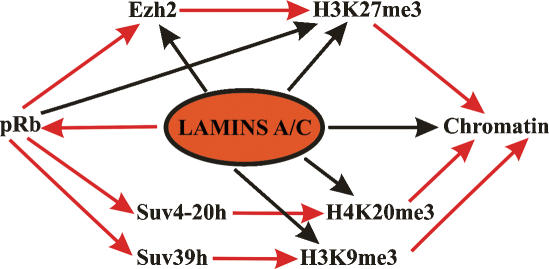
A potential mediator of the changes observed in histone methylation in the various laminopathies is the retinoblastoma gene product (pRb) (Fig. 5), which binds to A-type lamins and is altered in cells either expressing mutant lamins A/C or deficient in lamins A/C (for details, see below). For example, pRb regulates the expression of EZH2 and is required for H3K27 trimethylation (Bracken et al. 2003; Kotake et al. 2007). Furthermore, pRb associates with and regulates the HMTs SUV39H1 and Suv4-20h responsible for H3K9 methylation and H4K20me3, respectively (Nielsen et al. 2001; Gonzalo et al. 2005; Isaac et al. 2006). In addition, lamins may also interact directly with HMTs and histone demethylases and/or regulate the state of histone methylation by direct interaction with histones. These interactions may be impaired due to the expression of mutant lamins, leading to alterations in histone methylation patterns.
Figure 5.
Schematic diagram depicting potential pathways in the epigenetic regulation of chromatin by lamins A/C. Red arrows indicate a direct regulation and/or interaction, whereas black arrows indicate indirect evidences for a role of lamins A/C in the regulation of the respective factors. The retinoblastoma protein may be an important link in these processes (for details, see the text).
Lamins in DNA replication
Metazoan cells contain ∼30,000 replicons (Keller et al. 2002), which are spatially and temporally regulated during DNA replication in S phase. There is a significant amount of data indicating that lamins play an important role in the regulation of this complex process. For example, lamin B1 colocalizes with the replication factor proliferating cell nuclear antigen (PCNA) in replication factories during late S phase in 3T3 cells (Moir et al. 1994), and lamin A is associated with sites of early replication in primary fibroblasts (Kennedy et al. 2000). The role of lamins in DNA replication has been most extensively studied in nuclei assembled in vitro in Xenopus egg interphase extracts. If the major lamin in Xenopus eggs, XLB3, is immunodepleted from these extracts prior to nuclear assembly or its assembly state is disrupted by introducing a dominant-negative N-terminally deleted mutant lamin, DNA replication is inhibited (Newport et al. 1990; Meier et al. 1991; Ellis et al. 1997; Spann et al. 1997; Moir et al. 2000a). The addition of this dominant-negative mutant causes the redistribution of endogenous lamins into intranuclear foci along with the DNA replication elongation factors PCNA and replication factor C (RFC). Under these conditions, the DNA replication initiation factors, DNA polymerase α, minichromosome maintenance protein 3 (MCM3), and the origin recognition complex (ORC) protein Orc2 appear to function normally, as supported by the synthesis of short pieces of DNA that correspond to Okazaki fragments (Spann et al. 1997; Moir et al. 2000a). When these lamin-disrupted nuclei are transferred to fresh untreated extracts, the chain elongation phase of replication is rapidly reinitiated as the abnormal lamin foci disappear and normal lamin organization is re-established (Moir et al. 2000a). An association between lamins and PCNA during replication is further supported by the finding that PCNA is more readily extracted from nuclei assembled in lamin-depleted extracts (Jenkins et al. 1993). These data suggest that nuclear lamins play a role in organizing a nucleoplasmic scaffold that is necessary for the elongation phase of replication. Support for this hypothesis comes from nuclei assembled in Xenopus extracts containing Zn2+/Fe2+ chelators. These nuclei have assembled NEs and NPCs but are deficient in lamins and are unable to replicate their DNA (Shumaker et al. 1998).
Lamins in transcription
Evidence for a role of lamins in transcription comes from studies using N-terminally deleted lamins, which disrupt the head-to-tail assemblies of lamin dimers and higher-order lamin structures. When these mutant lamins are introduced into nuclei of either BHK 21 cells or of Xenopus embryos, they drive the disassembly of the endogenous lamin network, leading to an impairment of transcription as monitored by a reduction in BrUTP incorporation (Spann et al. 2002). Specifically, the activity of RNA polymerase II (pol II), but not of pol I or pol III, was found to be inhibited. These mutants also induce the formation of lamin aggregates, which recruit the TATA-binding protein (TBP), a factor known to play an important role in the initiation of transcription (Spann et al. 2002). However, the specific inhibition of pol II activity by the mutant lamin cannot be attributed to the recruitment of TBP to lamin aggregates, as TBP associates with all three RNA polymerases (Gill and Tjian 1992). Therefore, the possibility that pol II itself or pol II-specific binding partners are also recruited to these aggregates has to be investigated in the future. In agreement with the above findings overexpression of either lamin A or C leads to a dramatic reduction in pol II transcription in HeLa cells (Kumaran et al. 2002). Further evidence for a role of lamins in transcription comes from the finding that lamins A/C interact with the zinc finger transcription factor MOK2 (Dreuillet et al. 2002) and that MOK2 mislocalizes into abnormal nuclear aggregates upon the expression of pathogenic lamin A/C mutants (Dreuillet et al. 2008).
Lamins in cell proliferation
There is also evidence that lamins A/C are involved in gene expression related to the regulation of the cell cycle. Most notably the A-type lamins appear to interact in vitro with pRb (Ozaki et al. 1994; Markiewicz et al. 2005). pRb is a tumor suppressor and major cell cycle regulator that in its hypophosphorylated state binds and inhibits the E2 factor (E2F) family of transcription factors required for cell cycle progression (Giacinti and Giordano 2006). Upon hyperphosphorylation of pRb by cyclin/cyclin-dependent kinase (CDK) complexes, E2F is released to initiate S phase. A role for lamins in this process is further suggested by the finding that hypophosphorylated pRb is tightly associated with lamin A/C-enriched nucleoskeletal preparations of early G1 cells (Mancini et al. 1994). Furthermore, primary cultures of fibroblasts contain intranuclear foci associated with both lamins A/C and pRb from early G1 until early S phase (Kennedy et al. 2000). In addition, overexpression of lamin mutants that form nuclear aggregates also recruit pRb (Markiewicz et al. 2002; Hubner et al. 2006).
Lamins also seem to be involved in regulating the state of pRb phosphorylation. This is supported by the decrease in pRb dephosphorylation found in lamin A/C-deficient cells in response to the growth factor-β1 (TGF-β1) and by the evidence that PP2A phosphatase forms a complex with lamins A/C and pRb (Van Berlo et al. 2005). In addition, there is evidence for a significant reduction in hyperphosphorylated pRb in HGPS fibroblasts, although total pRb levels appear unaltered (Dechat et al. 2007).
Immunoprecipitation, in vitro binding, and differential extraction assays suggest that pRb is retained in the nucleus in a complex containing lamins A/C and LAP2α (Markiewicz et al. 2002; Johnson et al. 2004). In addition, there is a dramatic reduction in pRb levels in LMNA−/− fibroblasts, suggesting that lamins are involved in regulating the turnover and proteasomal degradation of pRb (Johnson et al. 2004). Similar decreases in pRb levels are also found in the liver of ZMPSTE24−/− mice, most likely related to the accumulation of abnormally farnesylated and carboxymethylated lamin A (Varela et al. 2005). Based on these findings, it has been suggested that the increased rate of cell proliferation and the decreased capacity to undergo cell cycle arrest reported for lamin A/C-deficient cells is attributable to the destabilization of pRb (Johnson et al. 2004; Van Berlo et al. 2005). This destabilization is independent of gankyrin and MDM2, two oncoproteins involved in pRb degradation (Nitta et al. 2007). In further support of a role for lamin A/C/LAP2α/pRb complexes in cell cycle control, it has been shown that down-regulation of LAP2α is accompanied by an acceleration of cell growth and impairment of cell cycle exit upon serum starvation (Dorner et al. 2006). In contrast to these results, it has also been shown that the down-regulation of either lamins A/C or LAP2α can cause G1 cell cycle arrest in human dermal fibroblasts (Pekovic et al. 2007). These latter results conflict with the other studies showing accelerated proliferation in lamin A- or Lap2α-deficient cells. This conflict may be attributable to the different cell types used in the individual studies. Obviously the resolution of these differences is an important topic for future investigations.
Based on these findings, it is interesting to speculate that LMNA mutations could impair the interactions between lamins A/C and pRb. Such mutations could alter the tumor-suppressing functions of pRb, leading to tumorigenesis. Indeed, lamins appear to play a role in cancer (Prokocimer et al. 2006), although it is not clear yet if they are directly involved in the development and progression of cancer or if the changes found in lamin expression levels in various cancers (for example, see Gaedtke et al. 2007; Hudson et al. 2007) are an indirect consequence of cellular transformation.
Lamins in differentiation
The developmental regulation of lamin expression has led various laboratories to suggest that these IF proteins play a role in differentiation. As indicated above, B-type lamins are constitutively expressed, while lamins A/C are first expressed when differentiation is initiated during early development. More specifically, in the case of muscle, there are pRb- and cyclin D3-dependent changes in lamin A/C organization and solubility during myoblast differentiation (Muralikrishna et al. 2001; Mariappan and Parnaik 2005; Markiewicz et al. 2005). Cyclin D3—which is a regulatory subunit of CDK4, CDK6, and CDK2 and is critical for G1 progression—has also been reported to interact directly with lamins A/C (Mariappan et al. 2007). Furthermore, expression of an EDMD lamin A mutant in C2C12 myoblasts, which induces elevated levels of hyperphosphorylated pRb, leads to an inhibition of the differentiation of myoblasts into myotubes (Favreau et al. 2003). Lamin A also appears to form a complex with the muscle-specific transcription factor MyoD (Bakay et al. 2006) and muscular dystrophy mutations in lamin A and emerin that inhibit skeletal muscle regeneration have been related to a disruption in the pRb–MyoD pathway (Bakay et al. 2006; Frock et al. 2006; Melcon et al. 2006). In mice engineered to express a homozygous mutation in LMNA linked to human AD-EDMD or deficient in emerin, the mitogen-activated protein kinase (MAPK) signaling pathway appears to be abnormally activated (Muchir et al. 2007a, b). These findings, as well as those derived from studies of other laminopathies (e.g., HGPS, MAD, FPLD) (Capanni et al. 2003; Robinson et al. 2003; Amati et al. 2004; Csoka et al. 2004; Scaffidi and Misteli 2005) suggest that global changes in gene expression that can lead to inhibition of cell differentiation are a common feature of the laminopathies. With respect to this, two similar models for the development of laminopathies have been proposed recently based on an impairment of adult stem cell differentiation involved in tissue regeneration (Gotzmann and Foisner 2006; Halaschek-Wiener and Brooks-Wilson 2007).
