Abstract
Germline genomic methylation is essential for gamete identity and integrity in mammals. The study by Kuramochi-Miyagawa and colleagues (908–917) in the previous issue of Genes & Development links the process of DNA methylation-dependent repression of retrotranspons with the presence of piwi-interacting RNAs (piRNAs) in fetal male germ cells undergoing de novo methylation.
[Keywords: Piwi, piRNA, retrotransposon, DNA methylation, spermatogenesis]
Cytosine methylation provides a stable, heritable, and reversible mark for transcriptional repression. Reprogramming of the methylation repertoire at each passage through the germline is a mammalian characteristic. An erasure step allows the acquisition of an epigenetic state unique to the germ cell lineage and central to the transmission of totipotency over generations. Subsequent initiation of gametic methylation has a major impact on fertility, by silencing parasitic transposable elements (TEs) and providing a parental mark for the monoallelic expression of imprinted genes. To control the overwhelming genomic contribution of TEs and their deleterious effects on gamete integrity, a variety of repressive mechanisms, including DNA methylation in mammals, have been developed over the course of evolution to lead to their suppression.
Gametic de novo methylation involves the activity of the DNA methyltransferases Dnmt3A and Dnmt3B, assisted by their regulatory factor Dnmt3L. Studies based on structural, biochemical, and DNA analyses provided information as to how these proteins functionally interact to add methyl groups on cytosines within CG dinucleotides. Besides this general recognition of CG sites, additional signals targeting DNA methylation to specific genomic sequences are progressively elucidated. Specific histone modifications and energy-dependent chromatin remodeling enzymes have been shown to regulate the accessibility of DNA methyltransferases to their substrates. A specific class of small RNAs, the piwi-interacting RNAs (piRNAs), was most recently proposed to guide DNA methylation in male germ cells in the mouse. The developmental analysis performed by Kuramochi-Miyagawa et al. (2008) in the context of normal and Piwi-mutant spermatogenesis, published in the previous issue of Genes & Development, definitely supports the existence of an RNA-directed pathway triggering the initiation of DNA methylation in the mammalian germline. The different pieces of the puzzle now lay on the table. Deciphering how these different pieces connect together to control the establishment of gametic methylation patterns at the right time and at the right place will be the next exciting challenge in the field.
Ontogeny of methylation patterns in the mammalian germline
DNA methylation mostly concerns cytosines involved in CG dinucleotides in mammals (Goll and Bestor 2005). Transposable elements (TEs) and pericentromeric repeats represent the most prominently methylated compartments of the genome (Yoder et al. 1997; Rollins et al. 2006). Allele-specific methylation is a characteristic of control regions of imprinted genes and genes subject to X inactivation, while CpG islands associated with nonimprinted autosomal genes are mostly unmethylated. An erasure and subsequent re-establishment of genomic methylation occur at each generation in the mammalian germline (Trasler 2006). This reprogramming prevents the inheritance of epimutations and ensures the resetting of gametic imprinted methylation marks according to the sex of the individual.
A small cohort of pluripotent epiblast cells is set aside early during mammalian development to become the founder population of primordial germ cells (PGCs). The germline compartimentalization occurs ∼7 d post- coitum (dpc) in the mouse (McLaren 2003). At this stage, PGCs exhibit a somatic-like pattern of genomic methylation that is maintained during their proliferating and migratory phase, ending in the genital ridges at 10.5–11.5 dpc. The proximity of the genital ridges triggers a global loss of DNA methylation, and PGCs colonizing the gonads at 12.5–13.5 dpc are globally hypomethylated (Yamazaki et al. 2005). This period provides a developmental window where TEs are not silenced by DNA methylation; however, some level of methylation is conserved at a particularly aggressive subclass of TEs (LTR-retrotransposon IAP) within the demethylated genome (Hajkova et al. 2002).
At 13.5 dpc, the genetic sex imposes a specific developmental and epigenetic fate to female and male PGCs. In the developing mouse ovary, germ cells synchronously enter prophase of the first meiotic division and progress until birth when they undergo a specialized arrest stage known as dictyate. The diameter of the oocyte increases dramatically during the subsequent maturating phase, reaching a critical size that triggers de novo methylation just before ovulation (Trasler 2006). In males, PGCs continue to proliferate until 14.5 dpc, where they enter mitotic G0/G1 arrest as prospermatogonia. Quiescent prospermatogonia undergo de novo methylation within a few days from 15.5 dpc until birth (Kato et al. 2007), where they become spermatogonial stem cells (SSCs) and resume mitosis. SSCs provide a continuous supply of male gametes throughout life. Male methylation patterns are maintained during the whole spermatogenetic cycle, beginning with a SSC and ending with the release of mature spermatozoa.
