Abstract
The enzymes catalyzing lysine and arginine methylation of histones are essential for maintaining transcriptional programs and determining cell fate and identity. Until recently, histone methylation was regarded irreversible. However, within the last few years, several families of histone demethylases erasing methyl marks associated with gene repression or activation have been identified, underscoring the plasticity and dynamic nature of histone methylation. Recent discoveries have revealed that histone demethylases take part in large multiprotein complexes synergizing with histone deacetylases, histone methyltransferases, and nuclear receptors to control developmental and transcriptional programs. Here we review the emerging biochemical and biological functions of the histone demethylases and discuss their potential involvement in human diseases, including cancer.
[Keywords: Cancer, differentiation, demethylases, epigenetic, histone, JmjC]
Histones constitute the basic scaffold proteins around which DNA is wound to form the highly ordered structure of chromatin. Histones, and in particular their tails, are subjected to a plethora of post-translational modifications that have been implicated in chromatin remodeling and closely linked to transcriptional regulation, DNA replication, and DNA repair (for recent reviews, see Berger 2007; Kouzarides 2007). Histone acetylation and methylation represent the most common modifications of the histone tails. These modifications differ in two ways: Histone acetylation results in a negative charge of the modified lysine residue, causing a decreased interaction between the histone and DNA that is generally associated with active transcription. In contrast, methylation of histones occurs at both arginine and lysine residues, and does not influence the net charge of the affected residues, and hence, has no effect on DNA–histone interactions. Rather, the effect of histone methylation impacts on the transcriptional activity of the underlying DNA by acting as a recognition template for effector proteins modifying the chromatin environment and leading to either repression or activation. Thus, histone methylation can be associated with either activation or repression of transcription depending on which effector protein is being recruited. It should be noted that the unmodified residues can also serve as a binding template for effector proteins leading to specific chromatin states (Lan et al. 2007b).
Arginine residues can be modified by one or two methyl groups; the latter form in either a symmetric or asymmetric conformation (Rme1, Rme2s, and Rme2a), permitting a total of four states: one unmethylated and three methylated forms.
Similarly, lysine residues can be unmethylated, mono-, di-, or trimethylated (Kme1, Kme2, and Kme3), and the extent of methylation at a specific residue is important for the recognition of effector proteins and has therefore impact on chromatin and the transcriptional outcome.
Histone methylation is involved in the regulation of a variety of nuclear processes essential for cellular regulation, homeostasis, and fate. Previously, methylation has been considered to constitute a permanent and irreversible histone modification that defined epigenetic programs in concert with DNA methylation. Recently, however, a large number of enzymes have been discovered with the ability to demethylate methylated histone lysine residues as well as methylated arginines via amine oxidation, hydroxylation or deimination. Here we review the current knowledge on histone demethylases, with a special focus on the Jumonji (JmjC) proteins, their role in chromatin regulation, cellular differentiation, and involvement in human diseases.
The identification of histone demethylases
Methylated lysine and arginine residues on histone tails are believed to constitute important regulatory marks delineating transcriptionally active and inactive chromatin. For instance, tri- and dimethylated Lys 9 on histone H3 (H3K9) is associated with silenced chromatin, whereas tri- and dimethylated Lys 4 on the same histone tail (H3K4) is often connected to euchromatin and active transcription.
In contrast to other histone modifications such as acetylation and phosphorylation, methylation, and in particular trimethylation, has been regarded as irreversible because of the high thermodynamic stability of the N–CH3 bond. Hence, for several years the consensus in the epigenetic field was that the only way to revert histone methylation was by histone exchange or by cleavage of the methylated histone tail. The identification of the amine oxidase LSD1 (KDM1 according to the newly suggested nomenclature) (Allis et al. 2007) as a histone demethylase changed this perception (Shi et al. 2004).
This enzyme demethylates the histone substrate through a flavin adenine dinucleotide (FAD)-dependent amine oxidase reaction. However, due to the requirement of a protonated methyl ε-ammonium group for LSD1-catalyzed oxidation, and thus, for LSD1-mediated demethylation, this and other related (and as-yet-undiscovered) enzymes are unable to catalyze the demethylation of trimethylated lysine residues. However, recently we and others have identified and characterized histone demethylases containing the Jumonji (JmjC) catalytic domain, which can demethylate trimethylated lysines (Cloos et al. 2006; Klose et al. 2006b; Tsukada et al. 2006; Whetstine et al. 2006; Yamane et al. 2006).
The JmjC-driven demethylase reaction is compatible with demethylation of mono-, di-, and trimethylated lysines and, indeed, most of the JmjC histone demethylases characterized so far are capable of demethylating trimethylated lysines, and in most cases favor a trimethylated substrate (e.g., see Couture et al. 2007; Ng et al. 2007). JmjC proteins can also demethylate arginine residues (Chang et al. 2007), and, at least in theory, other protein substrates or nucleotides. Certainly, the fact that JmjC domain-containing proteins are also found in bacteria that do not contain histones suggests that these proteins also serve other purposes in addition to being histone demethylases.
The amine oxidase LSD1 (KDM1)
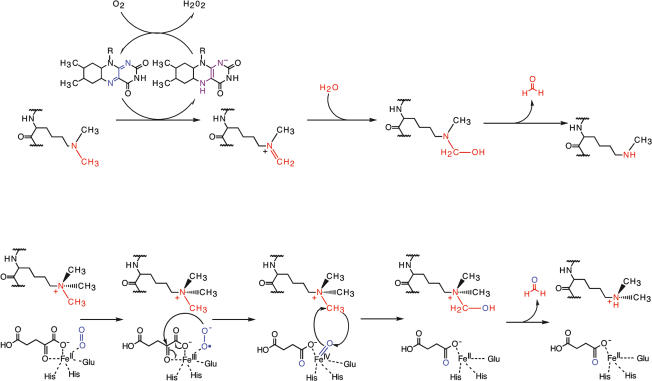
The idea that oxidases could function as histone demethylases was originally proposed by Kouzarides and coworkers (Bannister et al. 2002). Subsequently, in a groundbreaking study, the Shi lab showed (Shi et al. 2004) that the amine oxidase LSD1 (synonyms: BHC110, KIAA0601, p110b, and KDM1), which had been identified previously as a component of a number of corepressor complexes (You et al. 2001), could demethylate di- and monomethylated H3K4 (H3K4me2/me1) (Shi et al. 2004). LSD1 belongs to the class of FAD-dependent monoamine oxidases that typically catalyze the oxidation of amine-containing substrates utilizing molecular oxygen as electron acceptor. In LSD1-catalyzed histone demethylation, the amino group of the methylated lysine in question is oxidized, presumably to generate an imine intermediate that will spontaneously hydrolyze to produce formaldehyde and the corresponding amine residue (Fig. 1A). Substrate oxidation leads to the two-electron reduction of the cofactor FAD, which is reoxidized by molecular oxygen to produce hydrogen peroxide (Fig. 1A). The formation of the imine intermediate, and hence the LSD1-mediated demethylation, is as mentioned above, critically dependent on the protonation of the nitrogen. Thus, this enzyme can only catalyze demethylation of mono- and dimethylated lysines.
Figure 1.
Mechanisms of lysine demethylation by LSD1 and JMJC proteins. (A) LSD1 demethylates H3K4me2/me1 via an amine oxidation reaction using FAD as a cofactor. The imine intermediate is hydrolyzed to an unstable carbinolamine that subsequently degrades to release formaldehyde. (B) The JMJC proteins use αKG and iron (Fe) as cofactors to hydroxylate the methylated histone substrate. Fe(II) in the active site activates a molecule of dioxygen to form a highly reactive oxoferryl [Fe(IV) = O] species to react with the methyl group. The resulting carbinolamine intermediate spontaneously degrades to release formaldehyde. Throughout the figure, the wavy line indicates attachment to the peptide backbone.
LSD1 has been identified as a part of both repressive and activating complexes. Intriguingly, the demethylase activity and specificity of LSD1 appear to be determined by its binding partners, including CoREST, the androgen receptor (AR), and BHC80 (Lee et al. 2005; Metzger et al. 2005; Shi et al. 2005), as well as by the neighboring histone marks surrounding the substrate (Forneris et al. 2005, 2006). Thus, when associated with the repressive CoREST complex, it acts as an H3K4me2/me1 demethylase (Shi et al. 2004). In contrast, when in a complex with the AR it acts as a transcriptional coactivator, demethylating H3K9me2/me1 (Metzger et al. 2005). Here, inhibition of LSD1 expression resulted in an increase of H3K9 methylation of AR target genes and a concomitant decreased transcription of these, directly linking LSD1 to AR-dependent transcriptional activation.
In another study, LSD1 has been linked to estrogen receptor (ER) signaling (Garcia-Bassets et al. 2007). Garcia-Bassets et al. (2007) investigated the occupancy of estrogen-responsive promoters by chromatin modifiers in estradiol-stimulated MCF-7 breast cancer cells. Surprisingly, of 580 promoters found to be occupied by ERα, 58% were co-occupied by LSD1. RNAi-mediated inhibition of LSD1 expression led to decreased expression of ERα target genes co-occupied by LSD1, but not by those not occupied by LSD1. Moreover, LSD1, dependent on its catalytically activity, was able to restore ERα-induced expression of LSD1-positive genes (Garcia-Bassets et al. 2007). Taken together, these findings suggest that LSD1, by demethylating H3K9me2/me1, acts as an essential coactivator of a significant number of ERα-responsive genes.
Studies performed in Schizosacchaomyces pombe and Drosophilia melanogaster have implicated LSD1 orthologs in the organization of higher-order structure of chromatin (Nicolas et al. 2006; Lan et al. 2007c; Rudolph et al. 2007). These studies have corroborated the notion that LSD1 can act as a H3K9 demethylase associated with heterochromatin boundaries and euchromatic promoters (Nicolas et al. 2006; Lan et al. 2007c) or as a H3K4 demethylase (Rudolph et al. 2007). Thus, in S. pombe the LSD1 ortholog SWIRM1/2 (also referred to as spLsd1/2) has been demonstrated to demethylate H3K9 and depletion of SWIRM1/2 induces heterochromatin spreading beyond normal heterochromatin regions (Lan et al. 2007c). Consistent with a role as a transcriptional corepressor, transcription is decreased at adjacent genomic sites, correlating with an increase in H3K9 methylation. In contrast, the D. melanogaster LSD1 ortholog Su(var)3-3 (dLsd1) has been shown to demethylate H3K4me2 and to be important for subsequent H3K9 methylation and heterochromatin formation (Rudolph et al. 2007). Deletion of dLsd1 strongly reduces viability of mutant flies and leads to sterility and defects in ovary development (Di Stefano et al. 2007). Interestingly, mutant alleles of dLsd1 suppress positional effect variegation, indicating the involvement of dLsd1 in balancing euchromatin and heterochromatin (Di Stefano et al. 2007). In concert, these studies suggest an important role for LSD1 in the regulation of chromatin boundaries in these organisms (for review, see Chosed and Dent 2007). Mouse knockout studies have demonstrated that deletion of LSD1 leads to embryonic lethality, and that LSD1 is required for late cell lineage determination and differentiation (Wang et al. 2007b). Moreover, these studies suggest that LSD1 primarily affects gene activation or repression programs by recruiting specific coactivator or corepressor complexes (Wang et al. 2007b). In addition to the roles as a histone demethylase, a recent study reported that LSD1 demethylates a methylated form of Lys 370 of the transcription factor p53 (J. Huang et al. 2007). Whereas in vitro studies demonstrate that LSD1 can demethylate both mono- and dimethylated K370 on p53, the enzyme shows a strong preference for dimethylated K370 in vivo, repressing p53 function by inhibiting its interaction with 53BP1 (J. Huang et al. 2007).
In summary, LSD1 can function as both a histone demethylase specific for H3K4me2/me1 and H3K9me2/me1 and for nonhistone substrates, such as p53. The specific activity of the enzyme is determined partially by its association with different complexes, thereby allowing it to participate in the regulation of transcriptional programs, heterochromatin spreading and stress-induced responses.
The Jumonji (JmjC) domain protein family
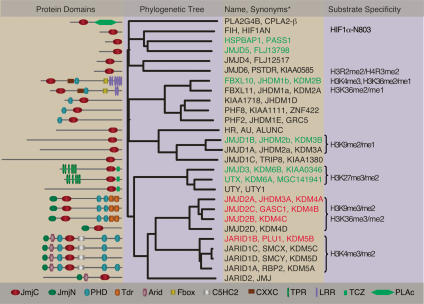
Similar to the discovery of LSD1, a JmJC domain-driven demethylase reaction was proposed in a review before the actual experimental characterization of the demethylases (Trewick et al. 2005). The Jumonji protein family contains the conserved JmjC domain that was first identified in the Jumonji protein (JARID2). Jumonji means cruciform in Japanese, and the gene was so named because mice with a genetrap inserted in the Jumonji locus develop an abnormal cross-like neural tube (Takeuchi et al. 1995). There are 27 different JmjC domain proteins within the human genome, of which 15 have been published to demethylate specific lysines or arginines in the H3 tail (Fig. 2). The first JmjC domain demethylase described, was FBXL11 (synonyms: JHDM1a and KDM2A), which was shown to specifically demethylate mono- and dimethylated H3K36 in a Fe(II) and α-ketoglutarate (αKG)-dependent manner (Tsukada et al. 2006).
Figure 2.