The association of some LMNA mutations with the development of lipodystrophies has led to the suggestion that lamins play a role in adipocyte differentiation. Further evidence for this comes from the finding that the adipocyte differentiation factor, sterol response element-binding protein 1 (SREBP1), interacts with the C terminus of lamins A/C and that this interaction is impaired to various degrees by FPLD mutations in lamin A (Lloyd et al. 2002; Hubner et al. 2006). Another study shows impaired adipocyte differentiation in 3T3 cells overexpressing either wild-type lamin A or an FPLD mutant lamin A, while lamins A/C-deficient MEFs differentiated more readily into fat-containing cells than control cells (Boguslavsky et al. 2006). Taken together, these findings suggest that the ratio between lamin A and SREBP1 as well as their proper interaction are required for normal adipocyte differentiation and that alterations in this interaction contribute to the development of lipodystrophies.
Lamins in DNA repair
Evidence for a role for the nuclear lamins in DNA repair comes mainly from studies of premature aging diseases, as it has been shown that the HGPS mutant protein, LAΔ50/progerin, or farnesylated pre-lamin A lead to defects in DNA repair. For example, the recruitment of the repair factor p53-binding protein (53BP1) to sites of DNA damage is impaired and the presence of fragmented DNA after irradiation is prolonged in HGPS fibroblasts and in ZMPSTE24−/− MEFs (Liu et al. 2005). In addition, sensitivity to various DNA-damaging agents causing double-strand breaks (DSBs), to UV radiation, and to γ-irradiation is increased in ZMPSTE24−/− mice (Liu et al. 2005). These mice also show an activation of the p53 stress signaling pathway under normal conditions, most likely due to damaged DNA (Vergnes et al. 2004). Furthermore, phosphorylated histone H2AX (γ-H2AX), a hallmark of DSBs, is increased in HGPS patient fibroblasts, in RD patient fibroblasts carrying a mutation in ZMPSTE24, and in ZMPSTE24−/− mice (Liu et al. 2005, 2006; Varela et al. 2005). All these findings suggest that there is an accumulation of damaged DNA in premature aging disorders due to defects in the repair machinery.
Intriguingly, the nuclei of a subpopulation of skin fibroblasts from normal elderly donors, which express an increase in LAΔ50/progerin, display also a higher percentage of γ-H2AX foci compared with fibroblasts from young individuals (Scaffidi and Misteli 2006). Furthermore, the expression of HGPS mutant lamin A in HeLa cells increases the levels of γ-H2AX (Liu et al. 2006), and this is correlated with defects in DNA repair foci (Manju et al. 2006). These mutants also effect the localization and expression levels of ATR (for ataxia-telangiectasia-mutated [ATM] and Rad3-related) kinase, a key regulator in the DNA damage signaling pathway (Manju et al. 2006). Changes in the normal localization of ATM and ATR kinases, reflecting an activation of these enzymes, were also observed in HGPS and RD fibroblasts (Liu et al. 2006). Interestingly, knockdown of the ATR and ATM kinases by siRNA rescued the early replication arrest phenotype seen in HGPS and RD fibroblasts (Liu et al. 2006). On the other hand, treatment of these cells with an FTI, which restores normal nuclear shape, does not appear to reverse the abnormalities seen in the DNA repair system (Liu et al. 2006).
Other insights into the mechanisms underlying the accumulation of DNA damage in HGPS and RD fibroblasts come from the finding that the DSB repair factors, Rad50 and Rad51, do not colocalize with the increased number of γtH2AX foci present in these cells (Liu et al. 2007). This suggests that the targeting of these essential factors to the DSB repair machinery is inhibited when mutant lamin A is present. Interestingly, the nucleotide excision repair factor, xeroderma pigmentosum group A (XPA), which is not involved in DSB repair, is found at these abnormal γtH2AX foci, which appears to inhibit normal DNA repair (Liu et al. 2007).
Taken together, these findings support defects in the DNA repair machinery in patients suffering from progeroid syndromes due to expression of LAΔ50/progerin or abnormal levels of farnesylated pre-lamin A. Similar defects in DNA repair have been reported in normal aging. Although all these studies clearly imply that nuclear lamins are involved in DNA repair, their precise role in this process remains to be elucidated.
The emerging roles of lamins in viral infections
There is increasing evidence that lamins play a role in the infection cycles of viruses. Most information in support for this comes from studies of the replication cycle of cytomegalovirus (CMV) and herpes simplex virus type-1 (HSV-1), both members of the Herpesviridae family. These viruses assemble their capsids and encapsidate their replicated viral DNA in the nucleus, while the final steps in the maturation of virions takes place in the cytoplasm (Mettenleiter et al. 2006). The nucleocapsids exit the nucleus in a two-step process involving primary envelopment at the INM and fusion of their primary membrane with the ONM to facilitate their movement into the cytoplasm. In order to gain access to the INM, the nuclear lamina has to be destabilized. There is evidence that this destabilization involves the phosphorylation and disassembly of the lamins specifically in locations where viruses are accessing the INM (Radsak et al. 1991).
Viruses and the disassembly of lamins
In the search for candidate viral proteins playing a role in the lamin disassembly process two murine CMV proteins, M50/p35 and M53/p38, have been identified (Muranyi et al. 2002). When M50/p35, a type II transmembrane protein, is expressed in CV-1 cells, it localizes to the NE. Lamins A, B, and C appear to be excluded from areas where M50/p35 accumulates. Coexpression of M50/p35 and M53/p38 as well as CMV infection leads to the recruitment of Ca-dependent protein kinases C (PKCs) from the cytoplasm to the NE, resulting in increased phosphorylation of lamins A, B, and C as determined by 32P-incorporation (Muranyi et al. 2002). In agreement with these findings, there is evidence that pUL50, the human CMV counterpart of murine M50/p35, interacts directly with PKCε and PKCζ and can recruit PKCα to the NE (Milbradt et al. 2007). These findings suggest that the recruitment of PKCs by M50/p35 leads to the localized phosphorylation and disassembly of the lamins within the lamina. Besides PKCs, the human CMV kinase pUL97 has been shown to phosphorylate lamins A, C, and B1 in vitro and to lead to the redistribution of lamins A/C from the NE to intranuclear foci when overexpressed in cells (Marschall et al. 2005). Localization of pUL97 to the NE is mediated by direct interaction with the LBR-binding protein p32 (Marschall et al. 2005).
A local loss of lamins A/C from the nuclear lamina has also been observed in HSV-1 infected cells (Scott and O’Hare 2001). The HSV-1 proteins pUL31 and pUL34, homologs of M50/p35 and M53/p38, have been shown to be involved in the redistribution of lamins from the NE in infected cells (Reynolds et al. 2004; Simpson-Holley et al. 2005). In these studies, both proteins have been shown to localize to the nuclear lamina and to bind directly to lamins A/C in vitro. Furthermore, pUL31 and pUL32 recruit PKCδ, a proapototic kinase, and PKCα to the NE (Park and Baines 2006). This leads to the phosphorylation of lamin B as determined by 32P-incorporation. Uniform distribution of the PKCs along the NE appears to be dependent on the HSV-1 kinase US3. The US3 can also phosphorylate lamins A/C in vitro, and its overexpression leads to changes in nuclear shape and disruptions in lamins A, B, and C in the lamina region (Bjerke and Roller 2006; Mou et al. 2007). In addition, two Epstein-Barr virus (EBV) proteins, BFRF1 and BFLF2 (homologs of pUL31 and pUL34), required for envelopment and egress from the nucleus, are located in the NE region, where they are suggested to form a complex with lamin B (Gonnella et al. 2005). Taken together, it appears that the recruitment of either host cells or viral kinases to the NE, where they phosphorylate and disassemble lamin structures, represent major steps in the process of the primary envelopment and migration of Herpesviridae viruses from the nucleus to the cytoplasm. It will be of great interest in the future to determine the specific sites of phosphorylation and their impact on lamin structure and assembly states.
Lamins are also involved in human immunodeficiency virus 1 (HIV1) infections. The expression of HIV1 Vpr, a virion-associated protein, induces herniations of the NE lacking the A- and B-type lamins and NPCs (de Noronha et al. 2001). These herniations frequently rupture, permitting the movement of the Wee1 kinase from the nucleus to the cytoplasm and of the Cdc25C phosphatase and cyclin B1 in the reverse direction. All three proteins are regulators of the mitotic kinase CDK1, and changes in their subcellular distribution appear to be required for the G2/M-phase transition (Ferrari 2006; Trinkle-Mulcahy and Lamond 2006). Therefore it has been proposed that these disruptions in the nuclear lamins are related to mechanisms responsible for the G2 cell cycle arrest in HIV1-infected cells (de Noronha et al. 2001).
Lamin-binding partners and virus infectivity
The lamin A-binding proteins BAF, LAP2α, and emerin have also been implicated in viral infections. BAF was originally described as a facilitator of the preintegration complexes (PICs) involved in the incorporation of retroviral cDNA into the genome of host mammalian cells (Segura-Totten and Wilson 2004). Recently, it has been shown that LAP2α stabilizes the association of BAF with the PIC of the Moloney murine leukemia virus (MMLV) and that knockdown of LAP2α impairs viral replication in NIH3T3 cells (Suzuki et al. 2004). Emerin appears to be required for the efficient integration of viral cDNA into the host genome in HIV1 infections in primary macrophages (Jacque and Stevenson 2006). However, in HeLa cells silenced for emerin, BAF, or LAP2α or in emerin-deficient MEFs, both HIV1 and MMLV infectivity appears to be normal (Shun et al. 2007). The discrepancy in the various observations could be explained by different compensatory systems or factors associated with the nuclear envelope present in the various types of cells used. It is obvious, however, that lamins and several of their binding partners are involved in distinct processes of the infection cycles of various viruses.
Viral therapeutics and lamins
It is also interesting to consider changes in lamin function that may be attributable to therapeutic protocols used in virus infections. For example, HIV-infected patients treated with highly active anti-retroviral therapy (HAART) often develop lipodystrophies resembling those caused by mutations in LMNA (for example, see Behrens et al. 2000). In HAART, HIV patients are treated with a combination of several drugs, including specific HIV protease inhibitors (PIs) (Hoffmann and Mulcahy 2006). Recent findings suggest that defective pre-lamin A processing caused by some of the PIs used in the HAART treatment (Fig. 1B) contribute to the lipodystrophy phenotype (Clarke 2007). Altered distributions of lamins A/C and B, accumulation of pre-lamin A, nuclear deformations, and a down-regulation and abnormal accumulation at the NE of the adipocyte differentiation factor SREBP1 have all been reported in 3T3 preadipocytes treated with HAART PIs (Caron et al. 2003). These alterations accompany defects in the process of the differentiation of these cells into adipocytes.