The selective pressure for de novo methylation in the male germline in mammals
Mammalian male and female gametogenesis are very different in terms of developmental progression. Notably, the male germline contains life-long regenerating stem cells, while the female germline relies on meiotically arrested oocytes stored since prenatal life. The kinetics and targeting of de novo methylation are also subject to a profound sexual dimorphism. As outlined above, male gametic methylation is a premeiotic phenomenon occurring in fetal life. It targets the entire population of prospermatogonia, upstream of the formation of actively dividing and self-renewing SSCs. Female gametic methylation occurs after birth and at each reproductive cycle, in cohorts of meiotic oocytes committed to ovulation. Beside the sex-specific methylation found at maternally and paternally imprinted genes, TEs are also differentially methylated in the male and female germlines (Sanford et al. 1987; Lucifero et al. 2007). Retrotransposons, which are the main TE representatives in mammalian genomes, have a higher invading ability in actively replicating cells (Kubo et al. 2006). Germlines containing a source of continuously dividing stem cells would be predicted to require multiple robust repressive pathways to compensate for this vulnerability toward TE amplification. The existence of SSCs in males is indeed associated with a dense methylated state at TEs, while nondividing oocytes tolerate a partial methylated state for similar sequences. Coincidentally, repression of TEs via alternative transcriptional and post-transcriptional means is inherent to gametic differentiation in both sexes in Drosophila and Caenorhabditis elegans, where male and female germlines indistinctly contain a source of stem cells. Efficient and heritable TE silencing has been proposed to be the driving force for the evolution of gametic DNA methylation in mammals (Yoder et al. 1997). Genomic imprinting may be a side effect of this primary role in host defense, co-opting this existing system for the purpose of parent-specific expression of a subset of developmentally important genes (Barlow 1993).
The gametic de novo methyltransferase complex
Developmental and genetic studies have pointed to the major involvement of two proteins belonging to the family of DNA methyltransferases, Dnmt3A and Dnmt3L, in global gametic methylation. While Dnmt3B plays a role in somatic lineages in the early embryo, its contribution to germ cell methylation is limited (Kaneda et al. 2004; Kato et al. 2007). Dnmt3A and Dnmt3B catalyze the transfer of methyl groups preferentially on unmethylated DNA, a feature of de novo methylating enzymes. Dnmt3L is an inactive paralog (L for like) specifically expressed in embryonic stem (ES) cells and during gametogenesis at the exact time when de novo methylation is established (Bourc’his et al. 2001; Trasler 2006). Targeted inactivation of Dnmt3L or Dnmt3A in mouse induces a similar failure in gametic methylation establishment, with a dramatic effect on TEs in males, while maternally imprinted genes are affected mostly in females (Bourc’his et al. 2001; Bourc’his and Bestor 2004; Kaneda et al. 2004; Lucifero et al. 2007). A massive reactivation of different classes of TEs is observed in mutant male germ cells and is associated with a sterility phenotype. TE mobilization is known to induce DNA damage and chromosome rearrangements and can perturb developmental expression programs upon integration or by position effect (Deininger et al. 2003; Maksakova et al. 2006). An interruption of the spermatogenetic process at the pachytene stage due to pairing anomalies between homologous chromosomes, combined with a rapid exhaustion of the SSC pool, leads to a complete depletion in the germ cell complement of Dnmt3L−/− testes within a few weeks after birth. The primary role of Dnmt3L on gametic TE silencing in mammals is emphasized by the phylogenetic restriction of this protein to the mammalian clade and its high divergence rate compared with catalytically active Dnmts (Yokomine et al. 2006). Dnmt3L, as a regulatory factor, has been proposed to provide the adaptability of the de novo methylation system to counteract the rapid evolution of TE expansion strategies.