Phylogenetic tree of the JmjC family of demethylases. The names, synonyms, substrate specificities, and domain structures of the proteins are provided. The lists of synonyms may be longer, but due to space limitations only the most relevant are provided. Putative oncoproteins are in red and putative tumor suppressors are in green. (JmjC) Jumonji C domain; (JmjN) Jumonji N domain; (PHD) plant homeodomain; (Tdr) Tudor domain; (Arid) AT-rich interacting domain; (Fbox) F-box domain; (C5HC2) C5CHC2 zinc-finger domain; (CXXC) CXXC zinc-finger domain; (TPR) tetratricopeptide domain; (LRR) leucine-rich repeat domain; (TCZ) treble-clef zinc-finger domain; (PLAc) cytoplasmic phospholipase A2 catalytic subunit.
Although the reaction mechanism for the FBXL11mediated demethylation, at least in theory, could utilize trimethyl H3K36 as substrate, no such activity could be demonstrated. Thus, even though the identification of LSD1 and FBXL11 as histone demethylases represent important milestones for epigenetic research, by demonstrating the dynamic regulation of methyl marks, they did not resolve the question of the reversibility of trimethylated lysine marks.
However, only a few months after publication of the FBXL11 enzyme, a flurry of papers showed that the JMJD2 protein can demethylate H3K9me3/me2 and H3K36me3/2 (Cloos et al. 2006; Fodor et al. 2006; Klose et al. 2006b; Whetstine et al. 2006), formally demonstrating the reversibility of trimethylated lysine marks. Today, these discoveries have been followed by the identification of the JMJD1 protein group that demethylates H3K9me2/me1 (Yamane et al. 2006), the JARID1 group of H3K4me3/me2-specific demethylases (Christensen et al. 2007; Iwase et al. 2007; Klose et al. 2007; Lee et al. 2007a; Tahiliani et al. 2007), and lately, the UTX/JMJD3 group, capable of demethylating H3K27me3/me2/me1 (Agger et al. 2007; De Santa et al. 2007; Jepsen et al. 2007; Lan et al. 2007a; Lee et al. 2007b).
The JmjC proteins belong to the αKG-dependent oxygenase super family. A widespread feature of these proteins is the ability to bind Fe(II) ions. In addition, many of these proteins have the capability to hydroxylate protein substrates utilizing oxo-ferryl(IV) and αKG as cofactors. Moreover, some cupin dioxygenases, including members of the JmjC demethylase family, have the additional requirement of ascorbate for full catalytic activity. Various studies of αKG-dependent oxygenases including the Factor-Inhibiting HIF1 (FIH) suggest the following reaction mechanism: First, the enzyme binds iron (Fe) through its metal-binding motif HXD/EXnH, the so-called facial triad. Then, the Fe(II)–enzyme complex binds the cofactor αKG, and subsequently the substrate and oxygen. The binding of oxygen is followed by the oxidative decarboxylation of αKG to produce succinate, carbon dioxide, and ferryl. The latter is a highly reactive group and can potentially oxidize a C–N bond in a lysine ζ-methyl group, forming an unstable carbinolamine that will rapidly break down, leading to the release of formaldehyde and loss of a methyl group from lysine (Fig. 1B).
The mode of action of ascorbate is presently unclear but has been suggested to reduce Fe(III) to its active state Fe(II) or to function as a “surrogate reducing substrate” to “rescue” the dioxygenase enzyme in the event of the uncoupled production of a ferryl [Fe(IV) = O] intermediate.
When constructing a phylogenetic tree of the Jumonji proteins based on an alignment of their respective JmjC domains (Fig. 2), these proteins segregate in distinct clusters. In general, it appears that each cluster has specificity for demethylating a certain histone mark. So far, most subfamilies conform to this rule; however, there are exceptions—for instance, the FBXL cluster apparently comprises both H3K36me2/me1 and H3K4me3 demethylase activities (Tsukada et al. 2006; Frescas et al. 2007). Below we provide an overview of the features of the different JmjC subfamilies.
The FBXL (KDM2) cluster
Two proteins FBXL11 (synonyms: JHDM1a and KDM2A) and FBXL10 (synonyms: JHDM1b and KDM2B) constitute the FBXL cluster. These were the first JmjC histone demethylases published (Tsukada et al. 2006). Both proteins contain an F-box domain in addition to two leucine-rich repeat (LRR) domains (Fig. 2). FBXL11 was demonstrated to demethylate di- and monomethylated H3K36 (Tsukada et al. 2006). The di- and trimethylated variants of the H3K36 mark are enriched at the 3′-end of actively transcribed genes and are involved in regulating transcriptional elongation (Joshi and Struhl 2005). Thus, Set 2, the histone methyl transferase responsible for the methylation of H3K36 in Saccharmyces cerevisiae, binds the elongating form of RNA polymerase II (RNAPII). The Rpd3 histone deacetylase (HDAC) complex is recruited to H3K36me2 thereby preventing transcription from cryptic promoters (Li et al. 2007). These findings suggest that the H3K36 demethylases could have a role resetting this histone modification when active transcription is turned of. Apart from its role as an H3K36me2/me1 demethylase, little is currently known about the biological role of this enzyme.
As FBXL11, FBXL10 was also originally reported to demethylate H3K36me2/me1 (Tsukada et al. 2006). However, it was recently published that FBXL10 has H3K4me3-specific demethylase activity (Frescas et al. 2007). Frescas et al. (2007) showed that FBXL10 acts in the nucleolus, and is involved in the repression of ribosomal RNA genes. Consistent with a role for FBXL10 in repressing ribosomal RNA genes and thus RNA synthesis, FBXL10 negatively regulates cell proliferation (Frescas et al. 2007). FBXL10 features a DNA-binding CXXC zinc finger and the protein localizes together with the RNA polymerase I transcription factor UBF at nucleolar organizing regions, indicating a stable association with ribosomal DNA (rDNA). Indeed, genome localization analysis demonstrated that FBXL10 bound various regions of the human rDNA repeat and was especially enriched on transcribed CpG-rich areas (Frescas et al. 2007).
In agreement with a role of FBLX10 in transcriptional repression, the protein has been found in a complex comprising the Bcl6 corepressor (BcoR), CK2α, SKP1, YAF2, HP1γ, RING1, CBX8, BMI, and Nspc1/Pcgf1 (Gearhart et al. 2006; Sanchez et al. 2007). Moreover, in addition to repressing the transcription of ribosomal genes, recent studies also showed that FBXL10 functions as a transcriptional repressor of c-JUN target genes (Koyama-Nasu et al. 2007). Thus, FBXL10 is a candidate tumor suppressor gene. This notion has been supported by several studies. First, retroviral insertional mutagenesis within the FBXL10 gene has been shown to cause lymphoma in BLM (Bloom syndrome RecQ protein-like-3 DNA helicase)-deficient mice (Suzuki et al. 2006). Second, inhibition of FBXL10 expression increases cell proliferation (Koyama-Nasu et al. 2007), and third, FBXL10 expression is significantly decreased in various primary brain tumors, including glioblastoma multiforme (Frescas et al. 2007).
F-box domain proteins have foremost been characterized as the components of the SCF ubiquitin–ligase complexes, which recognize the substrate being targeted for ubiquitin-mediated proteolysis. The presence of an F-box domain in the FBXL family proteins could suggest that these proteins regulate transcription through the combined action of demethylation and ubiquitylation of transcription factors or other proteins associated with transcription. In agreement with this, a possible function of the FBXL10–Ring1B complex in H2A ubiquitylation, has been demonstrated (Sanchez et al. 2007).
The JMJD1 (KDM3) cluster
JMJD1A (synonyms: TSGA, JDHM2A, and KDM3A) was originally isolated as a male germ-specific transcript (Hoog et al. 1991). JMJD1A was later shown to be an H3K9me2/me1-specific demethylase (Yamane et al. 2006). The protein features an LXXLL motif that is a signature involved in nuclear receptor interactions (Heery et al. 1997). The expression of JMJD1A is most prominent in testes, and has been implicated in demethylation of H3K9me2 of AR target genes (Yamane et al. 2006). Thus, JMJD1A was found to interact with the AR in a ligand-dependent manner. Inhibition of JMJD1A expression in the prostate cancer cell line LnCaP led to an increase in H3K9me2 in a subset of AR target genes including PSA, NKX3.1, and TMPRSS22, and a decrease in their expression (Yamane et al. 2006). These results show that JMJD1A acts as a coactivator of AR-mediated transcription.
More recently, Jmjd1a genetrap mice were described (Okada et al. 2007); these mice apparently develop normally but are infertile (Okada et al. 2007). A careful analysis of the infertility phenotype revealed that sperm cells from Jmjd1a genetrap mice exhibited post-meiotic chromatin condensation defects, leading to a low number of mature sperm cells, of which all had abnormally shaped heads and the vast majority were immotile (Okada et al. 2007). Moreover, Jmjd1a was found to bind to, and positively regulate, the expression of the two genes, transition nuclear protein (Tnp1) and protamine 1 (Prm1), by removing the repressive H3K9 marks from their promoters. The gene products of Tnp1 and Prm1 are indispensable for the histone replacement process that takes place during the final stages of sperm chromatin condensation and maturation, supporting the notion that JMJD1A contributes to spermiogenesis by controlling the expression of these genes (Okada et al. 2007). In conclusion, JMJD1A appears to fulfill essential roles for spermiogenesis, and its disruption causes male infertility phenotypes reminiscent of human syndromes as azoospermia and globospermia, advancing JMJD1A as a candidate gene for these infertility conditions.
In another study, JMJD1A was found to associate with the cardiac and smooth muscle cell (SMC)-specific transcription factor myocardin and the related proteins MRTF-A and MRTF-B (Lockman et al. 2007). Moreover, genomic localization analysis and gene expression analysis showed that JMJD1A binds to SMC-specific promoters and is regulating TGFβ-mediated activation of these genes. These results suggest that JMJD1A is involved in regulating SMC differentiation.
Human JMJD1B (synonyms: JHDM2B, 5qCNA and KDM3B) has also been identified as an H3K9me2/me1 demethylase (Klose et al. 2006a). The human JMJD1B gene is located at 5q31,a chromosomal area that is often deleted in malignant myeloid disorders, including acute myeloid leukemia and myelodysplasia (Hu et al. 2001). Hu et al. (2001) showed that the enforced expression of JMJD1B in a cell line carrying a 5q deletion inhibits clonogenic growth, indicating that loss of JMJD1B may be involved in the pathogenesis of these malignancies, and that the protein may have tumor suppressor activities.
The protein JMJD1C (also known as TRIP8 or JHDM2C) was originally described a thyroid-hormone receptor (TR)-interacting protein and is closely related to JMJD1A and JMJD1B, but so far, no enzymatic activity has been reported. The protein features a conserved iron-binding HXD/EXnH motif, as well as a αKG-binding site, indicating that this protein might be an H3K9me2/me1-specific histone demethylase. Consistent with such a role, a short variant s-JMJD1C was reported recently to be a transcriptional coactivator interacting with the AR (Wolf et al. 2007). In the brain, s-JMJD1C is foremost present in AR-expressing neuronal population, and its concentration appears to vary according to the hormonal status in a region-specific manner. Of note, Castermans et al. (2007) reported the identification of a de novo balanced paracentric inversion 46,XY,inv(10) with a breakpoint in chromosome 10q21.3 located within the first intron of the JMJD1C gene in a boy with autism. This chromosomal aberration caused a twofold decrease in expression of two of three JMJD1C transcripts, implicating this JmjC family member as a candidate gene for autism. Finally, JMJD1C expression is reduced in breast cancer tissues compared with normal breast tissues, suggesting a possible role in tumor suppression (Wolf et al. 2007).
Hairless (HR) is one of the best-studied members of the JmjC protein family, but has so far not been ascribed histone demethylase activity. The protein has high homology with JMJD1A and JMJD1B, but its potential Fe-binding HXDXnH motif appears to align differently compared with catalytically active JmjC members, and the protein could therefore be devoid of demethylase activity.
Various studies point to a role of HR in hair formation; thus, the protein appears to be crucial for the normal regulation of several key cellular functions for hair growth including (1) the maintenance of the dermal papilla, (2) the disintegration of the inner root sheet, (3) club hair formation, and (4) apoptosis of keratinocytes in the hair follicle (Panteleyev et al. 1999). Numerous studies have underscored the importance of this JmjC member for normal hair growth, a few of which are mentioned below.
HR-null mice are born with fur, but loose their hairs within their first 3 wk after birth, indicating that HR is required for regenerating hair follicles (Beaudoin et al. 2005). The biological mechanisms underlying the reinitiation of hair growth are currently not fully elucidated, but probably require the regulation of several genes by HR. Candidate target genes, most likely repressed by HR, include (1) Soggy, encoding a protein related to Dickkopf-3, a member of the Dickkopf family of Wnt inhibitors; and (2) Wnt modulator in surface ectoderm (Wise), which is involved in the regulation of Wnt signaling in the hair follicle and in hair follicle regeneration (Beaudoin et al. 2005). Several other reports have also suggested HR as a transcriptional corepressor for various nuclear receptors including the vitamin D receptor (VDR), the thyroid hormone receptor (THR), as well as the retinoic acid receptor-related orphan receptors (ROR) α, β, γ (Potter et al. 2001, 2002).