Two recent studies have provided evidence in support of a more direct involvement of the PI-induced defects in pre-lamin A processing in the development of lipodystrophies in HIV patients. First, fibroblasts from lipodystrophy patients and control fibroblasts treated with PIs display similar phenotypes (Caron et al. 2007). These phenotypes include pre-lamin A accumulation, alterations in the NE, and premature replicative senescence, all of which can be reversed with FTIs. Secondly, inhibition of Zmpste24 by PIs used in the HAART treatment (Fig. 1B) leads to an accumulation of pre-lamin A in human and mouse fibroblasts similar to that observed in ZMPSTE24−/− fibroblasts (Coffinier et al. 2007). A recent report shows a reduction in lamin A but not lamin C mRNA levels in abdominal subcutaneous adipose tissues from HAART-treated HIV-positive patients whether these patients had clinical indications of lipodystrophy (Miranda et al. 2007). These findings suggest that HAART treatment affects lamin A processing both at the transcriptional and post-translational levels and raises the possibility that feedback control mechanisms connecting these two levels of lamin A regulation may be involved. Although these data are very interesting, there is no direct proof that defects in lamin A processing are responsible for the development of lipodystrophies in HIV patients treated with HAART. In fact, a recent study shows that the down-regulation of lamins A/C does not appear to alter the inhibitory effect on adipocyte differentiation by PIs (Kudlow et al. 2005).
Summary and conclusions
In summary, there has been an explosion in interest in the nuclear lamins over the past few years. Studies of normal cells and cells from the multitude of laminopathy patients demonstrate that these type V IF proteins play important and in some cases essential roles in a wide range of nuclear functions, including transcription, DNA replication, DNA repair, and cell proliferation and differentiation. However, the detailed mechanisms remain unknown and have to be determined in future studies. In addition, lamins are suggested to take part in the structural organization and epigenetic regulation of chromatin. Although it is clear that chromatin organization and histone methylation are altered in diseases caused by mutant lamins, virtually nothing is known about the specific functions of lamins in these processes under normal physiological conditions. Since lamins are expressed in a developmentally regulated manner, they may be involved in the regulation of the epigenetic changes responsible for cell differentiation (Surani et al. 2007), specifically in the differentiation steps of embryonic (which are devoid of lamins A/C) and adult stem cells.
The majority of the data available led us to propose a model in which lamins act as a dynamic molecular scaffold for chromatin and chromatin interacting/modifying proteins throughout the nucleus (Goldman et al. 2002; Shumaker et al. 2003). This scaffold may vary from cell type to cell type, dependent on the expression levels of the various lamin isoforms. In this way it may contribute to the determination of the specific functions of the respective cell. In addition, different disease-causing mutations appear to differ in their structural properties in vitro and in situ (Wiesel et al. 2008), suggesting that they affect the various lamin structures present in the nucleus in different ways. Taken together, cell-type- and mutant-dependent alterations in the lamin scaffold may be attributable to the variations in phenotypes and affected tissues observed in the various diseases caused by the enormous number of mutations in LMNA described so far.
Acknowledgments
We thank Dr. Stephen Adam (Northwestern University Medical School, Chicago, IL) for critically reading the manuscript. Support for our research program comes from the National Cancer Institute (no. CA031760), the National Institute on Aging (no. AG023776), and a Senior Scholar Award from the Ellison Medical Research Foundation (no. AG-SS-1344-04).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1652708.
References
- Aaronson R.P., Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc. Natl. Acad. Sci. 1975;72:1007–1011. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Amati F., Biancolella M., D’Apice M.R., Gambardella S., Mango R., Sbraccia P., D’Adamo M., Margiotti K., Nardone A., Lewis M., et al. Gene expression profiling of fibroblasts from a human progeroid disease (mandibuloacral dysplasia, MAD #248370) through cDNA microarrays. Gene Expr. 2004;12:39–47. doi: 10.3727/000000004783992189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora P., Muralikrishna B., Parnaik V.K. Cell-type-specific interactions at regulatory motifs in the first intron of the lamin A gene. FEBS Lett. 2004;568:122–128. doi: 10.1016/j.febslet.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Bakay M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C., et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb–MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- Barboro P., D’Arrigo C., Diaspro A., Mormino M., Alberti I., Parodi S., Patrone E., Balbi C. Unraveling the organization of the internal nuclear matrix: RNA-dependent anchoring of NuMA to a lamin scaffold. Exp. Cell Res. 2002;279:202–218. doi: 10.1006/excr.2002.5605. [DOI] [PubMed] [Google Scholar]
- Baricheva E.A., Berrios M., Bogachev S.S., Borisevich I.V., Lapik E.R., Sharakhov I.V., Stuurman N., Fisher P.A. DNA from Drosophila melanogaster β-heterochromatin binds specifically to nuclear lamins in vitro and the nuclear envelope in situ. Gene. 1996;171:171–176. doi: 10.1016/0378-1119(96)00002-9. [DOI] [PubMed] [Google Scholar]
- Behrens G.M., Lloyd D., Schmidt H.H., Schmidt R.E., Trembath R.C. Lessons from lipodystrophy: LMNA, encoding lamin A/C, in HIV therapy-associated lipodystrophy. AIDS. 2000;14:1854–1855. doi: 10.1097/00002030-200008180-00023. [DOI] [PubMed] [Google Scholar]
- Belmont A.S., Zhai Y., Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J. Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benavente R., Krohne G., Franke W.W. Cell type-specific expression of nuclear lamina proteins during development of Xenopus laevis. Cell. 1985;41:177–190. doi: 10.1016/0092-8674(85)90072-8. [DOI] [PubMed] [Google Scholar]
- Benetti R., Gonzalo S., Jaco I., Schotta G., Klatt P., Jenuwein T., Blasco M.A. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 2007;178:925–936. doi: 10.1083/jcb.200703081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke S.L., Roller R.J. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology. 2006;347:261–276. doi: 10.1016/j.virol.2005.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- Boguslavsky R.L., Stewart C.L., Worman H.J. Nuclear lamin A inhibits adipocyte differentiation: Implications for Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2006;15:653–663. doi: 10.1093/hmg/ddi480. [DOI] [PubMed] [Google Scholar]
- Bonne G., Di Barletta M.R., Varnous S., Becane H.M., Hammouda E.H., Merlini L., Muntoni F., Greenberg C.R., Gary F., Urtizberea J.A., et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Protein–protein and protein–DNA interactions in calf thymus nuclear matrix using cross-linking by ultraviolet irradiation. Biochem. Cell Biol. 1986;64:474–484. doi: 10.1139/o86-066. [DOI] [PubMed] [Google Scholar]
- Bouvier D., Hubert J., Seve A.P., Bouteille M. Characterization of lamina-bound chromatin in the nuclear shell isolated from HeLa cells. Exp. Cell Res. 1985;156:500–512. doi: 10.1016/0014-4827(85)90557-9. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Pasini D., Capra M., Prosperini E., Colli E., Helin K. EZH2 is downstream of the pRB–E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger J.M., Kill I.R., O’Farrell M., Hutchison C.J. Internal lamin structures within G1 nuclei of human dermal fibroblasts. J. Cell Sci. 1993;104:297–306. doi: 10.1242/jcs.104.2.297. [DOI] [PubMed] [Google Scholar]
- Broers J.L., Machiels B.M., Kuijpers H.J., Smedts F., van den Kieboom R., Raymond Y., Ramaekers F.C. A- and B-type lamins are differentially expressed in normal human tissues. Histochem. Cell Biol. 1997;107:505–517. doi: 10.1007/s004180050138. [DOI] [PubMed] [Google Scholar]
- Broers J.L., Machiels B.M., van Eys G.J., Kuijpers H.J., Manders E.M., van Driel R., Ramaekers F.C. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Broers J.L., Ramaekers F.C., Bonne G., Yaou R.B., Hutchison C.J. Nuclear lamins: Laminopathies and their role in premature ageing. Physiol. Rev. 2006;86:967–1008. doi: 10.1152/physrev.00047.2005. [DOI] [PubMed] [Google Scholar]
- Burke B. On the cell-free association of lamins A and C with metaphase chromosomes. Exp. Cell Res. 1990;186:169–176. doi: 10.1016/0014-4827(90)90223-w. [DOI] [PubMed] [Google Scholar]
- Cao K., Capell B.C., Erdos M.R., Djabali K., Collins F.S. A lamin A protein isoform overexpressed in Hutchinson-Gilford progeria syndrome interferes with mitosis in progeria and normal cells. Proc. Natl. Acad. Sci. 2007;104:4949–4954. doi: 10.1073/pnas.0611640104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capanni C., Cenni V., Mattioli E., Sabatelli P., Ognibene A., Columbaro M., Parnaik V.K., Wehnert M., Maraldi N.M., Squarzoni S., et al. Failure of lamin A/C to functionally assemble in R482L mutated familial partial lipodystrophy fibroblasts: Altered intermolecular interaction with emerin and implications for gene transcription. Exp. Cell Res. 2003;291:122–134. doi: 10.1016/s0014-4827(03)00395-1. [DOI] [PubMed] [Google Scholar]
- Capco D.G., Wan K.M., Penman S. The nuclear matrix: Three-dimensional architecture and protein composition. Cell. 1982;29:847–858. doi: 10.1016/0092-8674(82)90446-9. [DOI] [PubMed] [Google Scholar]
- Caron M., Auclair M., Sterlingot H., Kornprobst M., Capeau J. Some HIV protease inhibitors alter lamin A/C maturation and stability, SREBP-1 nuclear localization and adipocyte differentiation. AIDS. 2003;17:2437–2444. doi: 10.1097/00002030-200311210-00005. [DOI] [PubMed] [Google Scholar]
- Caron M., Auclair M., Donadille B., Bereziat V., Guerci B., Laville M., Narbonne H., Bodemer C., Lascols O., Capeau J., et al. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ. 2007;14:1759–1767. doi: 10.1038/sj.cdd.4402197. [DOI] [PubMed] [Google Scholar]