Recent crystallographic studies have provided insight into the mechanism of action of Dnmt3A and Dnmt3L (Jia et al. 2007). These proteins associate into a tetrameric complex composed of a central Dnmt3A dimer surrounded by Dnmt3L on each side. The interface Dnmt3L/Dnmt3A potentially stabilizes the conformation of the active site loop of Dnmt3A, in agreement with the significant stimulation induced by Dnmt3L on Dnmt3A formerly evidenced in biochemical assays (Chedin et al. 2002; Kareta et al. 2006). The dimeric association of Dnmt3A gives the potential to methylate two CG sites in one binding event, provided that they are separated by one helical turn of the DNA molecule (8–10 base pairs). This optimal CG spacing was not only tested in vitro on synthetic DNA fragments, but it was also confirmed to naturally occur in imprinting control regions methylated during oogenesis (Jia et al. 2007). A marked 8-nucleotide (nt) spacing between CG dinucleotides was also found to be overrepresented throughout the human genome (Ferguson-Smith and Greally 2007), which potentially reflects the genome-wide distribution of methylated TEs. It should be pointed out that the methylation patterning induced by DRM2 (Domains Rearranged Methylase 2), a plant homolog of Dnmt3A, shows the same periodicity on a genome-wide level (Cokus et al. 2008). These observations suggest that the interdependence between optimal target spacing and positioning of de novo DNA methyltransferases may be conserved in eukaryote species with heavily methylated genomes.
Manipulating chromatin states to regulate DNA methylation
The simple sequence information does not resolve the sex- and sequence-specific methylation of the male and female germlines. Moreover, Dnmt3L does not bind DNA per se, nor does it enhance the ability of Dnmt3A to bind DNA (Kareta et al. 2006). De novo methylation occurs in vivo in the context of an organized chromatin, which likely adds an additional layer of methylation regulation. The Dnmt3L protein indeed binds to the N-terminal tail of histone H3 in ES cells (Ooi et al. 2007). Peptide interaction assays showed that methylation at the H3 Lys 4 residue eliminates the interaction of the H3 tail with Dnmt3L by steric occlusion, suggesting that H3K4 methylation may protect against DNA methylation. In this model, Dnmt3L binds to genomic sequences devoid of H3K4 methylation through its PHD-like domain (Plant HomeoDomain) and interacts with Dnmt3A through its C-terminal region to locally stimulate de novo methylation.
Dnmt3A is a nonprocessive enzyme in vitro, meaning that it methylates where it lands on DNA (Gowher and Jeltsch 2001). Processive motors have been shown to cooperate with DNA methyltransferases. A favorable CG spacing or H3K4 methylation state are therefore not the sole regulators of de novo methylation. Lsh (lymphoid-specific helicase), a protein related to the SNF2 family of chromatin remodeling ATPases, is required for efficient DNA methylation in mammals, as is its homolog DDM1 (Decreased DNA Methylation 1) in plants (Dennis et al. 2001). The effect is particularly pronounced for TE methylation and silencing in somatic cells, and potentially in female germ cells as well (Dennis et al. 2001; De La Fuente et al. 2006). Cellular assays have indicated a direct association between Lsh function and de novo methylation via Dnmt3A (Zhu et al. 2006). However, the mechanism of action of Lsh in facilitating DNA methylation and its spatiotemporal position in the Dnmt3A–Dnmt3L system at the time of gametic de novo methylation are unknown. Another SNF2 protein, ATRX, confirms the link between chromatin remodeling and DNA methylation. Mutations in ATRX result in X-linked α-thalassemia with mental retardation in humans and correlate with hypomethylation as well as hypermethylation at repetitive sequences in patients (Gibbons et al. 2000). These opposite effects may indicate that remodeling enzymes can facilitate as well as impede access of DNA methyltransferases to DNA, depending on the chromatin context.
piRNA-directed DNA methylation
While recent studies have started to uncover the role of local chromatin states on Dnmt activity, it was still not clear how specific sequences were recognized and marked for de novo DNA methylation. A possible solution to this enigma came recently from an unexpected direction: Small RNAs might provide specificity to this process by serving as guides that direct DNA methylation to target genomic loci.
Small RNA or RNAi pathways operate in animals, plants, and protozoa to regulate gene expression. The effector complex consists of a 20- to 30-nt RNA that is tightly bound to members of the Argonaute protein family, equipped with an RNaseH-like domain that confers ssRNA cleavage activity (Song et al. 2004). Bound small RNAs provide a specific recognition system using complementary interactions followed by Argonaute-induced degradation or translational repression of target mRNAs. Although siRNAs and microRNAs are mostly involved in post-transcriptional repression, this pathway has also been shown to influence gene expression by controlling chromatin structure and transcriptional output. Particularly, a single Argonaute protein in fission yeast is involved in establishment and maintenance of repressive chromatin modifications of heterochromatic loci (Noma et al. 2004). In plants, small RNAs bound to the Argonaute member Ago4 direct DNA methylation through direct interaction with the RNA Polymerase IV (El-Shami et al. 2007).