The importance of HR for normal regulation of hair growth in human beings is underlined by the fact that several nonsense, missense, insertion, and deletion mutations of the human HR gene have been identified. All these mutations result in hair loss disorders, including alopecia universalis congenita (AUC; OMIM 203,655) and arthricia with papular lesions (APL; OMIM 209,500) (Table 1). Thompson et al. (2006) demonstrated that several mutations in rat HR corresponding to missense mutations in human APL reduced or abrogated the ability of HR to function as a corepressor activity together with the THR nuclear receptor. Similarly, a large number of HR mutants, previously described as the molecular cause of APL, and of which five resulted in amino acid changes within the JmjC domain of HR, caused loss of corepressor activity and defective interactions with HDAC1 (Wang et al. 2007a). In summary, HR is a transcriptional corepressor essential for hair growth, whose biochemical action may involve the removal of an “activatory” methylation mark.
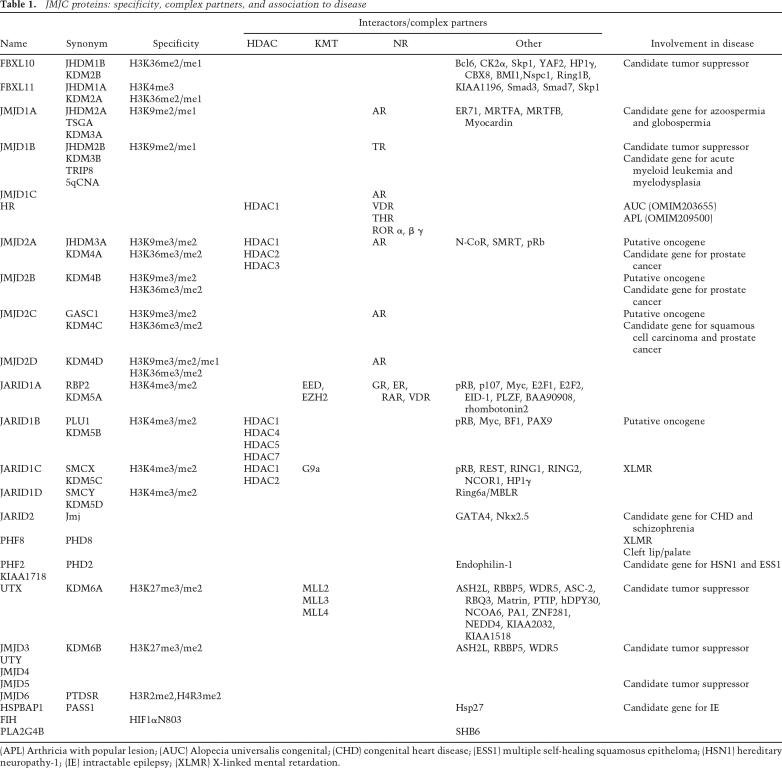
Table 1.
JMJC proteins: specificity, complex partners, and association to disease
(APL) Arthricia with popular lesion; (AUC) Alopecia universalis congenital; (CHD) congenital heart disease; (ESSI) mutliple self-healing squamosus epitheloma; (HSNI) hereditary neuropathy-1; (IE) intractable epilepsy; (XLMR) X-linked mental retardation.
The JMJD2 (KDM4) cluster
The JMJD2 cluster consists of four genes, JMJD2A (synonym: KDM4A), JMJD2B (synonym: KDM4B), JMJD2C (synonyms: GASC1 and KDM4C), and JMJD2D (synonym: KDM4D) (Fig. 2). They direct the expression of histone lysine demethylases capable of demethylating both the H3K9me3/me2 and H3K36me3/me2 marks (Cloos et al. 2006; Fodor et al. 2006; Klose et al. 2006b; Whetstine et al. 2006).
Whereas the H3K36me3/me2 mark is associated with transcriptional elongation (Joshi and Struhl 2005; Keogh et al. 2005; Lee and Shilatifard 2007) and possibly fulfills important roles in suppression of inappropriate transcription within the body of genes (Carrozza et al. 2005), the H3K9me3/me2 mark has generally been associated with transcriptional repression and formation of heterochromatin involving the recruitment of factors such as the HP1 proteins (Bannister et al. 2001; Lachner et al. 2001). Moreover, increased H3K9me3/me2 levels, present in so-called senescence-associated heterochromatic foci (SAHFs) together with HP1, have been associated with stress signals inducing senescence (Narita et al. 2003). Some E2F target genes have been found associated with SAHFs, resulting in permanent shutdown of transcription from these promoters during senescence (Narita et al. 2003). Thus, the idea is that the recruitment of specific H3K9me3 methyltransferases (KMTs) to active promoters or chromosomal regions can result in the generation of a heterochromatic environment and repression of transcription. Due to the apparent important role of the H3K9me3 mark in regulating cellular senescence and chromatin structure, Cloos et al. (2006) searched for proteins binding to this mark in the hope of finding proteins mediating its function. Among the identified proteins was JMJD2C, which originally was named gene amplified in squamous cell carcinoma 1 (GASC1), because of its amplification in esophagus cell lines (Yang et al. 2001).
Enforced expression of JMJD2A, JMJD2B, and JMJD2C significantly decreases H3K9me3 and H3K9me2 levels, and delocalizes HP1, which is consistent with an antagonistic role of the JMJD2 family to heterochromatin (Cloos et al. 2006; Fodor et al. 2006). The possible oncogenic potential of the JMJD2 family could be the result of H3K9me3/me2 demethylation, leading to the dissociation of the SAHFs and re-expression of E2F target genes. Additionally, Whetstine et al. (2006) demonstrated that depletion of the Caenorhabditis elegans JMJD2 homolog, CeJMJD2, resulted in a global increase of H3K9me3 levels, localized H3K36me3 to meiotic chromosomes and activation of p53-dependent germline apoptosis. In this organism, DNA damage-induced apoptosis, as opposed to physiological apoptosis, is dependent on the p53 ortholog CEP-1. Interestingly JMJD2-induced apoptosis was lost in CEP-1 deletion mutants indicating a functional link between JMJD2 loss-of-function and DNA damage-induced germline apoptosis. Moreover, depletion of CeJMJD2 increased RAD51-positive foci in the mid-pachytene nucleus, suggesting an increase in the levels of DNA double-strand breaks (DSB), or a delay in the progression of meiotic DSB repair.
Previous studies have established that disruption of Suv39h1/h2 leads to loss of H3K9 trimethylation at pericentric chromatin, impairment of heterochromatin structures, and genomic stability (Peters et al. 2001). The double-knockout mice also develop B cell lymphomas, suggesting that Suv39h1/h2 act as tumor suppressors. JMJD2A-C proteins are overexpressed in cancer, and inhibition of JMJD2A and JMJD2C affects cellular growth (Cloos et al. 2006). The enzymatic activity of JMJD2 proteins toward demethylation of the repressive marks H3K9me3/me2 indicates that these proteins work as transcriptional coactivators. Taken together, these data suggest that the JMJD2 proteins are oncogenes, which contribute to tumor formation by (1) inducing genomic instability, (2) suppressing of cellular senescence, or (3) activating transcription.
The involvement of the JMJD2 proteins in tumorigenesis has been supported further by a recent report demonstrating the functional interaction between JMJD2C and the AR in prostate carcinomas (Wissmann et al. 2007). Wissmann et al. (2007) showed that JMJD2C can bind to the AR and work as an essential coactivator of AR-induced transcription and cellular growth.
The JMJD2 proteins have also been proposed to work as transcriptional repressors: JMJD2A has been shown previously to be associated with the transcriptional repressor complex, N-CoR (Gray et al. 2005), and the inhibition of JMJD2A expression has been shown to trigger an increase in H3K9me3 levels correlating with expression of the N-CoR target gene ASCL (Klose et al. 2006b).
The fact that JMJD2 proteins can both catalyze the demethylation of H3K9 and H3K36 is surprising, and suggests multiple roles for these enzymes. The crystallization of the catalytical JmjC domain of JMJD2A together with its peptide substrates has provided insights into the specificity of substrate binding as well as the structural requirements for catalysis (Chen et al. 2007; Couture et al. 2007; Ng et al. 2007). The studies show that despite significant differences in the adjacent sequence motifs of lysine K9 and K36, the two substrates have a very similar, but not identical, binding mode. Moreover, two of the studies found the K9 lysine to be the favored substrate (Couture et al. 2007; Ng et al. 2007), whereas the last could not detect significant differences in affinity toward K9 and K36 (Chen et al. 2007).
In addition to the catalytic domain the JMJD2 proteins also contain PHD and Tudor domains (Fig. 2), which are expected to make significant contributions to the activity and specificity of these enzymes by guiding them to specific areas of chromatin. Thus, the tandem Tudor domains of JMJD2A has been found to bind to H3K4me3, H3K9me3, and H4K20me3, thereby potentially recruiting this demethylase to areas of active or inactive transcription (Huang et al. 2006; Kim et al. 2006; Lee et al. 2008).
Although we only have limited knowledge regarding the biological role of the JMJD2 proteins, recent studies have implicated JMJD1 and JMJD2 family members in the regulation of self-renewal capacity of mouse embryonic stem cells (ES): The stem cell transcription factor Oct4 was shown to partially control the expression of JMJD1A and JMJD2C, and depletion of the two demethylases by shRNA decreased the expression of stem cell markers and induced genes involved in differentiation (Loh et al. 2007). It is noteworthy that depletion of the two demethylases only modestly affected the expression of stem cell markers. This finding suggests that these demethylases are involved in fine-tuning expression levels during differentiation, rather than being instrumental for cell fate decisions and/or determining the balance between pluripotency and differentiation, such as, for example, the transcription factors Klf4, Sox2, and Oct4.
In summary, current data suggest that the JMJD2 proteins can act as oncogenes, and are essential for normal embryogenesis. They can function as both transcriptional corepressors and coactivators. There are still substantial gaps in our understanding of how the activity and specificity of these proteins is regulated and of the dual role of these proteins in transcriptional regulation. The activity of the JMJD2 proteins may be dictated, as demonstrated for LSD1, by their complex partners. Further studies are required to attain a deeper understanding of these questions.
The JARID1 (KDM5) cluster
The JARID1 subfamily of JmjC proteins encompasses four members: JARID1A, JARID1B, JARID1C, and JARID1D (also called KDM5A-D according to the novel nomenclature), which all can demethylate tri- and dimethylated H3K4 (Christensen et al. 2007; Iwase et al. 2007; Klose et al. 2007; Lee et al. 2007a; Tahiliani et al. 2007; Yamane et al. 2007).
Tri- and dimethylation on histone H3 at Lys 4 (H3K4me3/me2) is often found at transcribed genes. While H3K4me3 is highly enriched around transcriptional start sites, H3K4me2 seems to be present throughout the coding region of transcribed genes (Noma et al. 2001; Santos-Rosa et al. 2002; Liang et al. 2004; R. Schneider et al. 2004; Bernstein et al. 2005). Evidence indicates that trimethylation of K4 first appears after the assembly of the preinitiation complex (PIC), leading to the suggestion that this modification is involved in maintenance of transcription rather than initiation of transcription (Pavri et al. 2006). However, another study has shown that the basal transcription factor TFIID binds to the H3K4me3 mark via the PHD finger of TAF3, indicating that the interaction between TFIID and H3K4me3 may be important for PIC assembly and subsequent activation of genes in mammalian cells (Vermeulen et al. 2007).
Consistent with a contribution of H3K4 methylation to transcription, the bulk of data currently indicates that JARID1 proteins function as transcriptional repressors through the demethylation of H3K4me3/me2, and through the recruitment of other repressive chromatin modifiers (Christensen et al. 2007; Iwase et al. 2007; Klose et al. 2007; Lee et al. 2007a; Tahiliani et al. 2007).
The JARID1 proteins are highly homologous, and contain a similar domain structure comprising a JmjN, Arid/Bright, and C5HC2 zinc-finger domain, in addition to two or three plant homeodomains (PHD) and the catalytic JmjC domain (Fig. 2). Moreover, this group features various conserved motifs, which are probably involved in pRB modulation—among those a leukemia-associated protein (LAP), a rhombotin-2 (RBTN2, LMO2)-binding domain, and a pRB-binding motif (LXCXE). Indeed, JARID1A (synonyms: RBP2 and KDM5A) was originally identified in a screen for proteins binding to the retinoblastoma protein, pRB (Defeo-Jones et al. 1991; Fattaey et al. 1993). It has been reported that the interaction with pRB converts JARID1A from a transcriptional repressor to a transcriptional activator (Benevolenskaya et al. 2005).
In a recent study, Christensen et al. (2007) identified JARID1A and its related proteins JARID1B and JARID1C as demethylases specific for H3K4me3/me2. Consistent with previous reports linking JARID1A to the control of cellular differentiation (Benevolenskaya et al. 2005) JARID1A was found to be an important regulator of HOX transcription (Christensen et al. 2007). Hence, JARID1A occupies several homeotic gene promoters including Hoxa1, Hoxa5, and Hoxa7, and is displaced from Hox genes during differentiation of mouse ES cells in response to retinoic acid, correlating with an increase of their H3K4 trimethylation and expression.
C. elegans and D. melanogaster both encode a single ortholog of the JARID1 demethylases that share significant homology and domain structure with the mammalian JARID1 proteins. The C. elegans JARID1 ortholog rbr-2 is a bona fide H3K4me3 demethylase, which is essential for development. Thus, the C. elegans mutant strain (tm1231) expressing a mutant RBR-2 protein lacking the entire catalytic JmjC domain, displayed a highly penetrant vulval phenotype with 80% of the animals being either multivulval or vulvaless (Christensen et al. 2007). In addition, mutant animals displayed a significant increase in global H3K4 methylation in all larval stages as well as in the adult worm, showing that ongoing H3K4 demethylation by RBR-2 is essential for the correct regulation of H3K4 methylation and for normal development in this organism.