- Clarke S.G. HIV protease inhibitors and nuclear lamin processing: Getting the right bells and whistles. Proc. Natl. Acad. Sci. 2007;104:13857–13858. doi: 10.1073/pnas.0706529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements L., Manilal S., Love D.R., Morris G.E. Direct interaction between emerin and lamin A. Biochem. Biophys. Res. Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- Coffinier C., Hudon S.E., Farber E.A., Chang S.Y., Hrycyna C.A., Young S.G., Fong L.G. HIV protease inhibitors block the zinc metalloproteinase ZMPSTE24 and lead to an accumulation of prelamin A in cells. Proc. Natl. Acad. Sci. 2007;104:13432–13437. doi: 10.1073/pnas.0704212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Columbaro M., Capanni C., Mattioli E., Novelli G., Parnaik V.K., Squarzoni S., Maraldi N.M., Lattanzi G. Rescue of heterochromatin organization in Hutchinson-Gilford progeria by drug treatment. Cell. Mol. Life Sci. 2005;62:2669–2678. doi: 10.1007/s00018-005-5318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinescu D., Gray H.L., Sammak P.J., Schatten G.P., Csoka A.B. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells. 2006;24:177–185. doi: 10.1634/stemcells.2004-0159. [DOI] [PubMed] [Google Scholar]
- Corrigan D.P., Kuszczak D., Rusinol A.E., Thewke D.P., Hrycyna C.A., Michaelis S., Sinensky M.S. Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem. J. 2005;387:129–138. doi: 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I., Fakan S. Chromosome territories—A functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Crisp M., Liu Q., Roux K., Rattner J.B., Shanahan C., Burke B., Stahl P.D., Hodzic D. Coupling of the nucleus and cytoplasm: Role of the LINC complex. J. Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft J.A., Bridger J.M., Boyle S., Perry P., Teague P., Bickmore W.A. Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csoka A.B., English S.B., Simkevich C.P., Ginzinger D.G., Butte A.J., Schatten G.P., Rothman F.G., Sedivy J.M. Genome-scale expression profiling of Hutchinson-Gilford progeria syndrome reveals widespread transcriptional misregulation leading to mesodermal/mesenchymal defects and accelerated atherosclerosis. Aging Cell. 2004;3:235–243. doi: 10.1111/j.1474-9728.2004.00105.x. [DOI] [PubMed] [Google Scholar]
- Dahl K.N., Scaffidi P., Islam M.F., Yodh A.G., Wilson K.L., Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 2006;103:10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf A., van Bergenen Henegouwen P.M., Meijne A.M., van Driel R., Verkleij A.J. Ultrastructural localization of nuclear matrix proteins in HeLa cells using silver-enhanced ultra-small gold probes. J. Histochem. Cytochem. 1991;39:1035–1045. doi: 10.1177/39.8.1856453. [DOI] [PubMed] [Google Scholar]
- de Noronha C.M., Sherman M.P., Lin H.W., Cavrois M.V., Moir R.D., Goldman R.D., Greene W.C. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science. 2001;294:1105–1108. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., Lyonnet S., Stewart C.L., Munnich A., Le Merrer M., et al. Lamin a truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- Dechat T., Gotzmann J., Stockinger A., Harris C.A., Talle M.A., Siekierka J.J., Foisner R. Detergent-salt resistance of LAP2α in interphase nuclei and phosphorylation-dependent association with chromosomes early in nuclear assembly implies functions in nuclear structure dynamics. EMBO J. 1998;17:4887–4902. doi: 10.1093/emboj/17.16.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T., Korbei B., Vaughan O.A., Vlcek S., Hutchison C.J., Foisner R. Lamina-associated polypeptide 2α binds intranuclear A-type lamins. J. Cell Sci. 2000;113:3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- Dechat T., Gajewski A., Korbei B., Gerlich D., Daigle N., Haraguchi T., Furukawa K., Ellenberg J., Foisner R. LAP2α and BAF transiently localize to telomeres and specific regions on chromatin during nuclear assembly. J. Cell Sci. 2004;117:6117–6128. doi: 10.1242/jcs.01529. [DOI] [PubMed] [Google Scholar]
- Dechat T., Shimi T., Adam S.A., Rusinol A.E., Andres D.A., Spielmann H.P., Sinensky M.S., Goldman R.D. Alterations in mitosis and cell cycle progression caused by a mutant lamin A known to accelerate human aging. Proc. Natl. Acad. Sci. 2007;104:4955–4960. doi: 10.1073/pnas.0700854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhe-Paganon S., Werner E.D., Chi Y.I., Shoelson S.E. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 2002;277:17381–17384. doi: 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Dorner D., Vlcek S., Foeger N., Gajewski A., Makolm C., Gotzmann J., Hutchison C.J., Foisner R. Lamina-associated polypeptide 2α regulates cell cycle progression and differentiation via the retinoblastoma–E2F pathway. J. Cell Biol. 2006;173:83–93. doi: 10.1083/jcb.200511149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner D., Gotzmann J., Foisner R. Nucleoplasmic lamins and their interaction partners, LAP2α, Rb, and BAF, in transcriptional regulation. FEBS J. 2007;274:1362–1373. doi: 10.1111/j.1742-4658.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Dreuillet C., Tillit J., Kress M., Ernoult-Lange M. In vivo and in vitro interaction between human transcription factor MOK2 and nuclear lamin A/C. Nucleic Acids Res. 2002;30:4634–4642. doi: 10.1093/nar/gkf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreuillet C., Harper M., Tillit J., Kress M., Ernoult-Lange M. Mislocalization of human transcription factor MOK2 in the presence of pathogenic mutations of lamin A/C. Biol. Cell. 2008;100:51–61. doi: 10.1042/BC20070053. [DOI] [PubMed] [Google Scholar]
- Ellis D.J., Jenkins H., Whitfield W.G., Hutchison C.J. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J. Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Eriksson M., Brown W.T., Gordon L.B., Glynn M.W., Singer J., Scott L., Erdos M.R., Robbins C.M., Moses T.Y., Berglund P., et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau C., Dubosclard E., Ostlund C., Vigouroux C., Capeau J., Wehnert M., Higuet D., Worman H.J., Courvalin J.C., Buendia B. Expression of lamin A mutated in the carboxyl-terminal tail generates an aberrant nuclear phenotype similar to that observed in cells from patients with Dunnigan-type partial lipodystrophy and Emery-Dreifuss muscular dystrophy. Exp. Cell Res. 2003;282:14–23. doi: 10.1006/excr.2002.5669. [DOI] [PubMed] [Google Scholar]
- Fawcett D.W. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 1966;119:129–145. doi: 10.1002/aja.1001190108. [DOI] [PubMed] [Google Scholar]
- Ferrari S. Protein kinases controlling the onset of mitosis. Cell. Mol. Life Sci. 2006;63:781–795. doi: 10.1007/s00018-005-5515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields A.P., Thompson L.J. The regulation of mitotic nuclear envelope breakdown: A role for multiple lamin kinases. Prog. Cell Cycle Res. 1995;1:271–286. doi: 10.1007/978-1-4615-1809-9_22. [DOI] [PubMed] [Google Scholar]
- Filesi I., Gullotta F., Lattanzi G., D’Apice M.R., Capanni C., Nardone A.M., Columbaro M., Scarano G., Mattioli E., Sabatelli P., et al. Alterations of nuclear envelope and chromatin organization in mandibuloacral dysplasia, a rare form of laminopathy. Physiol. Genomics. 2005;23:150–158. doi: 10.1152/physiolgenomics.00060.2005. [DOI] [PubMed] [Google Scholar]
- Fisher D.Z., Chaudhary N., Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foisner R., Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Fong L.G., Frost D., Meta M., Qiao X., Yang S.H., Coffinier C., Young S.G. A protein farnesyltransferase inhibitor ameliorates disease in a mouse model of progeria. Science. 2006a;311:1621–1623. doi: 10.1126/science.1124875. [DOI] [PubMed] [Google Scholar]
- Fong L.G., Ng J.K., Lammerding J., Vickers T.A., Meta M., Cote N., Gavino B., Qiao X., Chang S.Y., Young S.R., et al. Prelamin A and lamin A appear to be dispensable in the nuclear lamina. J. Clin. Invest. 2006b;116:743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Frock R.L., Kudlow B.A., Evans A.M., Jameson S.A., Hauschka S.D., Kennedy B.K. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes & Dev. 2006;20:486–500. doi: 10.1101/gad.1364906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Hotta Y. cDNA cloning of a germ cell specific lamin B3 from mouse spermatocytes and analysis of its function by ectopic expression in somatic cells. EMBO J. 1993;12:97–106. doi: 10.1002/j.1460-2075.1993.tb05635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa K., Inagaki H., Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp. Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- Furukawa K., Glass C., Kondo T. Characterization of the chromatin binding activity of lamina-associated polypeptide (LAP) 2. Biochem. Biophys. Res. Commun. 1997;238:240–246. doi: 10.1006/bbrc.1997.7235. [DOI] [PubMed] [Google Scholar]
- Gaedtke L., Thoenes L., Culmsee C., Mayer B., Wagner E. Proteomic analysis reveals differences in protein expression in spheroid versus monolayer cultures of low-passage colon carcinoma cells. J. Proteome Res. 2007;6:4111–4118. doi: 10.1021/pr0700596. [DOI] [PubMed] [Google Scholar]
- Galcheva-Gargova Z., Dessev G.N. Crosslinking of DNA to nuclear lamina proteins by UV irradiation in vivo. J. Cell. Biochem. 1987;34:163–168. doi: 10.1002/jcb.240340303. [DOI] [PubMed] [Google Scholar]
- Georgatos S.D., Stournaras C., Blobel G. Heterotypic and homotypic associations between the nuclear lamins: Site-specificity and control by phosphorylation. Proc. Natl. Acad. Sci. 1988;85:4325–4329. doi: 10.1073/pnas.85.12.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerace L., Blum A., Blobel G. Immunocytochemical localization of the major polypeptides of the nuclear pore complex–lamina fraction. Interphase and mitotic distribution. J. Cell Biol. 1978;79:546–566. doi: 10.1083/jcb.79.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacinti C., Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Gill G., Tjian R. Eukaryotic coactivators associated with the TATA box binding protein. Curr. Opin. Genet. Dev. 1992;2:236–242. doi: 10.1016/s0959-437x(05)80279-5. [DOI] [PubMed] [Google Scholar]
- Glass J.R., Gerace L. Lamins A and C bind and assemble at the surface of mitotic chromosomes. J. Cell Biol. 1990;111:1047–1057. doi: 10.1083/jcb.111.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn M.W., Glover T.W. Incomplete processing of mutant lamin A in Hutchinson-Gilford progeria leads to nuclear abnormalities, which are reversed by farnesyltransferase inhibition. Hum. Mol. Genet. 2005;14:2959–2969. doi: 10.1093/hmg/ddi326. [DOI] [PubMed] [Google Scholar]