Until recently, it was not clear if small RNAs functioned beyond the level of post-transcriptional regulation in mammals. The situation changed when one branch of the Argonaute protein family, the germline-specific Piwi proteins, was shown to bind a specific class of small RNAs. piRNAs are distinct and often slightly larger as compared with microRNAs and siRNAs and are described in Drosophila, C. elegans, zebrafish, and mammals (Kim 2006). Two classes of piRNAs have been identified in the mouse. The first described piRNA species originated from male germ cells at pachytene stage of meiosis, ∼14 d post-partum (dpp), and were composed of amazingly diverse sequence populations that were relatively TE-poor and mapped mostly to unannotated genomic regions (Aravin et al. 2006; Girard et al. 2006). Their function is currently unknown. On the other side, piRNAs expressed before pachynema, ∼10 dpp, consist of different types of TEs (Aravin et al. 2007b) and appear to be related to repeat-associated siRNAs bound to Piwi members in Drosophila and zebrafish (Aravin et al. 2003; Vagin et al. 2006; Brennecke et al. 2007; Houwing et al. 2007). By their nature, repeat-derived piRNAs are able to target TEs, which is supported by the activation of several elements observed in Piwi mutants in Drosophila and the mouse (Vagin et al. 2006; Aravin et al. 2007a; Carmell et al. 2007). Piwi proteins, similarly to conventional Argonautes, have RNA cleavage activity pointing toward a transcript degradation mode of TE repression. However, mutations in two mouse Piwi members, Mili and Miwi2, also lead to elimination of TE DNA methylation in testes and to a sterility phenotype strikingly identical to Dnmt3L−/− males (Aravin et al. 2007a; Carmell et al. 2007). These data suggested for the first time the existence of an endogenous and developmentally regulated small RNA-directed pathway influencing DNA methylation in the mammalian germline. However, it was not clear whether the piRNA pathway cooperated with the Dnmt3A–Dnmt3L de novo methylating system or was involved in the maintenance of methylation patterns established previously by this complex.
The discovery of repeat-derived piRNAs revealed a system that was ideally suited to the recognition of TEs: piRNAs working as specific guides that have enough sequence complexity to find small pieces of TEs scattered throughout the genome. Furthermore, Piwi proteins and piRNAs in Drosophila and the mouse operate in a cycle that amplifies piRNA sequences matching actively transcribed TEs (Aravin et al. 2007b). The findings of Kuramochi-Miyagawa et al. (2008) demonstrate that piRNAs matching TEs are produced abundantly during the window of de novo methylation in fetal prospermatogonia and are engaged in the so-called ping-pong amplification cycle. By finally showing that the methylation defect was present as early as 16.5 dpc in Mili and Miwi2 mutant prospermatogonia, and therefore at the time of de novo methylation initiation, Kuramochi-Miyagawa et al. (2008) genetically proved that fetal piRNAs were involved in de novo methylation rather than in maintenance of pre-existing methylation. It should be noted, however, that some gametic de novo DNA methylation may be independent of the Piwi/piRNA pathway. Sequences from imprinted or other methylation-regulated genes were not found in deeply sequenced fetal testis piRNA libraries (A.A. Aravin and G. Hannon, unpubl.), suggesting that the role of this pathway may be restricted to the purpose of TE methylation and repression.