The D. melanogaster JARID1 ortholog, Lid (short for little imaginal disc), is also an H3K4 demethylase, and mutation of Lid results in a global increase of this mark (Secombe et al. 2007). This was rather surprising, because Lid was originally classified as a Trithorax (Trx) gene (Gildea et al. 2000) and the protein was therefore anticipated to maintain H3K4 methylation, rather than removing it. Two proposals have been put forward to rationalize these findings. One model holds that a global increase of H3K4me3 levels in Lid mutants may displace Trx proteins from their appropriate HOX loci, resulting in decreased expression of these genes and a Trx phenotype (Secombe et al. 2007). Another proposition is based on the observation that the transcription factor Myc binds to the JmjC domain of Lid and inhibits its demethylation activity. According to this, Myc could sequester Lid preventing its demethylase activity triggering a local increase in H3K4 trimethylation at Myc target genes. In turn, this could lead to recruitment of activating complexes promoting transcriptional activation at these sites, resulting in a Trx phenotype (Secombe and Eisenman 2007).
Because of the high conservation of the JmjC domain among JARID1 proteins between species, one would predict that the binding between the JARID1 proteins and MYC is conserved during evolution. That this indeed might be the case is supported by data showing that endogenous JARID1A and JARID1B can bind to MYC in ES cells (Secombe et al. 2007).
Whereas mutation of Lid is fatal and mutation of RBR-2 leads to severe developmental phenotypes, Jarid1a-null mice only display minor phenotypes, probably reflecting functional redundancy among mammalian JARID1 proteins (Klose et al. 2007). Thus, apart from neutrophillia and behavioral abnormalities when held upside-down by the tail, these mice appear to be grossly normal. However, a detailed analysis documented decreased apoptosis and increased entry into the G1 phase of the cell cycle in hematopoietic stem cells and myeloid progenitor compartments (Klose et al. 2007). In addition, expression arrays indicated that the levels of several genes were affected in Jarid1a-null mice: The mRNA levels of genes such as Sdf1, Cxlc5, and Foxp2 were increased compared with wild-type littermates, whereas Kcnd3, Sema4a, and Sorbs1 expression was decreased. Genomic location analysis suggests that Sdf1 is a direct target gene for Jarid1a, and that increased Jarid1a binding to the Sdf1 promoter decreased H3K4 trimethylation at this site.
JARID1B (synonyms: PLU1 or KDM5B) was originally identified in a screen for genes regulated by the oncogene c-ErbB2. JARID1B is highly expressed in 90% of ductal breast carcinomas, and the gene has been associated with the malignant phenotype of breast (Lu et al. 1999; Barrett et al. 2002) and prostate cancer (Xiang et al. 2007b). Based on this link to cancer and to its rather restricted expression pattern in normal adult tissues, where only ovary, testes, and the mammary gland of the pregnant female display high levels of the protein, JARID1B was suggested to belong to the family of testis cancer antigens (Chen and Old 1999; Barrett et al. 2002). Consistent with a putative role in promoting cancer, JARID1B is required for the proliferation of the breast cancer cell line MCF-7 and for the tumor growth of mammary carcinoma cells in nude mice (Yamane et al. 2007).
JARID1B target genes have been identified by genomic location analysis, and several of these have been implicated in breast cancer proliferation including 14–3–3σ, BRCA1, CAV1, and HOXA5 (Yamane et al. 2007). In agreement with a role of JARID1B as a transcriptional repressor, inhibition of its expression leads to increased H3K4 trimethylation at these target genes and increased transcription of these genes (Yamane et al. 2007).
Previously, other studies have linked JARID1B to repression of developmental genes. Here, JARID1B was found to bind the developmental transcription factors such as brain factor-1 (BF-1) and paired box 9 (PAX9) via a conserved sequence motif (AXAAXVPX4VPX4VPX8P), termed the VP motif (Tan et al. 2003). Mutation of the VP motif in BF-1 and PAX9 abrogated JARID1B corepression activity (Tan et al. 2003). Both BF-1 and PAX are known to interact with members of the Groucho corepressor family potentially supporting a role for JARID1B in Groucho-mediated transcriptional repression.
JARID1C (synonyms: SMCX and KDM5C) is transcribed from the X chromosome, and it is one of few genes that are not transcriptionally silenced during the X-inactivation process (Wu et al. 1994a). Interestingly, a large number of point mutations have been identified within the JARID1C gene in patients affected by X-linked mental retardation (XLMR) (Wu et al. 1994b; Brown et al. 1995; Jensen et al. 2005; Santos et al. 2006; Tzschach et al. 2006). One of these mutations (A388P) is located in the vicinity of the proteins N-terminal PHD domain. Importantly, in the context of the wild-type protein this PHD domain, but not the A388P mutant, can bind to H3K9me3 (Iwase et al. 2007). Moreover, introduction of this or other XLMR-linked mutations into wild-type JARID1C was found to reduce the in vitro demethylation activity of the protein (Iwase et al. 2007). Collectively, these findings suggest that XLMR-associated mutations in JARID1C may affect both chromatin association and the demethylating activity of the protein, and could be important for the pathophysiology of some forms of XLMR. Consistent with such a role, RNAi-mediated depletion of JARID1C in rat cerebellar granule neurons led to a significant decrease in the dendritic lengths of neurons that could be rescued by reintroduction of the wild-type protein, but not by XLMR-linked mutants (Iwase et al. 2007). The importance of JARID1C for brain development was further underscored by studies in zebrafish. Here, the expression of the JARID1C homolog is largely restricted to the brain during development, and its depletion suggests a role for the protein in neuronal survival (Iwase et al. 2007).
Another report has suggested a direct role for JARID1C in chromatin dynamics and REST-mediated repression (Tahiliani et al. 2007). In this study, JARID1C was copurified with HDAC1 and HDAC2, as well as the histone H3K9 methyltransferase G9a (synonym: KMT1C), and the transcriptional repressor REST. Chromatin immunoprecipitation (ChIP) analysis revealed that JARID1C and REST co-occupy the neuron restrictive silencing elements in the promoters of a subgroup of REST target genes. Moreover, depletion of JARID1C by RNAi led to the derepression of several of these target genes and a simultaneous increase in H3K4 trimethylation at the sodium channel type 2A (SCN2A) and synapsin I (SYN1) promoters (Tahiliani et al. 2007). This observation led to the interesting suggestion that loss of JARID1C activity could abrogate REST-mediated neuronal gene regulation, thus contributing to the pathogenesis of JARID1C-associated XLMR (Tahiliani et al. 2007).
Analogously, JARID1D (synonym: SMCY and KDM5D) was found to associate with the Polycomb-like ring finger-containing protein Ring6a/MBLR (Lee et al. 2007a). Ring6a/MBLR has been identified previously as a component of an E2F6-containing complex with the ability to ubiquitylate Lys 119 on histone H2A (Akasaka et al. 2002; Ogawa et al. 2002; H. Wang et al. 2004). JARID1D was reported to occupy the promoters of the Synapsin, Engrailed 1, and Engrailed 2 genes, and to be implicated in the regulation of the latter (Lee et al. 2007a). Consistent with such a role, depletion of JARID1D by RNAi led to an increase in H3K4me3/me2 levels at the Engrailed 2 promoter and a concurrent increase in binding of the RNAPII machinery and an increased transcription of Engrailed 2 (Lee et al. 2007a). In vitro reconstitution experiments indicated that Ring6a/MBLR could enhance the demethylase activity of JARID1D. Taken together, these findings support a model where recruitment of Ring6a-JARID1D to the transcription start sites of specific promoters leads to loss of K4 methylation, as well as NURF and RNAPII complex members leading to the repression of the affected gene.
In summary, the four JARID1 proteins are specific histone demethylases for H3K4me3 and H3K4me2, whose main function is to work as transcriptional corepressors. Results from C. elegans and D. melanogaster have shown that the proteins are required for normal development, and studies of human malignancies have, in particular, pointed to a role of JARID1B in the development of cancer and JARID1C in XLMR.
The UTX/JMJD3 (KDM6) cluster
This cluster consists of three proteins, UTX (synonym: KDM6A), UTY, and JMJD3 (synonym: KDM6B) (Fig. 2). UTX and JMJD3 are histone demethylases specific for H3K27me3/me2 (Agger et al. 2007; De Santa et al. 2007; Hong et al. 2007; Lan et al. 2007a, b; Xiang et al. 2007a), whereas no activity has been reported for UTY so far.
UTX and UTY are highly homologous and characterized by the presence of 6 tetratricopeptide repeat (TPR) domains in addition to the catalytic JmjC domain and a treble clef zinc-finger domain (Fig. 2). The TPR domain, is a structural motif present in a wide range of proteins in prokaryotes and eukaryotes, and is supposed to constitute an ancient protein–protein interaction module (Blatch and Lassle 1999; D’Andrea and Regan 2003). TPR proteins contain multiple copies of a degenerate 34-amino-acid motif and self-associate via a “knobs and holes” mechanism mediating the assembly of multiprotein complexes. Indeed, several recent studies have demonstrated that UTX forms part of different multiprotein complexes. One study identified UTX as part of an H3K4–methyltransferase complex containing MLL2 (ALR or KMT2B), PTIP, ASC2, ASH2, RBQ3, WDR5, and matrin (Issaeva et al. 2007). Three other studies, found UTX in association with two H3K4–methyltransferase Set1-like complexes containing MLL3 and MLL2, respectively (Cho et al. 2007; Lee et al. 2007b; Patel et al. 2007). In addition to ASH2L, RBBP5, and WDR5, three subunits shared by all human Set1-like complexes this complex also contained hDPY-30, the human homolog of the Sdc1 subunit of the yeast COMPASS/Set1 complex as well as NCOA6, PA1, and PTIP. Of interest, both UTX complexes contained the protein PTIP. PTIP is known to interact with 53BP1, a key regulator of the DNA damage response that translocates to DNA DSB foci upon ionizing radiation. This finding indicates that these UTX-containing complexes and their associated demethylase and methyltransferase activities may be involved in DNA damage response and/or repair. Analogously, De Santa et al. (2007) reported that JMJD3 can coimmunoprecipitate with all three K4-methyltransferase core subunits (ASH2L, RBBP5, and WDR5), providing further support for a general association of H3K27 demethylases with H3K4 KMTs.
Interestingly, in vitro demethylation studies using recombinant UTX have demonstrated the ability of this enzyme to convert trimethylated H3K27 to its unmethylated state (Agger et al. 2007; Lan et al. 2007a). Conversely, several parallel studies have independently found UTY to be enzymatically inactive (Hong et al. 2007; Lan et al. 2007a). This is surprising, given the high homology between UTX and UTY, and may indicate that the latter protein fulfills other functions than demethylation, or that additional factors are required for its activity. Moreover, the fact that UTX escapes X inactivation (Greenfield et al. 1998) would mean that males only have a half dose of UTX-like H3K27 demethylase activity.
In contrast to some of the other demethylases characterized so far, the UTX/JMJD3 group has no or little activity on the physiologically relevant substrate, the nucleosome in vitro (Lan et al. 2007a; J Christensen, K. Agger, P. Cloos, and K. Helin, unpubl.). This finding suggests that these enzymes require additional cofactors for their function; indeed, ectopic expression of UTX alone appears to have little effect on K27 methylation levels in most cell types.
UTX is recruited to the promoters of the anterior genes of the HOXA and HOXB clusters upon differentiation of NT2/D1 cells with retinoic acid (Agger et al. 2007; Lee et al. 2007b). The recruitment correlates with the loss of H3K27me3, SUZ12, and EZH2 from these promoters, and with activation of the genes. Moreover, depletion of UTX prevents H3K27 demethylation of the HOXB1 promoter and its induction in response to retinoic acid treatment (Agger et al. 2007). These results suggest that UTX is required for the activation of the HOX cluster during differentiation.
Similarly, De Santa et al. (2007) demonstrated that JMJD3 is transiently associated with the HOXA7 and HOXA11 promoters; however, the functional importance of this association was not addressed.
De Santa et al. (2007) identified JMJD3 as a gene highly induced in activated macrophages. Bone morphogenetic protein 2 (BMP2) was identified as a JMJD3 target gene by expression studies and ChIP analysis whose expression increased with LPS-induced loss of JMJD3 from its promoter (De Santa et al. 2007). These data indicate that JMJD3 has functional role as a transcriptional activator during differentiation (De Santa et al. 2007).
Various studies have identified H3K27 demethylases as essential for normal development. Depletion of the two zebrafish UTX orthologs, zUTX1 and zUTX2, by morpholino treatment caused a decreased expression of Hox genes, truncations of the posterior notocord, and abnormal development of the posterior structures (Lan et al. 2007a). The phenotypes observed in these animals with disturbed development of the posterior trunk, resemble thrithorax-like phenotypes and are consistent with an antagonistic role of UTX to PcG-mediated silencing (Lan et al. 2007a). Human UTX could partially rescue the phenotype, whereas a catalytically inactive form could not, demonstrating that the H3K27-demethylating activity of the enzyme was responsible for the observed phenotype (Lan et al. 2007a).