- Goldberg M., Harel A., Brandeis M., Rechsteiner T., Richmond T.J., Weiss A.M., Gruenbaum Y. The tail domain of lamin Dm0 binds histones H2A and H2B. Proc. Natl. Acad. Sci. 1999;96:2852–2857. doi: 10.1073/pnas.96.6.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A.E., Maul G., Steinert P.M., Yang H.Y., Goldman R.D. Keratin-like proteins that coisolate with intermediate filaments of BHK-21 cells are nuclear lamins. Proc. Natl. Acad. Sci. 1986;83:3839–3843. doi: 10.1073/pnas.83.11.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A.E., Moir R.D., Montag-Lowy M., Stewart M., Goldman R.D. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J. Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R.D., Gruenbaum Y., Moir R.D., Shumaker D.K., Spann T.P. Nuclear lamins: Building blocks of nuclear architecture. Genes & Dev. 2002;16:533–547. doi: 10.1101/gad.960502. [DOI] [PubMed] [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonnella R., Farina A., Santarelli R., Raffa S., Feederle R., Bei R., Granato M., Modesti A., Frati L., Delecluse H.J., et al. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: Interactions with BFRF1 and with the nuclear lamina. J. Virol. 2005;79:3713–3727. doi: 10.1128/JVI.79.6.3713-3727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalo S., Garcia-Cao M., Fraga M.F., Schotta G., Peters A.H., Cotter S.E., Eguia R., Dean D.C., Esteller M., Jenuwein T., et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Gotzmann J., Foisner R. A-type lamin complexes and regenerative potential: A step towards understanding laminopathic diseases? Histochem. Cell Biol. 2006;125:33–41. doi: 10.1007/s00418-005-0050-8. [DOI] [PubMed] [Google Scholar]
- Gruber J., Lampe T., Osborn M., Weber K. RNAi of FACE1 protease results in growth inhibition of human cells expressing lamin A: Implications for Hutchinson-Gilford progeria syndrome. J. Cell Sci. 2005;118:689–696. doi: 10.1242/jcs.01652. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Goldman R.D., Meyuhas R., Mills E., Margalit A., Fridkin A., Dayani Y., Prokocimer M., Enosh A. The nuclear lamina and its functions in the nucleus. Int. Rev. Cytol. 2003;226:1–62. doi: 10.1016/s0074-7696(03)01001-5. [DOI] [PubMed] [Google Scholar]
- Guilly M.N., Kolb J.P., Gosti F., Godeau F., Courvalin J.C. Lamins A and C are not expressed at early stages of human lymphocyte differentiation. Exp. Cell Res. 1990;189:145–147. doi: 10.1016/0014-4827(90)90267-e. [DOI] [PubMed] [Google Scholar]
- Haithcock E., Dayani Y., Neufeld E., Zahand A.J., Feinstein N., Mattout A., Gruenbaum Y., Liu J. Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2005;102:16690–16695. doi: 10.1073/pnas.0506955102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halaschek-Wiener J., Brooks-Wilson A. Progeria of stem cells: Stem cell exhaustion in Hutchinson-Gilford progeria syndrome. J. Gerontol. A Biol. Sci. Med. Sci. 2007;62:3–8. doi: 10.1093/gerona/62.1.3. [DOI] [PubMed] [Google Scholar]
- Haque F., Lloyd D.J., Smallwood D.T., Dent C.L., Shanahan C.M., Fry A.M., Trembath R.C., Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol. Cell. Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J., Elbashir S.M., Bechert K., Tuschl T., Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- Hegele R.A., Cao H., Liu D.M., Costain G.A., Charlton-Menys V., Rodger N.W., Durrington P.N. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am. J. Hum. Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H., Foisner R. Intermediate filaments: Novel assembly models and exciting new functions for nuclear lamins. Cell. Mol. Life Sci. 2003;60:1607–1612. doi: 10.1007/s00018-003-3004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer A.C., Tran R.T., Aziz O.Z., Wright W., Novelli G., Shay J., Lewis M. Shared phenotypes among segmental progeroid syndromes suggest underlying pathways of aging. J. Gerontol. A Biol. Sci. Med. Sci. 2005;60:10–20. doi: 10.1093/gerona/60.1.10. [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Mulcahy F. ART 2006. In: Hoffmann C., et al., editors. HIV Medicine 2006. Flying Publisher; Paris: 2006. pp. 89–278. [Google Scholar]
- Hoger T.H., Krohne G., Kleinschmidt J.A. Interaction of Xenopus lamins A and LII with chromatin in vitro mediated by a sequence element in the carboxyterminal domain. Exp. Cell Res. 1991;197:280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- Holaska J.M., Lee K.K., Kowalski A.K., Wilson K.L. Transcriptional repressor germ cell-less (GCL) and barrier to autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 2003;278:6969–6975. doi: 10.1074/jbc.M208811200. [DOI] [PubMed] [Google Scholar]
- Houben F., Ramaekers F.C., Snoeckx L.H., Broers J.L. Role of nuclear lamina–cytoskeleton interactions in the maintenance of cellular strength. Biochim. Biophys. Acta. 2007;1773:675–686. doi: 10.1016/j.bbamcr.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Hozak P., Sasseville A.M., Raymond Y., Cook P.R. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- Huang S., Chen L., Libina N., Janes J., Martin G.M., Campisi J., Oshima J. Correction of cellular phenotypes of Hutchinson-Gilford progeria cells by RNA interference. Hum. Genet. 2005;118:444–450. doi: 10.1007/s00439-005-0051-7. [DOI] [PubMed] [Google Scholar]
- Huang S., Risques R.A., Martin G.M., Rabinovitch P.S., Oshima J. Accelerated telomere shortening and replicative senescence in human fibroblasts overexpressing mutant and wild-type lamin A. Exp. Cell Res. 2008;314:82–91. doi: 10.1016/j.yexcr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner S., Eam J.E., Hubner A., Jans D.A. Laminopathy-inducing lamin A mutants can induce redistribution of lamin binding proteins into nuclear aggregates. Exp. Cell Res. 2006;312:171–183. doi: 10.1016/j.yexcr.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Hudson M.E., Pozdnyakova I., Haines K., Mor G., Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C.E., Francis S.M., Martens A.L., Julian L.M., Seifried L.A., Erdmann N., Binne U.K., Harrington L., Sicinski P., Berube N.G., et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isobe K., Gohara R., Ueda T., Takasaki Y., Ando S. The last twenty residues in the head domain of mouse lamin A contain important structural elements for formation of head-to-tail polymers in vitro. Biosci. Biotechnol. Biochem. 2007;71:1252–1259. doi: 10.1271/bbb.60674. [DOI] [PubMed] [Google Scholar]
- Jacque J.M., Stevenson M. The inner-nuclear-envelope protein emerin regulates HIV-1 infectivity. Nature. 2006;441:641–645. doi: 10.1038/nature04682. [DOI] [PubMed] [Google Scholar]
- Jagatheesan G., Thanumalayan S., Muralikrishna B., Rangaraj N., Karande A.A., Parnaik V.K. Colocalization of intranuclear lamin foci with RNA splicing factors. J. Cell Sci. 1999;112:4651–4661. doi: 10.1242/jcs.112.24.4651. [DOI] [PubMed] [Google Scholar]
- Janaki Ramaiah M., Parnaik V.K. An essential GT motif in the lamin A promoter mediates activation by CREB-binding protein. Biochem. Biophys. Res. Commun. 2006;348:1132–1137. doi: 10.1016/j.bbrc.2006.07.171. [DOI] [PubMed] [Google Scholar]
- Jenkins H., Holman T., Lyon C., Lane B., Stick R., Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J. Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Johnson B.R., Nitta R.T., Frock R.L., Mounkes L., Barbie D.A., Stewart C.L., Harlow E., Kennedy B.K. A-type lamins regulate retinoblastoma protein function by promoting subnuclear localization and preventing proteasomal degradation. Proc. Natl. Acad. Sci. 2004;101:9677–9682. doi: 10.1073/pnas.0403250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller C., Hyrien O., Knippers R., Krude T. Site-specific and temporally controlled initiation of DNA replication in a human cell-free system. Nucleic Acids Res. 2002;30:2114–2123. doi: 10.1093/nar/30.10.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy B.K., Barbie D.A., Classon M., Dyson N., Harlow E. Nuclear organization of DNA replication in primary mammalian cells. Genes & Dev. 2000;14:2855–2868. doi: 10.1101/gad.842600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketema M., Wilhelmsen K., Kuikman I., Janssen H., Hodzic D., Sonnenberg A. Requirements for the localization of nesprin-3 at the nuclear envelope and its interaction with plectin. J. Cell Sci. 2007;120:3384–3394. doi: 10.1242/jcs.014191. [DOI] [PubMed] [Google Scholar]
- Kotake Y., Cao R., Viatour P., Sage J., Zhang Y., Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4α tumor suppressor gene. Genes & Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krimm I., Ostlund C., Gilquin B., Couprie J., Hossenlopp P., Mornon J.P., Bonne G., Courvalin J.C., Worman H.J., Zinn-Justin S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure. 2002;10:811–823. doi: 10.1016/s0969-2126(02)00777-3. [DOI] [PubMed] [Google Scholar]
- Kudlow B.A., Jameson S.A., Kennedy B.K. HIV protease inhibitors block adipocyte differentiation independently of lamin A/C. AIDS. 2005;19:1565–1573. doi: 10.1097/01.aids.0000186827.91408.90. [DOI] [PubMed] [Google Scholar]
- Kudlow B.A., Kennedy B.K., Monnat R.J. Werner and Hutchinson-Gilford progeria syndromes: Mechanistic basis of human progeroid diseases. Nat. Rev. Mol. Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- Kumaran R.I., Spector D.L. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J. Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaran R.I., Muralikrishna B., Parnaik V.K. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 2002;159:783–793. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupper K., Kolbl A., Biener D., Dittrich S., von Hase J., Thormeyer T., Fiegler H., Carter N.P., Speicher M.R., Cremer T., et al. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma. 2007;116:285–306. doi: 10.1007/s00412-007-0098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanctot C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lebel S., Lampron C., Royal A., Raymond Y. Lamins A and C appear during retinoic acid-induced differentiation of mouse embryonal carcinoma cells. J. Cell Biol. 1987;105:1099–1104. doi: 10.1083/jcb.105.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Hale C.M., Panorchan P., Khatau S.B., George J.P., Tseng Y., Stewart C.L., Hodzic D., Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys. J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner C.F., Stick R., Eppenberger H.M., Nigg E.A. Differential expression of nuclear lamin proteins during chicken development. J. Cell Biol. 1987;105:577–587. doi: 10.1083/jcb.105.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Worman H.J. Expression of nuclear lamins in human tissues and cancer cell lines and transcription from the promoters of the lamin A/C and B1 genes. Exp. Cell Res. 1997;236:378–384. doi: 10.1006/excr.1997.3735. [DOI] [PubMed] [Google Scholar]