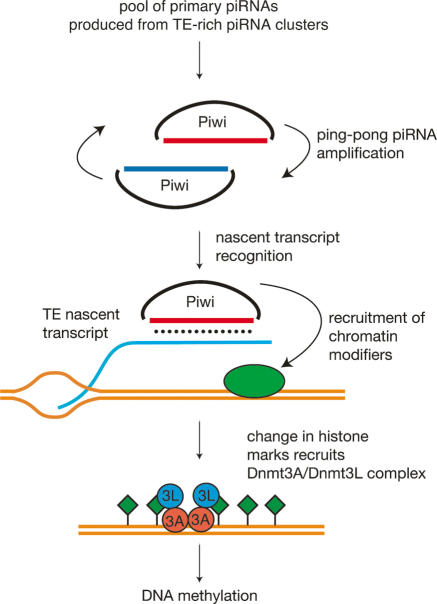
Many questions are still awaiting answers; most importantly, the molecular mechanism underlying piRNA-induced DNA methylation is not known. In order to find their genomic targets, Piwi/piRNA complexes should be present in the nucleus. Nuclear localization was indeed shown for one of the Drosophila Piwi proteins (Brennecke et al. 2007), but not for mammalian Piwi members. By analogy with the situation in fission yeast, it is conceivable that Piwi–piRNA complexes recognize nascent TE transcripts and interact with chromatin-modification factors at target loci in cis (Zofall and Grewal 2006). In this scenario, Piwi–piRNA complexes may first recruit histone-modifying proteins affecting H3K9 and H3K4 methylation states and then promote de novo DNA methylation through the activity of the Dnmt3A–Dnmt3L complex in mammals (Fig. 1). Future studies should also address the specific roles of Piwi members linked to fetal piRNAs Mili and Miwi2 in TE repression and de novo DNA methylation. Through these studies, we may uncover the fascinating links that bring the worlds of small RNAs and de novo methylation together in mammalian germ cells.
Figure 1.
Proposed mechanism for piRNA-induced DNA methylation in male germ cells. Primary piRNAs produced from TE-rich piRNA clusters join Piwi (Mili and/or Miwi2) proteins. Sense (blue) and antisense (red) piRNAs are amplified in the ping-pong cycle. Piwi–piRNAs complexes recognize nascent TE transcripts in the nucleus and recruit chromatin modifiers to TE genomic loci. Subsequent changes in histone marks (H3K4 methylation loss, H3K9 methylation gain, green diamonds) induce the de novo activity of the Dnmt3A–Dnmt3L tetrameric complex to establish methylation patterns.
Concluding remarks
The synergic actions of the Dnmt3A–Dnmt3L complex, chromatin remodeling proteins, histone modifications, and piRNA pathway lead to de novo methylation and repression of TEs in the mammalian male germline. At this point, it is unclear how piRNAs fit into the whole process. Considering the evolutionarily conserved role of piRNAs in transcriptional TE silencing in organisms devoid of genomic methylation, piRNAs might serve as guides for histone-modifying and chromatin remodeling proteins to genomic targets. De novo methylation then mediates a heritable transcriptional repression at these tagged loci in mammals, provided that the CG spacing is compatible with the Dnmt3A substrate preference. Combined genetic inactivation and biochemical experiments will shed light on the epistatic relationships between these different protagonists and their order of involvement in the process of gametic de novo methylation. In vitro models of gametic differentiation should, moreover, provide important clues about the contribution of intrinsic timing mechanisms and external signals produced by the gonadal environment for de novo methylation induction in mammalian germ cells.
Acknowledgments
We thank Christopher Schaefer for critical reading of the manuscript and discussions. Research in D.B.’s laboratory is supported by EURYI grants. We apologize to colleagues who were not directly cited due to the size limitation of the perspective format.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1669408.
References
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophilia melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M.J., Kuramochi-Miyagawa S., Nakano T., et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G.J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007a;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Hannon G.J., Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007b;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Barlow D.P. Methylation and imprinting: From host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T.H. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–2539. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Girard A., van de Kant H.J., Bourc’his D., Bestor T.H., de Rooij D.G., Hannon G.J. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Chedin F., Lieber M.R., Hsieh C.L. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc. Natl. Acad. Sci. 2002;99:16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S.J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C.D., Pradhan S., Nelson S.F., Pellegrini M., Jacobsen S.E. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008 doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- De La Fuente R., Baumann C., Fan T., Schmidtmann A., Dobrinski I., Muegge K. Lsh is required for meiotic chromosome synapsis and retrotransposon silencing in female germ cells. Nat. Cell Biol. 2006;8:1448–1454. doi: 10.1038/ncb1513. [DOI] [PubMed] [Google Scholar]