Similarly, analysis of mutant strains and the use of RNAi have shown that F18E9.5, one of four UTX/JMJD3 orthologs encoded by C. elegans, which have high homology with human JMJD3, is required for normal gonadal development (Agger et al. 2007).
As discussed above the mammalian UTX and JMJD3 proteins have also been shown to have a role in various differentiation processes in mammalian cells, and in agreement with this notion, JMJD3 is activated by the retinoic acid receptor in neural stem cells (Jepsen et al. 2007).
Finally, because of the implication of these enzymes in cell fate decisions, counteracting pluripotency and because of their putative involvement in the transcriptional regulation of the INK4A-ARF locus these enzymes are putative tumor suppressors (see further below).
JARID2 (Jumonji)
The JARID2 protein also known as Jumonji (JMJ) is phylogeneticly closely related to the JARID1 family. As the JARID1 members, JARID2 features an Arid/Bright domain in addition to a JmjC and JmjN domain (Fig. 2).
The Jarid2 gene was originally identified in a genetrap screen for genes involved in mouse embryonic development (for review, see Jung et al. 2005b). Depending on the genetic background of the mouse strain, Jarid2-null mice die in utero (Takeuchi et al. 1995, 1997, 1999) or neonatally (Lee et al. 2000). The likely cause of death appears to be various heart abnormalities similar to human congenital heart diseases (CHD), including ventricular septal defects, noncompaction of the ventricular wall, double outlet right ventricle, and dilated atria (Lee et al. 2000). In addition, Jarid2-null mice have an abnormal cruciform neural tube morphology (Takeuchi et al. 1995). In situ hybridization studies of markers for cardiac development indicate that cardiomyocytes in Jarid2-null mice are differentiated, but that the transcriptional regulation of genes involved in the formation of heart chambers are defective in late-stage embryos (Lee et al. 2000; Toyoda et al. 2003). Interestingly, Volcik et al. (2004) found a threefold increased risk for CHD in individuals carrying single-nucleotide polymorphisms in exon 6 of the JARID2 gene, suggesting it as a candidate gene for this condition.
At the molecular level, JARID2 appears to function as a repressor of cardiomyocyte proliferation through interaction with pRB (Jung et al. 2005a), which along with its relative p130, are key regulators of cell cycle progression in these cells (MacLellan et al. 2005). JARID2 has been shown to repress the expression of Atrial natriuretic factor (ANF) through physical interaction with two cardiac-restricted transcription factors, the homeodomain protein Nkx2.5 and the zinc-finger protein GATA4 (Kim et al. 2004). More recently, a single-nucleotide polymorphism located in the Arid/Bright domain of JARID2 was associated to schizophrenia, identifying it as a candidate gene for this psychiatric disorder (Pedrosa et al. 2007). So far, no demethylation activity has been assigned to JARID2, but based on its biological function as a transcriptional repressor, JARID2 would be anticipated to remove an activating mark. However, the protein does not feature a fully conserved iron-binding site, and is therefore possibly devoid of demethylation activity.
The PHD finger (PHF) cluster
This cluster consists of three proteins that in addition to the JmjC domain contain a PHD finger domain: PHD finger protein 2 (PHF2), PHD finger protein 8 (PHF8), and KIAA1718 (Fig. 2). No enzymatic activities have been assigned to these proteins, and very little is known about their biology. Truncating mutations in the PHF8 gene are associated with XLMR and cleft lip/palate in affected patients (Siderius et al. 1999; Laumonnier et al. 2005; Koivisto et al. 2007), indicating an important function of PHF8 in the development of cognitive functions and midline formation. Alignment studies and phylogenetic analysis of the PHF cluster indicates that it is closely related to the FBXL and JMJD1 families. Moreover, the residues potentially involved in coordination of the iron and αKG cofactors in both PHF8 and KIAA1718, are identical to the ones found in FBXL11, making these proteins prime candidates for histone demethylases (Klose et al. 2006a). The last member of the PHF family is PHF2; this protein does not feature a conserved HXD/EXnH motif, and it is therefore unlikely that it has demethylase activity. Little is known about the biological function of this protein, but the PHF2 gene is located on human chromosome 9q22 within the candidate region for hereditary neuropathy I (HSN1) (Nicholson et al. 1996; Blair et al. 1997) and multiple self-healing squamosus epitheloma (ESS1) (Goudie et al. 1993).
Interestingly, deletion of the C. elegans PHF2/PHF8 ortholog 4F429 causes embryonic lethality with a low penetrance, suggesting an important developmental role of this protein (http://www.wormbase.org).
HSPBAP1
Heat-shock 27 (Hsp27)-associated protein 1 (HSPBAP1, also denoted PASS1), is a member of the JmjC family, most closely related to JMJD5 and FIH (Fig. 2). The protein features an HXDXnH motif and could potentially be a histone demethylase. HSPBAP1 is expressed in most tissues except the brain. The protein is most abundant in thymus, pancreas, and testis (Liu et al. 2000; Jiang et al. 2001).
As suggested by its name, HSPBAP1 was initially identified in a screen for proteins associated with Hsp27 (Liu et al. 2000). Functionally, HSPBAP1 appears to be involved in the regulation of stress responses in cells by inhibiting the function of Hsp27 (Liu et al. 2000; Jiang et al. 2001).
Hsp27 has a neuroprotective effect during experimentally induced epileptic neuropathology. Interestingly, in this context, HSPBAP1 inhibits the ability of Hsp27 to protect cells against heat shock (Liu et al. 2000) and it is abnormally expressed in the anterior temporal neocortex of patients with intractable epilepsy (IE) (Xi et al. 2007). These results indicate that deregulation of HSPBAP1 may play a role in the development IE.
HSPBAP1 may also have a role in the development of cancer, since the gene has been found associated with a translocation, giving rise to a DIRC3-HSPBAP1 fusion protein in a family with renal cell cancer (Bodmer et al. 2003).
In conclusion, HSPBAP1 appears to be involved in regulating stress responses in cells by inhibiting Hsp27. At present, it is unclear how HSPBAP1 abrogates Hsp27 function, but it could potentially be involved in hydroxylation of some critical residues of Hsp27, similar to the inhibitory activity of FIH on HIF1α.
Arginine demethylases
As originally proposed by Bannister et al. (2002), deiminases might catalyze the reversal of arginine methylation. Members of the peptidyl arginine deiminase (PADI) family deiminate arginine residues by converting them into citrulline (Nakashima et al. 2002; Cloos and Christgau 2004). PADI4 is a nuclear member of the PADI family, and was therefore suggested and subsequently shown to deiminate histones (Cuthbert et al. 2004; Y. Wang et al. 2004). Strictly speaking, PADI4 does not cause demethylation as it catalyzes the conversion of methyl-arginine to citrulline and not an unmodified arginine; therefore, PADI4 rather works antagonistically to arginine methylation.
PADI4-catalyzed citrullination of histones has been associated with estrogen-regulated transcription of the pS2 promoter. This regulation occurs in a cyclic manner; after an initial phase with augmented transcription, arginine methylation is decreased together with a concomitant increase in histone citrullination and recruitment of PADI4 to the promoter (Bauer et al. 2002; Metivier et al. 2003). Hence, PADI4 action and appears to antagonize arginine methylation in vivo repressing transcription.
Recently, JMJD6 (previously believed to be the phosphatidyl serine receptor and therefore denoted PTDSR) was identified as a histone demethylase with specificity to H3R2me2 and H4R3me2, demonstrating that also arginine methylations are reversible (Chang et al. 2007). Although this study provided little insight into the biology of this demethylase, previous studies of the Jmjd6-null mouse have demonstrated that the gene is essential for the differentiation and maturation of a large variety of tissues during embryogenesis (Bose et al. 2004; J.E. Schneider et al. 2004). Hence, loss of Jmjd6 causes perinatal lethality, growth retardation, and a delay in terminal differentiation of multiple organs including the kidney, intestine, liver, and lungs (J.E. Schneider et al. 2004). Moreover, some Jmjd6-null mice display severe malformations of the forebrain and nasal structures and/or impaired eye formation, ranging from defects in the differentiation of the retina to complete absence of the eyes (J.E. Schneider et al. 2004). Jmjd6 also appears to be essential for development of the heart controlling ventricular septal, outflow tract, pulmonary artery, and thymus development (Bose et al. 2004). Consistent with a role for H4R3 demethylation in embryogenesis, H4R3 methylation is lost in metaphase during oocyte development and in preimplantation mouse development (Sarmento et al. 2004).
Several reports have addressed the importance of H3R2 methylation, providing possible clues to the biological role of this mark (Guccione et al. 2007; Kirmizis et al. 2007). Current data suggests that asymmetric dimethylation of H3R2 (H3R2me2a) is antagonistic to trimethylated H3K4; thus, H3R2me2a appears to be a repressive modification preventing gene transcription (Guccione et al. 2007; Kirmizis et al. 2007).
Given the opposing action of this mark to H3K4me3, it may be anticipated that JMJD6 will form a part of activating complexes, collaborating with or being integrated into Trithorax-like complexes catalyzing K4 methylation. Likewise, depletion of JMJD6 might cause “loss of derepression” during development explaining the multitude of severe phenotypes observed in the Jmjd6-null mouse.
Other JmjC proteins
In addition to the JmjC proteins reviewed above, additional JmjC proteins exist; most of these are, however, undescribed in terms of biological activity, and all except for one have not been ascribed any enzymatic activities. The one exception is the factor inhibiting HIF-1α (FIH), which has been identified as an asparaginyl hydroxylase targeting Asp 803 of the transcription factor hypoxia-inducible factor 1α (HIF-1α), the master switch of cellular hypoxia response (Lando et al. 2002a, b). FIH is active under normoxic conditions abrogating the interaction between HIF-1α- and p300-repressing HIF1α activity.
The biological impact of histone lysine demethylation
The versatile histone demethylases
It is widely accepted that epigenetic mechanisms, including signaling involving histone modifications, are essential for the control of gene expression programs and cell fate decisions. Although we are only just starting to uncover the biology of these exciting enzymes, histone demethylases are rapidly surfacing as essential players in an array of important biological processes.
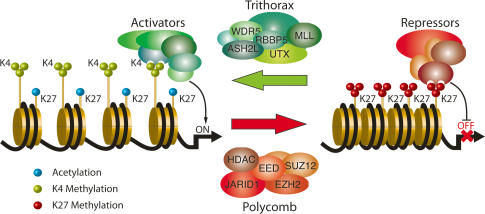
By catalyzing the removal of a histone methyl mark, these enzymes constitute an ideal and versatile regulatory instrument, ensuring the dynamic changes in global gene expression required for diverse cellular processes, such as transdifferentiation of macrophages in response to inflammatory cytokines (De Santa et al. 2007) or stem cell differentiation (Loh et al. 2007). In addition, the fact that both H3K4 and H3K27 KMTs (MLL2, MLL3, and EZH2) and demethylases (RBP2, UTX, and JMJD3) associate with “bivalent” homeotic target genes, suggest that histone demethylation is required for “fine-tuning” transcription by maintaining specific epigenetic states (Fig. 3; Agger et al. 2007; Pasini et al. 2008). Moreover, mathematical modeling suggest that the concerted action of opposing histone-modifying activities will permit a highly dynamic control of the chromatin state without compromising stability (Dodd et al. 2007).
Figure 3.
Model for the involvement of demethylases/methyltransferases in transcriptional regulation of developmental genes. Histone methyltransferases and demethylases are found in the same complex, which methylates one mark while removing the opposing mark. The methylation pattern at a specific gene is determined by the equilibrium between activities of the two opposing complexes, exemplified here by the activating MLL2/UTX complex and the repressive PRC2/RBP2 complex (Agger et al. 2007; Pasini et al. 2008). Analogously, it has been shown that repressive complexes carrying K9 methyltransferase activity (G9a) may also contain H3K4 demethylase activities (Tahiliani et al. 2007). Correspondingly, it may be envisioned that H3K9 demethylases may form a part of activating complexes carrying methyltransferases to activating marks as H3K4.
Since the degree of methylation of the histone tail appears to be an important factor for the regulation of a number of cellular processes such as X inactivation, differentiation, cellular senescence, and DNA damage repair, and histone demethylases take an active part in this, it is anticipated that the expression levels of these enzymes are carefully controlled and their localization exquisitely targeted. Indeed, many demethylases and/or their binding partners feature histone mark-binding modules as Tudor, PHD, or Chromo domains, opening the possibility for specific recruitment of demethylases to certain chromatin areas and for spreading of demethylation to neighboring nucleosomal areas. As an example of this, the PHD domain of JMJD2A has been demonstrated to bind specifically to H3K4me3 H3K9me3 and H4K20me3 (Huang et al. 2006; Kim et al. 2006; Lee et al. 2008). Although the biological significance of this observation is still unclear, this could be one example of demethylase targeting to specific chromatin areas. Another report has suggested that the interplay between the histone demethylase LSD1 and its complex partner BHC80 may be involved in spreading of histone demethylation. Remarkably, the PHD finger of BHC80 was shown to bind unmethylated H3K4, a binding that is specifically abrogated by methylation of H3K4 (Lan et al. 2007b). Gene location analysis demonstrated that LSD1 and BHC80 binding were interdependent, leading to the suggestion that the binding of BHC80 to demethylated K4 may constitute a targeting system leading to the propagation of K4 demethylation and required for the binding of LSD1 (Lan et al. 2007b).