- Liu J., Rolef Ben-Shahar T., Riemer D., Treinin M., Spann P., Weber K., Fire A., Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Wang J., Chan K.M., Tjia W.M., Deng W., Guan X., Huang J.D., Li K.M., Chau P.Y., Chen D.J., et al. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005;11:780–785. doi: 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- Liu Y., Rusinol A., Sinensky M., Wang Y., Zou Y. DNA damage responses in progeroid syndromes arise from defective maturation of prelamin A. J. Cell Sci. 2006;119:4644–4649. doi: 10.1242/jcs.03263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang Y., Rusinol A.E., Sinensky M.S., Liu J., Shell S.M., Zou Y. Involvement of xeroderma pigmentosum group A (XPA) in progeria arising from defective maturation of prelamin A. FASEB J. 2007;22:603–611. doi: 10.1096/fj.07-8598com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D.J., Trembath R.C., Shackleton S. A novel interaction between lamin A and SREBP1: Implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- Loewinger L., McKeon F. Mutations in the nuclear lamin proteins resulting in their aberrant assembly in the cytoplasm. EMBO J. 1988;7:2301–2309. doi: 10.1002/j.1460-2075.1988.tb03073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi F., Gullotta F., Columbaro M., Filareto A., D’Adamo M., Vielle A., Guglielmi V., Nardone A.M., Azzolini V., Grosso E., et al. Compound heterozygosity for mutations in LMNA in a patient with a myopathic and lipodystrophic mandibuloacral dysplasia type A phenotype. J. Clin. Endocrinol. Metab. 2007;92:4467–4471. doi: 10.1210/jc.2007-0116. [DOI] [PubMed] [Google Scholar]
- Lourim D., Kempf A., Krohne G. Characterization and quantitation of three B-type lamins in Xenopus oocytes and eggs: Increase of lamin LI protein synthesis during meiotic maturation. J. Cell Sci. 1996;109:1775–1785. doi: 10.1242/jcs.109.7.1775. [DOI] [PubMed] [Google Scholar]
- Luderus M.E., den Blaauwen J.L., de Smit O.J., Compton D.A., van Driel R. Binding of matrix attachment regions to lamin polymers involves single-stranded regions and the minor groove. Mol. Cell. Biol. 1994;14:6297–6305. doi: 10.1128/mcb.14.9.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels B.M., Zorenc A.H., Endert J.M., Kuijpers H.J., van Eys G.J., Ramaekers F.C., Broers J.L. An alternative splicing product of the lamin A/C gene lacks exon 10. J. Biol. Chem. 1996;271:9249–9253. doi: 10.1074/jbc.271.16.9249. [DOI] [PubMed] [Google Scholar]
- Malhas A., Lee C.F., Sanders R., Saunders N.J., Vaux D.J. Defects in lamin B1 expression or processing affect interphase chromosome position and gene expression. J. Cell Biol. 2007;176:593–603. doi: 10.1083/jcb.200607054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M.A., Shan B., Nickerson J.A., Penman S., Lee W.H. The retinoblastoma gene product is a cell cycle-dependent, nuclear matrix-associated protein. Proc. Natl. Acad. Sci. 1994;91:418–422. doi: 10.1073/pnas.91.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manju K., Muralikrishna B., Parnaik V.K. Expression of disease-causing lamin A mutants impairs the formation of DNA repair foci. J. Cell Sci. 2006;119:2704–2714. doi: 10.1242/jcs.03009. [DOI] [PubMed] [Google Scholar]
- Margalit A., Brachner A., Gotzmann J., Foisner R., Gruenbaum Y. Barrier-to-autointegration factor—A BAFfling little protein. Trends Cell Biol. 2007;17:202–208. doi: 10.1016/j.tcb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Mariappan I., Parnaik V.K. Sequestration of pRb by cyclin D3 causes intranuclear reorganization of lamin A/C during muscle cell differentiation. Mol. Biol. Cell. 2005;16:1948–1960. doi: 10.1091/mbc.E04-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariappan I., Gurung R., Thanumalayan S., Parnaik V.K. Identification of cyclin D3 as a new interaction partner of lamin A/C. Biochem. Biophys. Res. Commun. 2007;355:981–985. doi: 10.1016/j.bbrc.2007.02.060. [DOI] [PubMed] [Google Scholar]
- Markiewicz E., Dechat T., Foisner R., Quinlan R.A., Hutchison C.J. Lamin A/C binding protein LAP2α is required for nuclear anchorage of retinoblastoma protein. Mol. Biol. Cell. 2002;13:4401–4413. doi: 10.1091/mbc.E02-07-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz E., Ledran M., Hutchison C.J. Remodelling of the nuclear lamina and nucleoskeleton is required for skeletal muscle differentiation in vitro. J. Cell Sci. 2005;118:409–420. doi: 10.1242/jcs.01630. [DOI] [PubMed] [Google Scholar]
- Marschall M., Marzi A., aus dem Siepen P., Jochmann R., Kalmer M., Auerochs S., Lischka P., Leis M., Stamminger T. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 2005;280:33357–33367. doi: 10.1074/jbc.M502672200. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Masny P.S., Bengtsson U., Chung S.A., Martin J.H., van Engelen B., van der Maarel S.M., Winokur S.T. Localization of 4q35.2 to the nuclear periphery: Is FSHD a nuclear envelope disease? Hum. Mol. Genet. 2004;13:1857–1871. doi: 10.1093/hmg/ddh205. [DOI] [PubMed] [Google Scholar]
- Mattia E., Hoff W.D., den Blaauwen J., Meijne A.M., Stuurman N., van Renswoude J. Induction of nuclear lamins A/C during in vitro-induced differentiation of F9 and P19 embryonal carcinoma cells. Exp. Cell Res. 1992;203:449–455. doi: 10.1016/0014-4827(92)90020-9. [DOI] [PubMed] [Google Scholar]
- Mattout A., Goldberg M., Tzur Y., Margalit A., Gruenbaum Y. Specific and conserved sequences in D. melanogaster and C. elegans lamins and histone H2A mediate the attachment of lamins to chromosomes. J. Cell Sci. 2007;120:77–85. doi: 10.1242/jcs.03325. [DOI] [PubMed] [Google Scholar]
- Mattout-Drubezki A., Gruenbaum Y. Dynamic interactions of nuclear lamina proteins with chromatin and transcriptional machinery. Cell. Mol. Life Sci. 2003;60:2053–2063. doi: 10.1007/s00018-003-3038-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock D., Gordon L.B., Djabali K. Hutchinson-Gilford progeria mutant lamin A primarily targets human vascular cells as detected by an anti-Lamin A G608G antibody. Proc. Natl. Acad. Sci. 2006;103:2154–2159. doi: 10.1073/pnas.0511133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock D., Ratner D., Lokuge M., Owens D.M., Gordon L.B., Collins F.S., Djabali K. The mutant form of lamin A that causes Hutchinson-Gilford progeria is a biomarker of cellular aging in human skin. PLoS ONE. 2007;2:e1269. doi: 10.1371/journal.pone.0001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon F.D., Kirschner M.W., Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Meaburn K.J., Cabuy E., Bonne G., Levy N., Morris G.E., Novelli G., Kill I.R., Bridger J.M. Primary laminopathy fibroblasts display altered genome organization and apoptosis. Aging Cell. 2007;6:139–153. doi: 10.1111/j.1474-9726.2007.00270.x. [DOI] [PubMed] [Google Scholar]
- Meier I. The plant nuclear envelope. Cell. Mol. Life Sci. 2001;58:1774–1780. doi: 10.1007/PL00000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier J., Campbell K.H., Ford C.C., Stick R., Hutchison C.J. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J. Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Melcer S., Gruenbaum Y., Krohne G. Invertebrate lamins. Exp. Cell Res. 2007;313:2157–2166. doi: 10.1016/j.yexcr.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Melcon G., Kozlov S., Cutler D.A., Sullivan T., Hernandez L., Zhao P., Mitchell S., Nader G., Bakay M., Rottman J.N., et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- Mettenleiter T.C., Klupp B.G., Granzow H. Herpesvirus assembly: A tale of two membranes. Curr. Opin. Microbiol. 2006;9:423–429. doi: 10.1016/j.mib.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Milbradt J., Auerochs S., Marschall M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 2007;88:2642–2650. doi: 10.1099/vir.0.82924-0. [DOI] [PubMed] [Google Scholar]
- Miranda M., Chacon M.R., Vidal F., Megia A., Richart C., Veloso S., Saumoy M., Olona C., Vendrell J. LMNA messenger RNA expression in highly active antiretroviral therapy-treated HIV-positive patients. J. Acquir. Immune Defic. Syndr. 2007;46:384–389. doi: 10.1097/qai.0b013e31815aba1b. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Montag-Lowy M., Goldman R.D. Dynamic properties of nuclear lamins: Lamin B is associated with sites of DNA replication. J. Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Spann T.P., Herrmann H., Goldman R.D. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J. Cell Biol. 2000a;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir R.D., Spann T.P., Lopez-Soler R.I., Yoon M., Goldman A.E., Khuon S., Goldman R.D. Review: The dynamics of the nuclear lamins during the cell cycle—Relationship between structure and function. J. Struct. Biol. 2000b;129:324–334. doi: 10.1006/jsbi.2000.4251. [DOI] [PubMed] [Google Scholar]
- Moir R.D., Yoon M., Khuon S., Goldman R.D. Nuclear lamins A and B1: Different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 2000c;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R., Lee K.K., Wilson K.L. Binding of barrier to autointegration factor (BAF) to histone H3 and selected linker histones including H1.1. J. Biol. Chem. 2005;280:42252–42262. doi: 10.1074/jbc.M509917200. [DOI] [PubMed] [Google Scholar]
- Mou F., Forest T., Baines J.D. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 2007;81:6459–6470. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulson C.L., Fong L.G., Gardner J.M., Farber E.A., Go G., Passariello A., Grange D.K., Young S.G., Miner J.H. Increased progerin expression associated with unusual LMNA mutations causes severe progeroid syndromes. Hum. Mutat. 2007;28:882–889. doi: 10.1002/humu.20536. [DOI] [PubMed] [Google Scholar]
- Muchir A., van Engelen B.G., Lammens M., Mislow J.M., McNally E., Schwartz K., Bonne G. Nuclear envelope alterations in fibroblasts from LGMD1B patients carrying nonsense Y259X heterozygous or homozygous mutation in lamin A/C gene. Exp. Cell Res. 2003;291:352–362. doi: 10.1016/j.yexcr.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Muchir A., Pavlidis P., Bonne G., Hayashi Y.K., Worman H.J. Activation of MAPK in hearts of EMD null mice: Similarities between mouse models of X-linked and autosomal dominant Emery Dreifuss muscular dystrophy. Hum. Mol. Genet. 2007a;16:1884–1895. doi: 10.1093/hmg/ddm137. [DOI] [PubMed] [Google Scholar]
- Muchir A., Pavlidis P., Decostre V., Herron A.J., Arimura T., Bonne G., Worman H.J. Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. J. Clin. Invest. 2007b;117:1282–1293. doi: 10.1172/JCI29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralikrishna B., Parnaik V.K. SP3 and AP-1 mediate transcriptional activation of the lamin A proximal promoter. Eur. J. Biochem. 2001;268:3736–3743. doi: 10.1046/j.1432-1327.2001.02281.x. [DOI] [PubMed] [Google Scholar]
- Muralikrishna B., Dhawan J., Rangaraj N., Parnaik V.K. Distinct changes in intranuclear lamin A/C organization during myoblast differentiation. J. Cell Sci. 2001;114:4001–4011. doi: 10.1242/jcs.114.22.4001. [DOI] [PubMed] [Google Scholar]
- Muralikrishna B., Thanumalayan S., Jagatheesan G., Rangaraj N., Karande A.A., Parnaik V.K. Immunolocalization of detergent-susceptible nucleoplasmic lamin A/C foci by a novel monoclonal antibody. J. Cell. Biochem. 2004;91:730–739. doi: 10.1002/jcb.10765. [DOI] [PubMed] [Google Scholar]
- Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U.H. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science. 2002;297:854–857. doi: 10.1126/science.1071506. [DOI] [PubMed] [Google Scholar]