- Dennis K., Fan T., Geiman T., Yan Q., Muegge K. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes & Dev. 2001;15:2940–2944. doi: 10.1101/gad.929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M., Pontier D., Lahmy S., Braun L., Picart C., Vega D., Hakimi M.A., Jacobsen S.E., Cooke R., Lagrange T. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes & Dev. 2007;21:2539–2544. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith A.C., Greally J.M. Epigenetics: Perceptive enzymes. Nature. 2007;449:148–149. doi: 10.1038/449148a. [DOI] [PubMed] [Google Scholar]
- Gibbons R.J., McDowell T.L., Raman S., O’Rourke D.M., Garrick D., Ayyub H., Higgs D.R. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 2000;24:368–371. doi: 10.1038/74191. [DOI] [PubMed] [Google Scholar]
- Girard A., Sachidanandam R., Hannon G.J., Carmell M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Gowher H., Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: The enzyme modifies DNA in a non-processive manner and also methylates non-CpG sites. J. Mol. Biol. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Houwing S., Kamminga L.M., Berezikov E., Cronembold D., Girard A., van den Elst H., Filippov D.V., Blaser H., Raz E., Moens C.B., et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Jia D., Jurkowska R.Z., Zhang X., Jeltsch A., Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M., Okano M., Hata K., Sado T., Tsujimoto N., Li E., Sasaki H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kareta M.S., Botello Z.M., Ennis J.J., Chou C., Chédin F. Reconstitution and mechanism of the stimulation of de novo methylation by human DNMT3L. J. Biol. Chem. 2006;281:25893–25902. doi: 10.1074/jbc.M603140200. [DOI] [PubMed] [Google Scholar]
- Kato Y., Kaneda M., Hata K., Kumaki K., Hisano M., Kohara Y., Okano M., Li E., Nozaki M., Sasaki H. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 2007;16:2272–2280. doi: 10.1093/hmg/ddm179. [DOI] [PubMed] [Google Scholar]
- Kim V.N. Small RNAs just got bigger: Piwi-interacting RNAs (piRNAs) in mammalian testes. Genes & Dev. 2006;20:1993–1997. doi: 10.1101/gad.1456106. [DOI] [PubMed] [Google Scholar]
- Kubo S., Seleme M.C., Soifer H.S., Perez J.L., Moran J.V., Kazazian H.H., Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl. Acad. Sci. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S., Watanabe T., Gotoh K., Totoki Y., Toyoda A., Ikawa M., Asada N., Kojima K., Yamaguchi Y., Ijiri T.W., et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes & Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifero D., La Salle S., Bourc’his D., Martel J., Bestor T.H., Trasler J.M. Coordinate regulation of DNA methyltransferase expression during oogenesis. BMC Dev. Biol. 2007;7:36. doi: 10.1186/1471-213X-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maksakova I.A., Romanish M.T., Gagnier L., Dunn C.A., de van Lagemaat L.N., Mager D.L. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen. 0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev. Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Noma K., Sugiyama T., Cam H., Verdel A., Zofall M., Jia S., Moazed D., Grewal S.I. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat. Genet. 2004;36:1174–1180. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- Ooi S.K., Qiu C., Bernstein E., Li K., Jia D., Yang Z., Erdjument-Bromage H., Tempst P., Lin S.-P., Allis C.D., et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins R.A., Haghighi F., Edwards J.R., Das R., Zhang M.Q., Ju J., Bestor T.H. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16:157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.P., Clark H.J., Chapman V.M., Rossant J. Differences in DNA methylation during oogenesis and spermatogenesis and their persistence during early embryogenesis in the mouse. Genes & Dev. 1987;1:1039–1046. doi: 10.1101/gad.1.10.1039. [DOI] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Trasler J.M. Gamete imprinting: Setting epigenetic patterns for the next generation. Reprod. Fertil. Dev. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- Vagin V.V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Yamazaki Y., Low E.W., Marikawa Y., Iwahashi K., Bartolomei M.S., McCarrey J.R., Yanagimachi R. Adult mice cloned from migrating primordial germ cells. Proc. Natl. Acad. Sci. 2005;102:11361–11366. doi: 10.1073/pnas.0504943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yokomine T., Hata K., Tsudzuki M., Sasaki H. Evolution of the vertebrate DNMT3 gene family: A possible link between existence of DNMT3L and genomic imprinting. Cytogenet. Genome Res. 2006;113:75–80. doi: 10.1159/000090817. [DOI] [PubMed] [Google Scholar]
- Zhu H., Geiman T.M., Xi S., Jiang Q., Schmidtmann A., Chen T., Li E., Muegge K. Lsh is involved in de novo methylation of DNA. EMBO J. 2006;25:335–345. doi: 10.1038/sj.emboj.7600925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M., Grewal S.I. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb. Symp. Quant. Biol. 2006;71:487–496. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]