Thus, demethylases appear to represent a fundamental piece of equipment in the cells chromatin toolbox, permitting a fast and robust regulation of transcriptional programs. Evidently, aberrant localization or deregulation of these enzymes may contribute to diseases such as cancer, for instance, by promoting a pluripotent-like state or by disruption of senescence induction as discussed below.
Interaction of histone demethylases with other histone-modifying enzymes and nuclear receptors
Several reports have documented that demethylases are often found in large histone-modifying complexes associated with other chromatin modifiers such as KMTs and HDACs. Thus, as mentioned above, the K27 demethylase UTX has been found in complex with K4 KMTs MLL2 and MML3, and JMJD3 coimmunoprecipitates with core subunits of K4–methyltransferase complexes.
Similarly, the K4 histone demethylase JARID1A is associated with the PRC2 complex, instilling the repressive K27 mark (Fig. 3; Pasini et al. 2008). The fact that depletion of JARID1A by RNAi decreases the ability of PRC2 target genes to remain repressed demonstrates the importance of the concerted action of these activities (Pasini et al. 2008). The apparent intimate association between several demethylases and KMTs affecting marks of complementary activity suggest that this is a general phenomenon, ensuring a fast and efficient shift in expression of the affected gene.
It remains to be established whether these examples constitute a general mechanism for gene regulation. The recent observation that the K4me3/me2 demethylase JARID1C is in a complex with the H3K9 methyltransferase G9a indicates that this may indeed be a general phenomenon (Tahiliani et al. 2007).
Several reports have highlighted the interaction of demethylases and HDACs, and have suggested a possible interplay or coregulation of such activities. As an example of this, one study reported that HR interacts with HDACs and is localized in matrix-associated deacetylase bodies, consistent with a role of HR as a transcriptional corepressor (Potter et al. 2001). Similarly, the JARID1 S. pombe ortholog Msc1 has been shown to interact with HDACs (Ahmed et al. 2004). Cells lacking Msc1 have a 20-fold increase in global acetylation of histone H3, indicating that it may be required for the recruitment or activity of certain HDACs. Moreover, JARID1A has been shown to be part of a complex that contains HDAC activity (Klose et al. 2007). The reintroduction of a catalytically dead JARID1A mutant into Jarid1a-null mice caused repression of the Sdf1 gene (Klose et al. 2007), demonstrating that JARID1A-mediated repression cannot entirely be explained by K4-demethylation activity.
Histone demethylases have also been implicated as essential players in nuclear receptor signaling. Nuclear receptors are transcription factors whose activity is dependent of the binding of lipophilic ligands, triggering their translocation to the nucleus. Most nuclear receptors function by activating transcription of specific target genes. Nuclear receptor-mediated transcriptional activation occurs through binding of the ligand to the C-terminal domain, triggering conformational changes of the receptor into an activator and favoring association with specific coactivator complexes (for review, see Rosenfeld et al. 2006). In addition to this, a subgroup of these receptors including the receptors for retinoic acid, vitamin D, and thyroid hormone have the additional capacity of functioning as transcriptional repressors when not bound to their ligands, primarily by recruiting repressor complexes through the so-called CORNR domain. Moreover, many nuclear receptors including the ERα and the AR can be converted to transcriptional activators even in the absence of a ligand, through other signaling pathways.
The transcriptional control exerted by nuclear receptors is critical for the development and homeostasis of all mammals and is mediated through their interaction with various corepressor complexes. Not surprisingly, given their important biological roles, deregulation of nuclear receptor function has been implicated in a variety of diseases. Indeed, inhibition or activation of nuclear receptors forms the central mechanisms of action for many current drugs, and more than a tenth of the world’s 100 best selling drugs are targeting nuclear receptors.
As mentioned previously, several reports have provided evidence linking demethylase function to the regulation of transcription by nuclear receptors (Table 1; Chan and Hong 2001; Metzger et al. 2005; Yamane et al. 2006; Garcia-Bassets et al. 2007; Wissmann et al. 2007; Wolf et al. 2007). Thus, LSD1, JMJD1A, JMJD1C, and JMJD2C have been involved in AR-mediated transcription (Fig. 4; Metzger et al. 2005; Yamane et al. 2006; Wissmann et al. 2007; Wolf et al. 2007). Accordingly, Garcia-Bassets et al. (2007) found that LSD1 occupied a significant fraction of estrogen-responsive genes, and that the occupancy of this promoter by LSD1 was inversely correlated to H3K9 dimethylation. Conversely, Garcia-Bassets et al. (2007) found that RNAi-mediated depletion of several H3K9 KMTs (RIZ1, ESET, and EuHMTase1) led to the activation of these promoters. This activation was independent of the presence of ligand (estrogen), but relied on the binding of the unligated receptor to the promoter. These data led Garcia-Bassets et al. (2007) to suggest a model where repressive methylations imposed by KMTs act as gatekeepers inhibiting inappropriate transcriptional activation of genes by unligated nuclear receptors. In contrast, the ligand-dependent recruitment of LSD1 (and possibly other demethylases) could act as an activator of ER-mediated transcription by reversing the repressive H3K9 methylation (Garcia-Bassets et al. 2007). This study, taken together with a number of other studies (Table 1), suggests a general regulatory mechanism for transcriptional regulation by nuclear receptors where a carefully coordinated interplay between histone methyltransferases and histone demethylases balance repressing and activating histone modifications, leading to either transcriptional activation or repression.
Figure 4.
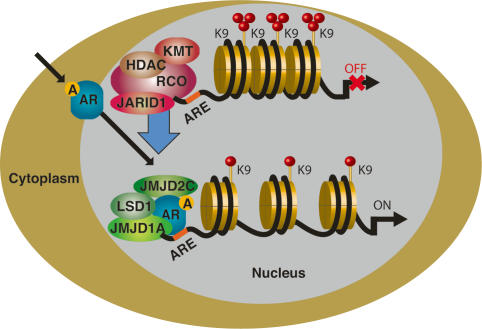
The involvement of demethylases in AR-mediated transcription. When bound to its ligands, androgen (A), the AR translocates to the nucleus to interact with histone demethylases on androgen-responsive elements (ARE) on specific genes. Through its interaction with JMJD2C, LSD1 or JMJD1A demethylation is triggered, removing the repressive H3K9 methylation and leading to the transcriptional induction of these androgen-responsive genes. Repressive complexes (RCO), possibly featuring H3K9-methyltransferase (KMT), HDAC, and H3K4 demethylase (JARID1) activities, may potentially act to prevent ligand-independent activation.
Roles of demethylases in development and differentiation
During cellular differentiation, the transcriptional program of cells is rapidly modulated to acquire new lineage-specific phenotypic states. Concurrently or prior to this, modulation of genome-wide chromatin changes occur with establishment of new histone marks and erasure of others. Chromatin areas featuring both H3K4 and H3K27 methylations are often seen in ES cells or early precursor cells, located at genomic regions encoding transcription factors such as HOX genes that are essential for development and differentiation. Due to the presence of both trimethylated H3K4 and H3K27, which are normally associated with transcriptionally active and silent chromatin, respectively, these areas have been coined bivalent domains, and been proposed to be poised for transcription (Bernstein et al. 2006) or poised for permanent silencing (Christensen et al. 2007). These bivalent marks may provide plasticity to the gene; i.e., the gene can be either activated (K4-methylated/K27-unmethylated) or maintained repressed (K27-methylated/K4-unmethylated) in specific cell lineages during differentiation. Indeed, upon differentiation of ES cells these bivalent domains are induced in a timed and spatially controlled manner or remain silent depending on the phenotypic characteristics of the lineage in question. ChIP studies of cells before, during, and after differentiation has documented that the status of various histone methyl marks is rapidly modulated on specific genes during these processes.
As described above, members of the JARID1 and UTX/JMJD3 groups appear to have specific roles in this regulation. In fact, the JARID1 and UTX/JMJD3 proteins are recruited to HOX promoters or displaced from them in response to retinoic acid differentiation, and they are essential for the correct regulation of the HOX genes (Klose et al. 2006a; Agger et al. 2007; Christensen et al. 2007; De Santa et al. 2007; Lan et al. 2007a; Lee et al. 2007b), revealing a causal relationship between H3K4 and H3K27 demethylation and transcriptional control during these processes.
Moreover, as also described previously, JMJD1A and JMJD2C are involved in regulating ES cell differentiation (Loh et al. 2007). Oct4 positively regulates both genes and inhibition of their expression induces ES cell differentiation and a concomitant increase of lineage-specific genes and decrease of ES cell-specific genes (Loh et al. 2007). Moreover, JMJD1A was shown to be a positive regulator of the pluripotency-associated genes Tcl1, Tcfcp2l1, and Zpf57, whereas JMJD2C acts as a positive regulator of Nanog, encoding a key transcription factor for self-renewal, presumably by removing repressive H3K9 methylations at the promoters of these genes. In addition, several other members of the JmjC family display developmental phenotypes, suggesting putative roles in cellular differentiation. Examples include PHF8, JARID2, and JMJD6.
In summary, several examples exist suggesting that demethylases are at the nexus of cellular differentiation and development. Importantly, since histone demethylases appear to take center stage in executing nuclear receptor function, this protein group may constitute excellent drug targets for novel therapies to cancer and other human diseases.
Putative roles in senescence
Cellular senescence constitutes a barrier against excessive cell proliferation, and is a specific form for irreversible growth arrest that can be induced by a number of different stress inducers, including activation of oncogenes and DNA damage, as well as telomere erosion. Several studies have revealed the importance of the senescence process in suppression of tumorigenesis in vivo (Braig et al. 2005; Collado et al. 2005; Lazzerini Denchi et al. 2005; Michaloglou et al. 2005). The two tumor suppressor proteins pRB and p53 play a central role in the induction of senescence. Stabilization of p53 leads to transcriptional induction of the CDK inhibitor p21, which negatively affects the cell cycle. pRB is found in its active hypophosphorylated form in complex with E2F family members. These complexes induce heterochromatin on E2F-responsive promoters during senescence, resulting in formation of SAHFs (Narita et al. 2003). The exact mechanism leading to SAHF formation is not clear at present. However, recent studies suggest a key role for trimethylation of H3K9 in the establishment of SAHFs and induction of the senescent state. For instance, the accelerated formation of lymphomas in transgenic mice expressing the activated version of the oncogene N-RAS (RASG12D) as a result of Suv39h1 loss correlates with a decreased number of SAHFs and ability to enter senescence in primary lymphocytes (Braig et al. 2005). As suggested above, demethylases removing the repressive H3K9me3 marks may, in theory, act to increase genomic instability, dissolve SAHFs, and prevent or override senescence induction, and may thus potentially provide cancer cells with the possibility to evade this important tumor suppressor mechanism (Cloos et al. 2006).
Another demethylase family, which potentially could be involved in the senescence process, is the UTX/JMJD3 family of H3K27 demethylases. Even though induction of heterochromatin and increased levels of repressive histone marks have been clearly linked to senescence, the gain of repressive K27 methylation at specific genetic loci and concomitant gene silencing has likewise been associated with immortalization and cancer. Thus, the polycomb-repressive complexes PRC2 and PRC1, which catalyze methylation of H3K27 and are involved in chromatin compaction, respectively, are overexpressed or amplified in cancer (Valk-Lingbeek et al. 2004). Moreover, a recent study has provided a direct mechanistic link between PcG activity, H3K27 methylation, and the transcription of the INK4A–ARF locus in senescence (Bracken et al. 2007). This study showed that the PRC2 members, including the H3K27-specific methyltransferase EZH2, and subsequently PRC1 members, are lost from the INK4A–ARF locus as cells undergo senescence, concomitant with the loss of H3K27 and increased transcription of INK4A and ARF. The INK4A–ARF locus is a sensor of signals inducing senescence and the deletion and/or inactivation of the locus, which occurs in a large number of cancers, prevents induction of cellular senescence. The loss of H3K27me3 at the INK4A–ARF locus as cells undergo senescence could, however, be a result of the concerted loss of H3K27 trimethylation activity and increased association of H3K27me3 demethylases (Fig. 5). In this context, it is noteworthy that UTX is generally down-regulated in tumors. Thus, a large number of studies providing normalized expression levels for UTX in tumors are available on the Oncomine online database, and the large majority of these show a significant decrease in UTX expression in diverse malignancies such as melanoma, prostate, brain, and breast carcinoma. Only a limited number of studies provide JMJD3 data, in the online databases, but the gene is significantly decreased in hepatocellular carcinoma.
Figure 5.
Demethylases and histone modifications in senescence. (A) JMJD3 and p53 are transcribed from the same genomic region: chromosome 17p13.1. In response to oncogenic, replicative, or other types of stress (DNA damage, drug treatment, etc.) the tumor suppressors p53 and INK4A–ARF are induced, triggering senescence. JMJD3 or UTX may be involved in the transcriptional activation of the INK4A–ARF tumor suppressor locus by removing repressive K27 methylation from the gene (see B). In turn, p16 and p14ARF lead to the induction of pRB and p53, respectively, triggering senescence and/or apoptosis. (B) The compacted, transcriptionally silent chromatin structure of the INK4A–ARF locus serves to ensure cell cycle progression in normal “unstressed” cells and in some malignant cells. (Left panel) This silent state is maintained by H3K27 methylation mediated by the PcG proteins, and possibly also by the repressive activities of JARID1 proteins and other chromatin modifiers. When cells with intact tumor suppressor pathways are subjected to oncogenic stimuli or other types of stress, PcG proteins are displaced from the INK4A–ARF locus and K27 demethylases are possibly recruited to remove the repressive K27 methylation, leading to the expression of the INK4A– ARF locus and the induction of senescence. (C, left panel) In normal “unstressed” cells the transcription factor E2F mediates transcription, while the tumor suppressor pRB is maintained in an inactive phosphorylated state as a result of low p16 levels. During senescence induction, p16 is induced, causing hypophoshorylation of pRB. (Right panel) pRB subsequently recruits chromatin modifiers as HDAC, SUV39H1 (setting H3K9 methylation), SUV4H20 (setting H4K20 methylation), HP1, and possibly JARID1 proteins to silence euchromatic E2F target promoters and form SAHFs.