- Newport J.W., Wilson K.L., Dunphy W.G. A lamin-independent pathway for nuclear envelope assembly. J. Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.J., Schneider R., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Nikolova V., Leimena C., McMahon A.C., Tan J.C., Chandar S., Jogia D., Kesteven S.H., Michalicek J., Otway R., Verheyen F., et al. Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Invest. 2004;113:357–369. doi: 10.1172/JCI19448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta R.T., Smith C.L., Kennedy B.K. Evidence that proteasome-dependent degradation of the retinoblastoma protein in cells lacking A-type lamins occurs independently of gankyrin and MDM2. PLoS ONE. 2007;2:e963. doi: 10.1371/journal.pone.0000963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K., Hosoe Y., Nakajima N. c-Jun and Sp1 family are critical for retinoic acid induction of the lamin A/C retinoic acid-responsive element. Biochem. Biophys. Res. Commun. 2004;320:487–492. doi: 10.1016/j.bbrc.2004.05.191. [DOI] [PubMed] [Google Scholar]
- Osouda S., Nakamura Y., de Saint Phalle B., McConnell M., Horigome T., Sugiyama S., Fisher P.A., Furukawa K. Null mutants of Drosophila B-type lamin Dm(0) show aberrant tissue differentiation rather than obvious nuclear shape distortion or specific defects during cell proliferation. Dev. Biol. 2005;284:219–232. doi: 10.1016/j.ydbio.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Ozaki T., Saijo M., Murakami K., Enomoto H., Taya Y., Sakiyama S. Complex formation between lamin A and the retinoblastoma gene product: Identification of the domain on lamin A required for its interaction. Oncogene. 1994;9:2649–2653. [PubMed] [Google Scholar]
- Padiath Q.S., Saigoh K., Schiffmann R., Asahara H., Yamada T., Koeppen A., Hogan K., Ptacek L.J., Fu Y.H. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- Padmakumar V.C., Libotte T., Lu W., Zaim H., Abraham S., Noegel A.A., Gotzmann J., Foisner R., Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J. Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Pajerowski J.D., Dahl K.N., Zhong F.L., Sammak P.J., Discher D.E. From the Cover: Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R., Baines J.D. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 2006;80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrizi G., Poger M. The ultrastructure of the nuclear periphery. The zonula nucleum limitans. J. Ultrastruct. Res. 1967;17:127–136. doi: 10.1016/s0022-5320(67)80025-x. [DOI] [PubMed] [Google Scholar]
- Pekovic V., Harborth J., Broers J.L., Ramaekers F.C., van Engelen B., Lammens M., von Zglinicki T., Foisner R., Hutchison C., Markiewicz E. Nucleoplasmic LAP2α–lamin A complexes are required to maintain a proliferative state in human fibroblasts. J. Cell Biol. 2007;176:163–172. doi: 10.1083/jcb.200606139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendas A.M., Zhou Z., Cadinanos J., Freije J.M., Wang J., Hultenby K., Astudillo A., Wernerson A., Rodriguez F., Tryggvason K., et al. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase-deficient mice. Nat. Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Pickersgill H., Kalverda B., de Wit E., Talhout W., Fornerod M., van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Prasanth K.V., Spector D.L. Encyclopedia of life sciences. John Wiley & Sons; Ltd, Chichester, UK: 2006. The cell nucleus. http://www.els.net. [DOI] [Google Scholar]
- Prokocimer M., Margalit A., Gruenbaum Y. The nuclear lamina and its proposed roles in tumorigenesis: Projection on the hematologic malignancies and future targeted therapy. J. Struct. Biol. 2006;155:351–360. doi: 10.1016/j.jsb.2006.02.016. [DOI] [PubMed] [Google Scholar]
- Radsak K.D., Brucher K.H., Georgatos S.D. Focal nuclear envelope lesions and specific nuclear lamin A/C dephosphorylation during infection with human cytomegalovirus. Eur. J. Cell Biol. 1991;54:299–304. [PubMed] [Google Scholar]
- Rahman-Roblick R., Roblick U.J., Hellman U., Conrotto P., Liu T., Becker S., Hirschberg D., Jornvall H., Auer G., Wiman K.G. p53 targets identified by protein expression profiling. Proc. Natl. Acad. Sci. 2007;104:5401–5406. doi: 10.1073/pnas.0700794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao L., Perez D., White E. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz V., Carlotti F., Vermolen B.J., van der Poel E., Sloos W.C., Knaan-Shanzer S., de Vries A.A., Hoeben R.C., Young I.T., Tanke H.J., et al. Changes in lamina structure are followed by spatial reorganization of heterochromatic regions in caspase-8-activated human mesenchymal stem cells. J. Cell Sci. 2006;119:4247–4256. doi: 10.1242/jcs.03180. [DOI] [PubMed] [Google Scholar]
- Reynolds A.E., Liang L., Baines J.D. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 2004;78:5564–5575. doi: 10.1128/JVI.78.11.5564-5575.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemer D., Stuurman N., Berrios M., Hunter C., Fisher P.A., Weber K. Expression of Drosophila lamin C is developmentally regulated: Analogies with vertebrate A-type lamins. J. Cell Sci. 1995;108:3189–3198. doi: 10.1242/jcs.108.10.3189. [DOI] [PubMed] [Google Scholar]
- Rober R.A., Weber K., Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: A developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Rober R.A., Sauter H., Weber K., Osborn M. Cells of the cellular immune and hemopoietic system of the mouse lack lamins A/C: Distinction versus other somatic cells. J. Cell Sci. 1990;95:587–598. doi: 10.1242/jcs.95.4.587. [DOI] [PubMed] [Google Scholar]
- Robinson L.J., Karlsson N.G., Weiss A.S., Packer N.H. Proteomic analysis of the genetic premature aging disease Hutchinson Gilford progeria syndrome reveals differential protein expression and glycosylation. J. Proteome Res. 2003;2:556–557. doi: 10.1021/pr034035k. [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Korfali N., Villa P., Kottke T.J., Dingwall C., Kaufmann S.H., Earnshaw W.C. Caspase-6 gene disruption reveals a requirement for lamin A cleavage in apoptotic chromatin condensation. EMBO J. 2002;21:1967–1977. doi: 10.1093/emboj/21.8.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinol A.E., Sinensky M.S. Farnesylated lamins, progeroid syndromes and farnesyl transferase inhibitors. J. Cell Sci. 2006;119:3265–3272. doi: 10.1242/jcs.03156. [DOI] [PubMed] [Google Scholar]
- Rzepecki R., Bogachev S.S., Kokoza E., Stuurman N., Fisher P.A. In vivo association of lamins with nucleic acids in Drosophila melanogaster. J. Cell Sci. 1998;111:121–129. doi: 10.1242/jcs.111.1.121. [DOI] [PubMed] [Google Scholar]
- Sabatelli P., Lattanzi G., Ognibene A., Columbaro M., Capanni C., Merlini L., Maraldi N.M., Squarzoni S. Nuclear alterations in autosomal-dominant Emery-Dreifuss muscular dystrophy. Muscle Nerve. 2001;24:826–829. doi: 10.1002/mus.1076. [DOI] [PubMed] [Google Scholar]
- Sarma K., Reinberg D. Histone variants meet their match. Nat. Rev. Mol. Cell Biol. 2005;6:139–149. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Sasse B., Aebi U., Stuurman N. A tailless Drosophila lamin Dm0 fragment reveals lateral associations of dimers. J. Struct. Biol. 1998;123:56–66. doi: 10.1006/jsbi.1998.4006. [DOI] [PubMed] [Google Scholar]
- Sasseville A.M., Raymond Y. Lamin A precursor is localized to intranuclear foci. J. Cell Sci. 1995;108:273–285. doi: 10.1242/jcs.108.1.273. [DOI] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat. Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P., Misteli T. Lamin A-dependent nuclear defects in human aging. Science. 2006;312:1059–1063. [Google Scholar]
- Schirmer E.C., Foisner R. Proteins that associate with lamins: Many faces, many functions. Exp. Cell Res. 2007;313:2167–2179. doi: 10.1016/j.yexcr.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Gerace L. The stability of the nuclear lamina polymer changes with the composition of lamin subtypes according to their individual binding strengths. J. Biol. Chem. 2004;279:42811–42817. doi: 10.1074/jbc.M407705200. [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Guan T., Gerace L. Involvement of the lamin rod domain in heterotypic lamin interactions important for nuclear organization. J. Cell Biol. 2001;153:479–489. doi: 10.1083/jcb.153.3.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Krohne G. In vivo assembly kinetics of fluorescently labeled Xenopus lamin A mutants. Eur. J. Cell Biol. 1995;68:345–354. [PubMed] [Google Scholar]
- Schumacher J., Reichenzeller M., Kempf T., Schnolzer M., Herrmann H. Identification of a novel, highly variable amino-terminal amino acid sequence element in the nuclear intermediate filament protein lamin B(2) from higher vertebrates. FEBS Lett. 2006;580:6211–6216. doi: 10.1016/j.febslet.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Scott E.S., O’Hare P. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 2001;75:8818–8830. doi: 10.1128/JVI.75.18.8818-8830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Totten M., Wilson K.L. BAF: Roles in chromatin, nuclear structure and retrovirus integration. Trends Cell Biol. 2004;14:261–266. doi: 10.1016/j.tcb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Shoeman R.L., Traub P. The in vitro DNA-binding properties of purified nuclear lamin proteins and vimentin. J. Biol. Chem. 1990;265:9055–9061. [PubMed] [Google Scholar]
- Shumaker D.K., Vann L.R., Goldberg M.W., Allen T.D., Wilson K.L. TPEN, a Zn2+/Fe2+ chelator with low affinity for Ca2+, inhibits lamin assembly, destabilizes nuclear architecture and may independently protect nuclei from apoptosis in vitro. Cell Calcium. 1998;23:151–164. doi: 10.1016/s0143-4160(98)90114-2. [DOI] [PubMed] [Google Scholar]
- Shumaker D.K., Lee K.K., Tanhehco Y.C., Craigie R., Wilson K.L. LAP2 binds to BAF.DNA complexes: Requirement for the LEM domain and modulation by variable regions. EMBO J. 2001;20:1754–1764. doi: 10.1093/emboj/20.7.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D.K., Kuczmarski E.R., Goldman R.D. The nucleoskeleton: Lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 2003;15:358–366. doi: 10.1016/s0955-0674(03)00050-4. [DOI] [PubMed] [Google Scholar]
- Shumaker D.K., Lopez-Soler R.I., Adam S.A., Herrmann H., Moir R.D., Spann T.P., Goldman R.D. Functions and dysfunctions of the nuclear lamin Ig-fold domain in nuclear assembly, growth, and Emery-Dreifuss muscular dystrophy. Proc. Natl. Acad. Sci. 2005;102:15494–15499. doi: 10.1073/pnas.0507612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumaker D.K., Dechat T., Kohlmaier A., Adam S.A., Bozovsky M.R., Erdos M.R., Eriksson M., Goldman A.E., Khuon S., Collins F.S., et al. Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc. Natl. Acad. Sci. 2006;103:8703–8708. doi: 10.1073/pnas.0602569103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shun M.C., Daigle J.E., Vandegraaff N., Engelman A. Wild-type levels of human immunodeficiency virus type 1 infectivity in the absence of cellular emerin protein. J. Virol. 2007;81:166–172. doi: 10.1128/JVI.01953-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Holley M., Colgrove R.C., Nalepa G., Harper J.W., Knipe D.M. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 2005;79:12840–12851. doi: 10.1128/JVI.79.20.12840-12851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T.P., Moir R.D., Goldman A.E., Stick R., Goldman R.D. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J. Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann T.P., Goldman A.E., Wang C., Huang S., Goldman R.D. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector D.L. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Stewart C., Burke B. Teratocarcinoma stem cells and early mouse embryos contain only a single major lamin polypeptide closely resembling lamin B. Cell. 1987;51:383–392. doi: 10.1016/0092-8674(87)90634-9. [DOI] [PubMed] [Google Scholar]
- Stewart M., Whytock S. The structure and interactions of components of nuclear envelopes from Xenopus oocyte germinal vesicles observed by heavy metal shadowing. J. Cell Sci. 1988;90:409–423. [Google Scholar]
- Stewart C.L., Kozlov S., Fong L.G., Young S.G. Mouse models of the laminopathies. Exp. Cell Res. 2007;313:2144–2156. doi: 10.1016/j.yexcr.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stick R., Hausen P. Changes in the nuclear lamina composition during early development of Xenopus laevis. Cell. 1985;41:191–200. doi: 10.1016/0092-8674(85)90073-x. [DOI] [PubMed] [Google Scholar]
- Stierle V., Couprie J., Ostlund C., Krimm I., Zinn-Justin S., Hossenlopp P., Worman H.J., Courvalin J.C., Duband-Goulet I. The carboxyl-terminal region common to lamins A and C contains a DNA binding domain. Biochemistry. 2003;42:4819–4828. doi: 10.1021/bi020704g. [DOI] [PubMed] [Google Scholar]
- Strelkov S.V., Schumacher J., Burkhard P., Aebi U., Herrmann H. Crystal structure of the human lamin A coil 2B dimer: Implications for the head-to-tail association of nuclear lamins. J. Mol. Biol. 2004;343:1067–1080. doi: 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- Stuurman N., Heins S., Aebi U. Nuclear lamins: Their structure, assembly, and interactions. J. Struct. Biol. 1998;122:42–66. doi: 10.1006/jsbi.1998.3987. [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani M.A., Hayashi K., Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Yang H., Craigie R. LAP2α and BAF collaborate to organize the Moloney murine leukemia virus preintegration complex. EMBO J. 2004;23:4670–4678. doi: 10.1038/sj.emboj.7600452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori Y., Tamura Y., Kataoka Y., Cui Y., Seo S., Kanazawa T., Kurokawa K., Yamada H. Differential expression of nuclear lamin, the major component of nuclear lamina, during neurogenesis in two germinal regions of adult rat brain. Eur. J. Neurosci. 2007;25:1653–1662. doi: 10.1111/j.1460-9568.2007.05450.x. [DOI] [PubMed] [Google Scholar]
- Takata H., Uchiyama S., Nakamura N., Nakashima S., Kobayashi S., Sone T., Kimura S., Lahmers S., Granzier H., Labeit S., et al. A comparative proteome analysis of human metaphase chromosomes isolated from two different cell lines reveals a set of conserved chromosome-associated proteins. Genes Cells. 2007;12:269–284. doi: 10.1111/j.1365-2443.2007.01051.x. [DOI] [PubMed] [Google Scholar]
- Taniura H., Glass C., Gerace L. A chromatin binding site in the tail domain of nuclear lamins that interacts with core histones. J. Cell Biol. 1995;131:33–44. doi: 10.1083/jcb.131.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L.J., Bollen M., Fields A.P. Identification of protein phosphatase 1 as a mitotic lamin phosphatase. J. Biol. Chem. 1997;272:29693–29697. doi: 10.1074/jbc.272.47.29693. [DOI] [PubMed] [Google Scholar]
- Tilli C.M., Ramaekers F.C., Broers J.L., Hutchison C.J., Neumann H.A. Lamin expression in normal human skin, actinic keratosis, squamous cell carcinoma and basal cell carcinoma. Br. J. Dermatol. 2003;148:102–109. doi: 10.1046/j.1365-2133.2003.05026.x. [DOI] [PubMed] [Google Scholar]
- Toth J.I., Yang S.H., Qiao X., Beigneux A.P., Gelb M.H., Moulson C.L., Miner J.H., Young S.G., Fong L.G. Blocking protein farnesyltransferase improves nuclear shape in fibroblasts from humans with progeroid syndromes. Proc. Natl. Acad. Sci. 2005;102:12873–12878. doi: 10.1073/pnas.0505767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Lamond A.I. Mitotic phosphatases: No longer silent partners. Curr. Opin. Cell Biol. 2006;18:623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Van Berlo J.H., Voncken J.W., Kubben N., Broers J.L., Duisters R., van Leeuwen R.E., Crijns H.J., Ramaekers F.C., Hutchison C.J., Pinto Y.M. A-type lamins are essential for TGF-β1 induced PP2A to dephosphorylate transcription factors. Hum. Mol. Genet. 2005;14:2839–2849. doi: 10.1093/hmg/ddi316. [DOI] [PubMed] [Google Scholar]
- van Engelen B.G., Muchir A., Hutchison C.J., van der Kooi A.J., Bonne G., Lammens M. The lethal phenotype of a homozygous nonsense mutation in the lamin A/C gene. Neurology. 2005;64:374–376. doi: 10.1212/01.WNL.0000149763.15180.00. [DOI] [PubMed] [Google Scholar]
- Varela I., Cadinanos J., Pendas A.M., Gutierrez-Fernandez A., Folgueras A.R., Sanchez L.M., Zhou Z., Rodriguez F.J., Stewart C.L., Vega J.A., et al. Accelerated ageing in mice deficient in Zmpste24 protease is linked to p53 signalling activation. Nature. 2005;437:564–568. doi: 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Varga R., Eriksson M., Erdos M.R., Olive M., Harten I., Kolodgie F., Capell B.C., Cheng J., Faddah D., Perkins S., et al. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. 2006;103:3250–3255. doi: 10.1073/pnas.0600012103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecerova J., Koberna K., Malinsky J., Soutoglou E., Sullivan T., Stewart C.L., Raska I., Misteli T. Formation of nuclear splicing factor compartments is independent of lamins A/C. Mol. Biol. Cell. 2004;15:4904–4910. doi: 10.1091/mbc.E04-07-0645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnes L., Peterfy M., Bergo M.O., Young S.G., Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc. Natl. Acad. Sci. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten V.L., Broers J.L., Ramaekers F.C., van Steensel M.A. The nuclear envelope, a key structure in cellular integrity and gene expression. Curr. Med. Chem. 2007;14:1231–1248. doi: 10.2174/092986707780598032. [DOI] [PubMed] [Google Scholar]
- Vlcek S., Foisner R. Lamins and lamin-associated proteins in aging and disease. Curr. Opin. Cell Biol. 2007a;19:298–304. doi: 10.1016/j.ceb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Vlcek S., Foisner R. A-type lamin networks in light of laminopathic diseases. Biochim. Biophys. Acta. 2007b;1773:661–674. doi: 10.1016/j.bbamcr.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Wagner N., Krohne G. LEM-domain proteins: New insights into lamin-interacting proteins. Int. Rev. Cytol. 2007;261:1–46. doi: 10.1016/S0074-7696(07)61001-8. [DOI] [PubMed] [Google Scholar]
- Warren D.T., Zhang Q., Weissberg P.L., Shanahan C.M. Nesprins: Intracellular scaffolds that maintain cell architecture and coordinate cell function? Expert Rev. Mol. Med. 2005;7:1–15. doi: 10.1017/S1462399405009294. [DOI] [PubMed] [Google Scholar]
- Wiche G. Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 1998;111:2477–2486. doi: 10.1242/jcs.111.17.2477. [DOI] [PubMed] [Google Scholar]
- Wiesel N., Mattout A., Melcer S., Melamed-Book N., Herrmann H., Medalia O., Aebi U., Gruenbaum Y. Laminopathic mutations interfere with the assembly, localization, and dynamics of nuclear lamins. Proc. Natl. Acad. Sci. 2008;105:180–185. doi: 10.1073/pnas.0708974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 2005;171:799–810. doi: 10.1083/jcb.200506083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin S.L., Krohne G., Kirschner M.W. A new lamin in Xenopus somatic tissues displays strong homology to human lamin A. EMBO J. 1987;6:3809–3818. doi: 10.1002/j.1460-2075.1987.tb02717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Bonne G. ‘Laminopathies’: A wide spectrum of human diseases. Exp. Cell Res. 2007;313:2121–2133. doi: 10.1016/j.yexcr.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman H.J., Evans C.D., Blobel G. The lamin B receptor of the nuclear envelope inner membrane: A polytopic protein with eight potential transmembrane domains. J. Cell Biol. 1990;111:1535–1542. doi: 10.1083/jcb.111.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Meta M., Qiao X., Frost D., Bauch J., Coffinier C., Majumdar S., Bergo M.O., Young S.G., Fong L.G. A farnesyltransferase inhibitor improves disease phenotypes in mice with a Hutchinson-Gilford progeria syndrome mutation. J. Clin. Invest. 2006;116:2115–2121. doi: 10.1172/JCI28968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.H., Qiao X., Farber E., Chang S.Y., Fong L.G., Young S.G. Eliminating the synthesis of mature lamin A reduces disease phenotypes in mice carrying a Hutchinson-Gilford progeria syndrome allele. J. Biol. Chem. 2008 doi: 10.1074/jbc.M708138200. [DOI] [PubMed] [Google Scholar]
- Ye Q., Worman H.J. Primary structure analysis and lamin B and DNA binding of human LBR, an integral protein of the nuclear envelope inner membrane. J. Biol. Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- Ye Q., Worman H.J. Protein–protein interactions between human nuclear lamins expressed in yeast. Exp. Cell Res. 1995;219:292–298. doi: 10.1006/excr.1995.1230. [DOI] [PubMed] [Google Scholar]
- Ye Q., Worman H.J. Interaction between an integral protein of the nuclear envelope inner membrane and human chromodomain proteins homologous to Drosophila HP1. J. Biol. Chem. 1996;271:14653–14656. doi: 10.1074/jbc.271.25.14653. [DOI] [PubMed] [Google Scholar]
- Young S.G., Fong L.G., Michaelis S. Prelamin A, Zmpste24, misshapen cell nuclei, and progeria—New evidence suggesting that protein farnesylation could be important for disease pathogenesis. J. Lipid Res. 2005;46:2531–2558. doi: 10.1194/jlr.R500011-JLR200. [DOI] [PubMed] [Google Scholar]
- Yuan J., Simos G., Blobel G., Georgatos S.D. Binding of lamin A to polynucleosomes. J. Biol. Chem. 1991;266:9211–9215. [PubMed] [Google Scholar]
- Zastrow M.S., Vlcek S., Wilson K.L. Proteins that bind A-type lamins: Integrating isolated clues. J. Cell Sci. 2004;117:979–987. doi: 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- Zhao K., Harel A., Stuurman N., Guedalia D., Gruenbaum Y. Binding of matrix attachment regions to nuclear lamin is mediated by the rod domain and depends on the lamin polymerization state. FEBS Lett. 1996;380:161–164. doi: 10.1016/0014-5793(96)00034-8. [DOI] [PubMed] [Google Scholar]
- Zheng R., Ghirlando R., Lee M.S., Mizuuchi K., Krause M., Craigie R. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl. Acad. Sci. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]