Of particular interest, the JMJD3 gene is located on chromosome 17 in close proximity to the p53 tumor suppressor gene. Allele loss of the short arm of chromosome 17, where both p53 and JMJD3 map, is among the most frequently observed genetic lesions in cancer and is found in a variety of human malignancies (Nigro et al. 1989). The allelic losses at 17p13 are significantly correlated with high proliferative activity and associated with more aggressive tumor behavior. Detailed mapping of deletion patterns at this locus and the lack of good correlation between allelic loss at this locus and mutation at p53 has lead to the suggestion that additional, and as of yet undiscovered tumor suppressor genes, may be found in this area that could be responsible for at least a part of the observed malignant phenotype.
It is pertinent to note that a large part of the genetic lesions leading to p53 loss likewise causes the deletion of JMJD3. Thus, given the possibility that JMJD3 may be at the heart of p16 regulation, it could be envisioned that allelic loss at 17p13.1 may cause inactivation of both the p53 and p16 tumor suppressor pathways (Fig. 5A) explaining the aggressive tumor behavior in these cancers.
In addition to the putative involvement of K27 demethylases in the induction of senescence through activation of the INK4A–ARF locus, these proteins may have further tumor suppressor activities by maintaining cells in a nonpluripotent state. Thus, PcG proteins involved in setting the repressive methylation at K27 have been demonstrated to be key players in regulating “stemness” preserving cellular pluripotency by establishing and maintaining repression of specific genes implicated in differentiation (Boyer et al. 2006; Bracken et al. 2006; T.I. Lee et al. 2006). Correspondingly, it could be envisioned that loss of activity, or aberrant localization of K27 demethylases could potentially drive cells toward a more pluripotent and tumorigenic state.
Finally, other demethylases may have roles in senescence, either counteracting it or being involved in its induction. Remarkably, several JmjC genes, including JMJD3, JMJD6, and KIAA1718, are transcriptionally induced in response to ectopic expression of the oncogenic RAS in primary human epithelial cell lines (http://www.oncomine.org). RAS is known to trigger oncogene-induced senescence in primary mammalian cells, indicating that the three JmjC proteins may have roles in senescence induction.
Putative roles in genomic stability and cancer
JmjC proteins are deleted, translocated, mutated, and aberrantly expressed in human cancers. The JMJD3 and JMJD1B genes are located at 17p13.1 and 5q31, respectively. These are genomic areas that are frequently lost in various malignancies (17p13.1), and in myeloid leukemias (5q31). As described previously, HSBAP1 has been identified in a family with renal cell cancer having a t(2;3)(q35;q21) translocation, resulting in a DIRC3-HSPBAP1 fusion transcript (Bodmer et al. 2003). van Zutven et al. (2006) described a patient with NUP98 translocations with AML-M7 with a cryptic t(11;21;12)(p15;p13;p13) complex variant resulting in a NUP98-JARID1A fusion. The resulting NUP98-JARID1A fusion protein, including the first 13 exons of NUP98 and exons 28–31 of JARID1A contains the Phe–Gly (FG) repeats of the N-terminal part of NUP98. Moreover, the fusion protein features the sequence encoding the C-terminal PHD domain of JARID1A, which may be involved in securing the correct localization of the wild-type JARID1A protein. It may be envisioned that the mutant protein could contribute to the leukemic phenotype of AML-M7 patients, perhaps by working as a dominant-negative mutant. In this context, it is noteworthy that ectopic expression of a catalytic inactive mutant of JARID1A results in an increase in global H3K4me3 levels in human cell lines (Christensen et al. 2007).
Another recent study links mutations of JmjC demethylases to hematopoetic cancer. Hence, Suzuki et al. (2006) identified two JmjC proteins, FBXL10 and JMJD5, as candidate tumor suppressors in a mouse model of leukemia. In agreement with this notion an earlier study identified two JmjC genes T26A5.5 and C06H2.3 orthologs of the human FBXL10 and JMJD5 genes, respectively, in a RNAi screen designed to identify genes that contribute to genomic stability in C. elegans somatic cells (Pothof et al. 2003). While the exact mechanisms causing oncogenesis in response to loss-of-function mutations in FBXL10 and JMJD5 remain unclear, the observation that depletion of JMJD5 decreased the survival of cells exposed to N-methyl-N0-nitro-N-nitrosoguanidine (MNNG), suggests that it might fulfill functions in DNA mismatch repair. Finally, the JMJD2 genes JMJD2A, JMJD2B, and JMJD2C are all highly expressed in prostate cancer (Cloos et al. 2006), and the fact that JMJD2C is required for the proliferation of cancer cells (Cloos et al. 2006; Wissmann et al. 2007) point to a role of JMJD2 proteins to the development of cancer. In summary, the observation that genetic alterations of the histone demethylases are often found in cancer, and that several of these have been found to be regulation of cancer cell lines, suggest that activation or inactivation of these proteins contribute to tumor development (Table 1).
Putative roles of demethylases in X inactivation
One of the two X chromosomes becomes inactivated in the developing female embryo. In this way gene products expressed from the X chromosome can be dosage-compensated (Heard 2005). Central to the mechanism of X inactivation is the noncoding RNA Xist, which is expressed from a locus within a region of the X chromosome called the X-inactivation center (Heard 2005). Xist is induced 1 or 2 d following ES cell differentiation, and the Xist transcript subsequently covers the future inactive X chromosome. Xist induction and coating represent an essential and initial event in the X-inactivation process. One of the earliest chromosomal changes that occur during X-chromosome inactivation is the loss of euchromatic histone modifications. Immediately following Xist RNA coating, a reduction in H3K4me3/me2 and H3K9ac is observed at the X-inactivation center as detected by ChIP and immunofluoresence (Heard et al. 2001; Okamoto et al. 2004). Subsequently, the PRC2 complex is transiently recruited to the affected X chromosome, which is accompanied by an increase in the repressive H3K27me3 mark. The molecular mechanisms mediating these early changes in H3K4me3/me2 and H3K9ac are currently unknown, but it has been speculated that enzymes such as HDACs and histone demethylases could be involved in the process (Fig. 6; Heard 2005).
Figure 6.
Demethylases and histone modifications in X inactivation. Xist RNA associates with the X chromosome through unknown factor(s), and recruits an initial silencing activity. Demethylation of H3K4 is an early event in X inactivation, and most likely is mediated by JARID1 family members. HDAC activity is recruited, possibly through JARID1 members to deacetylate H3K9 and H3K27, probably in preparation for the subsequent repressive methylation of these marks. The PRC2 complex transiently associates with the inactivating X to methylate H3K27 and perhaps H3K9. The recruitment of PRC2 could be mediated through JARID1 members. Subsequently, PRC1 proteins transiently associate with the inactivating X chromosome, mediating histone H2A ubiquitinylation. Finally, localized Xist is indispensable for the accumulation of the histone variants macroH2A1 and macroH2A2 on the inactive X chromosome to form the so-called macrochromatin body.
Likewise, little is known about how the PRC2 complex is recruited to the X chromosome. As illustrated in Figure 6, several features point to an involvement of H3K4me3/me2 demethylases in establishing these epigenetic events. The JARID1 proteins are strong candidates as the enzymes mediating the initial K4 demethylation wave. Moreover, as mentioned previously, various JARID1 members have been reported to interact with HDACs. This could be the mechanism by which HDACs are recruited, leading to the secondary deacetylation events, which remove transcriptionally permissive acetyl groups from K9 and K27 positions on histone H3. Finally, the JARID1 members might also be instrumental for the subsequent recruitment of PRC2 members setting the repressive K27me3 mark. Thus, JARID1A has been shown to bind the PRC2 complex and to be required for its full repressive activity on gene transcription (Pasini et al. 2008). An intriguing possibility is that JARID1 family members could be initially recruited to Xist covering the X chromosome destined for inactivation. Hence, all JARID1 members feature a so-called AT-rich interacting domain (ARID) believed to be involved in the binding of DNA or RNA.
Perspectives and future directions
Considering the involvement of histone demethylases in essential cellular processes and their putative implication in various human diseases (Table 1), these enzymes may constitute attractive drug targets. Recently, several studies have reported the inhibitory effect of various small molecules on LSD1 (Culhane et al. 2006; M.G. Lee et al. 2006; Yang et al. 2006; Y. Huang et al. 2007). Significantly, some of these molecules were shown to reactivate transcription of otherwise silenced tumor suppressor genes, raising hopes for the success of such compounds for cancer therapies (Y. Huang et al. 2007). Correspondingly, inhibitors targeting specific members of the JmjC group of histone demethylases may be envisioned with potential for treatment.
While the recent years have provided and revealed many aspects of histone demethylase biology, several interesting questions remain to be answered: One question is whether LSD1 is unique, or whether additional FAD-type demethylases exist. The human genome features a dozen proteins related to LSD1, which potentially could be involved in demethylation of histones; however, so far, no activities have been reported.
Another open question is whether additional classes of histone demethylases exist. The cupin protein superfamily comprises several other enzymes that could potentially function as histone demethylases, including the recently characterized fat mass and obesity-associated (FTO) gene (Gerken et al. 2007) and the previously identified AlkbH enzymes (Falnes et al. 2002; Trewick et al. 2002) that catalyze the removal of methyl groups from alkylated nucleotides. In addition, other types of enzymes may exist with the ability to demethylate histones. Candidate enzymes include NADH/NADPH-dependent oxidoreductases, which could demethylate histones by cycling the NAD+/NADH cofactor in a mechanism similar to that used by LSD1; radical S-adenosyl-methionine (SAM) proteins; or enzymes with two iron centers as ribonucleotide reductases or methane mono-oxygenase-like proteins, which may hydroxylate a carbon backbone by using various reactive radicals and which therefore, in principle, could use a trimethylated lysine as substrate (Chinenov 2002; Shi and Whetstine 2007).
Yet another central question is whether 5-methylcytosine demethylases exist. Given the large importance of CpG methylation in epigenetic signaling and cancer biology, DNA demethylases have long been sought. Various observations indicate the existence of such enzymatic activities. Thus, methylation of CpG islands in imprinted genes is erased during early embryogenesis (embryonic day 10.5–12.5) in primordial germ cells to permit the subsequent setting of gender-specific methylation, suggesting that such enzymes exist. Theoretically, such demethylation could occur “indirectly“ through the exchange of methylated nucleotides by DNA repair mechanisms (for review, see Reik 2007). Considering the speed with which 5-methylcytosine is eliminated from DNA and the problems in a “genome-wide DNA repair mechanism,” direct demethylation seems more plausible. Direct demethylation of 5-methylcytosine would involve breaking a carbon–carbon bond that is energetically less favorable than the scission of a carbon–nitrogen bond as used in histone demethylation. However, it is still a possibility that iron and αKG-dependent enzymes as the JmjC proteins, or as those described above, could catalyze such reactions.
A final question we believe is important to address is if the histone demethylases target other nonhistone substrates. A recent report has demonstrated the involvement of LSD1 in demethylation of the transcription factor and tumor suppressor p53 (J. Huang et al. 2007). Similarly, JmjC proteins may be involved in demethylation or other hydroxylation-derived modifications of nonhistone proteins and/or nucleotides contributing to an increased regulatory complexity of these systems.
In conclusion, there is little doubt that the near future will provide detailed insights into the function and regulation of these enzymes contributing to unravel pathogenesis of various diseases and eventually pave the way for the discovery of novel therapeutic targets and treatment modalities.
Acknowledgments
The work in the Helin laboratory is supported by grants from the Danish Cancer Society, the Danish National Research Foundation, the Danish Medical Research Council, the Danish Natural Science Research Council, the Novo Nordisk Foundation, the Villum Kann Rasmussen Foundation, the European Union Framework 6 program (INTACT and DIAMONDS), and the Association for International Cancer Research. P.A.C.C. is supported by a grant from the Benzon Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1652908.
References
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- Ahmed S., Palermo C., Wan S., Walworth N.C. A novel protein with similarities to Rb binding protein 2 compensates for loss of Chk1 function and affects histone modification in fission yeast. Mol. Cell. Biol. 2004;24:3660–3669. doi: 10.1128/MCB.24.9.3660-3669.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka T., Takahashi N., Suzuki M., Koseki H., Bodmer R., Koga H. MBLR, a new RING finger protein resembling mammalian Polycomb gene products, is regulated by cell cycle-dependent phosphorylation. Genes Cells. 2002;7:835–850. doi: 10.1046/j.1365-2443.2002.00565.x. [DOI] [PubMed] [Google Scholar]
- Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Schneider R., Kouzarides T. Histone methylation: Dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Barrett A., Madsen B., Copier J., Lu P.J., Cooper L., Scibetta A.G., Burchell J., Taylor-Papadimitriou J. PLU-1 nuclear protein, which is upregulated in breast cancer, shows restricted expression in normal human adult tissues: A new cancer/testis antigen? Int. J. Cancer. 2002;101:581–588. doi: 10.1002/ijc.10644. [DOI] [PubMed] [Google Scholar]
- Bauer U.M., Daujat S., Nielsen S.J., Nightingale K., Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin G.M., Sisk J.M., Coulombe P.A., Thompson C.C. Hairless triggers reactivation of hair growth by promoting Wnt signaling. Proc. Natl. Acad. Sci. 2005;102:14653–14658. doi: 10.1073/pnas.0507609102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benevolenskaya E.V., Murray H.L., Branton P., Young R.A., Kaelin W.G. Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol. Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Blair I.P., Dawkins J.L., Nicholson G.A. Fine mapping of the hereditary sensory neuropathy type I locus on chromosome 9q22.1 → q22.3: Exclusion of GAS1 and XPA. Cytogenet. Cell Genet. 1997;78:140–144. doi: 10.1159/000134649. [DOI] [PubMed] [Google Scholar]
- Blatch G.L., Lassle M. The tetratricopeptide repeat: A structural motif mediating protein–protein interactions. Bioessays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Bodmer D., Schepens M., Eleveld M.J., Schoenmakers E.F., van Geurts Kessel A. Disruption of a novel gene, DIRC3, and expression of DIRC3-HSPBAP1 fusion transcripts in a case of familial renal cell cancer and t(2;3)(q35;q21) Genes Chromosomes Cancer. 2003;38:107–116. doi: 10.1002/gcc.10243. [DOI] [PubMed] [Google Scholar]
- Bose J., Gruber A.D., Helming L., Schiebe S., Wegener I., Hafner M., Beales M., Kontgen F., Lengeling A. The phosphatidylserine receptor has essential functions during embryogenesis but not in apoptotic cell removal. J. Biol. 2004;3:15. doi: 10.1186/jbiol10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Dietrich N., Pasini D., Hansen K.H., Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken A.P., Kleine-Kohlbrecher D., Dietrich N., Pasini D., Gargiulo G., Beekman C., Theilgaard-Monch K., Minucci S., Porse B.T., Marine J.C., et al. The Polycomb group proteins bind throughout the INK4A–ARF locus and are disassociated in senescent cells. Genes & Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dorken B., Jenuwein T., Schmitt C.A. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–665. doi: 10.1038/nature03841. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Miller A.P., Carrel L., Rupert J.L., Davies K.E., Willard H.F. The DXS423E gene in Xp11.21 escapes X chromosome inactivation. Hum. Mol. Genet. 1995;4:251–255. doi: 10.1093/hmg/4.2.251. [DOI] [PubMed] [Google Scholar]
- Carrozza M.J., Li B., Florens L., Suganum T., Swanson S.K., Lee K.K., Shia W.J., Anderson S., Yates J., Washburn M.P., et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Castermans D., Vermeesch J.R., Fryns J.P., Steyaert J.G., de Van Ven W.J., Creemers J.W., Devriendt K. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur. J. Hum. Genet. 2007;15:422–431. doi: 10.1038/sj.ejhg.5201785. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Hong W. Retinoblastoma-binding protein 2 (Rbp2) potentiates nuclear hormone receptor-mediated transcription. J. Biol. Chem. 2001;276:28402–28412. doi: 10.1074/jbc.M100313200. [DOI] [PubMed] [Google Scholar]
- Chang B., Chen Y., Zhao Y., Bruick R.K. JMJD6 is a histone arginine demethylase. Science. 2007;318:444–447. doi: 10.1126/science.1145801. [DOI] [PubMed] [Google Scholar]
- Chen Y.T., Old L.J. Cancer-testis antigens: Targets for cancer immunotherapy. Cancer J. Sci. Am. 1999;5:16–17. [PubMed] [Google Scholar]
- Chen Z., Zang J., Kappler J., Hong X., Crawford F., Wang Q., Lan F., Jiang C., Whetstine J., Dai S., et al. Structural basis of the recognition of a methylated histone tail by JMJD2A. Proc. Natl. Acad. Sci. 2007;104:10818–10823. doi: 10.1073/pnas.0704525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: A candidate for histone demethylase activity? Trends Biochem. Sci. 2002;27:115–117. doi: 10.1016/s0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M., et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chosed R., Dent S.Y. A two-way street: LSD1 regulates chromatin boundary formation in S. pombe and Drosophila. Mol. Cell. 2007;26:160–162. doi: 10.1016/j.molcel.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christgau S. Post-translational modifications of proteins: Implications for aging, antigen recognition, and autoimmunity. Biogerontology. 2004;5:139–158. doi: 10.1023/B:BGEN.0000031152.31352.8b. [DOI] [PubMed] [Google Scholar]
- Cloos P.A., Christensen J., Agger K., Maiolica A., Rappsilber J., Antal T., Hansen K.H., Helin K. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature. 2006;442:307–311. doi: 10.1038/nature04837. [DOI] [PubMed] [Google Scholar]
- Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A.J., Barradas M., Benguria A., Zaballos A., Flores J.M., Barbacid M., et al. Tumour biology: Senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Couture J.F., Collazo E., Ortiz-Tello P.A., Brunzelle J.S., Trievel R.C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Culhane J.C., Szewczuk L.M., Liu X., Da G., Marmorstein R., Cole P.A. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 2006;128:4536–4537. doi: 10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- Cuthbert G.L., Daujat S., Snowden A.W., Erdjument-Bromage H., Hagiwara T., Yamada M., Schneider R., Gregory P.D., Tempst P., Bannister A.J., et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- D’Andrea L.D., Regan L. TPR proteins: The versatile helix. Trends Biochem. Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Defeo-Jones D., Huang P.S., Jones R.E., Haskell K.M., Vuocolo G.A., Hanobik M.G., Huber H.E., Oliff A. Cloning of cDNAs for cellular proteins that bind to the retinoblastoma gene product. Nature. 1991;352:251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- DeSanta F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Di Stefano L., Ji J.Y., Moon N.S., Herr A., Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr. Biol. 2007;17:808–812. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I.B., Micheelsen M.A., Sneppen K., Thon G. Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell. 2007;129:813–822. doi: 10.1016/j.cell.2007.02.053. [DOI] [PubMed] [Google Scholar]
- Falnes P.O., Johansen R.F., Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- Fattaey A.R., Helin K., Dembski M.S., Dyson N., Harlow E., Vuocolo G.A., Hanobik M.G., Haskell K.M., Oliff A., Defeo-Jones D., et al. Characterization of the retinoblastoma binding proteins RBP1 and RBP2. Oncogene. 1993;8:3149–3156. [PubMed] [Google Scholar]
- Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes & Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneris F., Binda C., Vanoni M.A., Battaglioli E., Mattevi A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 2005;280:41360–41365. doi: 10.1074/jbc.M509549200. [DOI] [PubMed] [Google Scholar]
- Forneris F., Binda C., Dall’Aglio A., Fraaije M.W., Battaglioli E., Mattevi A. A highly specific mechanism of histone H3-K4 recognition by histone demethylase LSD1. J. Biol. Chem. 2006;281:35289–35295. doi: 10.1074/jbc.M607411200. [DOI] [PubMed] [Google Scholar]
- Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- Garcia-Bassets I., Kwon Y.S., Telese F., Prefontaine G.G., Hutt K.R., Cheng C.S., Ju B.G., Ohgi K.A., Wang J., Escoubet-Lozach L., et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearhart M.D., Corcoran C.M., Wamstad J.A., Bardwell V.J. Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 2006;26:6880–6889. doi: 10.1128/MCB.00630-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken T., Girard C.A., Tung Y.C., Webby C.J., Saudek V., Hewitson K.S., Yeo G.S., McDonough M.A., Cunliffe S., McNeill L.A., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318:1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea J.J., Lopez R., Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudie D.R., Yuille M.A., Leversha M.A., Furlong R.A., Carter N.P., Lush M.J., Affara N.A., Ferguson-Smith M.A. Multiple self-healing squamous epitheliomata (ESS1) mapped to chromosome 9q22-q31 in families with common ancestry. Nat. Genet. 1993;3:165–169. doi: 10.1038/ng0293-165. [DOI] [PubMed] [Google Scholar]
- Gray S.G., Iglesias A.H., Lizcano F., Villanueva R., Camelo S., Jingu H., Teh B.T., Koibuchi N., Chin W.W., Kokkotou E., et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J. Biol. Chem. 2005;280:28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- Greenfield A., Carrel L., Pennisi D., Philippe C., Quaderi N., Siggers P., Steiner K., Tam P.P., Monaco A.P., Willard H.F., et al. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 1998;7:737–742. doi: 10.1093/hmg/7.4.737. [DOI] [PubMed] [Google Scholar]
- Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Luscher B., Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Heard E. Delving into the diversity of facultative heterochromatin: The epigenetics of the inactive X chromosome. Curr. Opin. Genet. Dev. 2005;15:482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Heard E., Rougeulle C., Arnaud D., Avner P., Allis C.D., Spector D.L. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven E., Hoare S., Parker M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hong S., Cho Y.W., Yu L.R., Yu H., Veenstra T.D., Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoog C., Schalling M., Grunder-Brundell E., Daneholt B. Analysis of a murine male germ cell-specific transcript that encodes a putative zinc finger protein. Mol. Reprod. Dev. 1991;30:173–181. doi: 10.1002/mrd.1080300302. [DOI] [PubMed] [Google Scholar]
- Hu Z., Gomes I., Horrigan S.K., Kravarusic J., Mar B., Arbieva Z., Chyna B., Fulton N., Edassery S., Raza A., et al. A novel nuclear protein, 5qNCA (LOC51780) is a candidate for the myeloid leukemia tumor suppressor gene on chromosome 5 band q31. Oncogene. 2001;20:6946–6954. doi: 10.1038/sj.onc.1204850. [DOI] [PubMed] [Google Scholar]
- Huang Y., Fang J., Bedford M.T., Zhang Y., Xu R.M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- Huang J., Sengupta R., Espejo A.B., Lee M.G., Dorsey J.A., Richter M., Opravil S., Shiekhattar R., Bedford M.T., Jenuwein T., et al. 2007p53 is regulated by the lysine demethylase LSD1 Nature 449 :105–108. [DOI] [PubMed] [Google Scholar]
- Huang Y., Greene E., Murray Stewart T., Goodwin A.C., Baylin S.B., Woster P.M., Casero R.A.2007Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes Proc. Natl. Acad. Sci. 104 :8023–8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I., Zonis Y., Rozovskaia T., Orlovsky K., Croce C.M., Nakamura T., Mazo A., Eisenbach L., Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Jensen L.R., Amende M., Gurok U., Moser B., Gimmel V., Tzschach A., Janecke A.R., Tariverdian G., Chelly J., Fryns J.P., et al. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am. J. Hum. Genet. 2005;76:227–236. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K., Solum D., Zhou T., McEvilly R.J., Kim H.J., Glass C.K., Hermanson O., Rosenfeld M.G. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Jiang M., Ma Y., Cheng H., Ni X., Guo L., Xie Y., Mao Y. Molecular cloning and characterization of a novel human gene (HSPBAP1) from human fetal brain. Cytogenet. Cell Genet. 2001;95:48–51. doi: 10.1159/000057016. [DOI] [PubMed] [Google Scholar]
- Joshi A.A., Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Jung J., Kim T.G., Lyons G.E., Kim H.R., Lee Y. Jumonji regulates cardiomyocyte proliferation via interaction with retinoblastoma protein. J. Biol. Chem. 2005a;280:30916–30923. doi: 10.1074/jbc.M414482200. [DOI] [PubMed] [Google Scholar]
- Jung J., Mysliwiec M.R., Lee Y. Roles of JUMONJI in mouse embryonic development. Dev. Dyn. 2005b;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- Keogh M.C., Kurdistani S.K., Morris S.A., Ahn S.H., Podolny V., Collins S.R., Schuldiner M., Chin K., Punna T., Thompson N.J., et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim T.G., Chen J., Sadoshima J., Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 2004;24:10151–10160. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M.T. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A., Santos-Rosa H., Penkett C.J., Singer M.A., Vermeulen M., Mann M., Bahler J., Green R.D., Kouzarides T. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R.J., Kallin E.M., Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006a;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006b;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yan Q., Tothova Z., Yamane K., Erdjument-Bromage H., Tempst P., Gilliland D.G., Zhang Y., Kaelin W.G. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Koivisto A.M., Ala-Mello S., Lemmela S., Komu H.A., Rautio J., Jarvela I. Screening of mutations in the PHF8 gene and identification of a novel mutation in a Finnish family with XLMR and cleft lip/cleft palate. Clin. Genet. 2007;72:145–149. doi: 10.1111/j.1399-0004.2007.00836.x. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Koyama-Nasu R., David G., Tanese N. The F-box protein Fbl10 is a novel transcriptional repressor of c-Jun. Nat. Cell Biol. 2007;9:1074–1080. doi: 10.1038/ncb1628. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lan F., Bayliss P.E., Rinn J.L., Whetstine J.R., Wang J.K., Chen S., Iwase S., Alpatov R., Issaeva I., Canaani E., et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007a;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lan F., Collins R.E., De Cegli R., Alpatov R., Horton J.R., Shi X., Gozani O., Cheng X., Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007b;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F., Zaratiegui M., Villen J., Vaughn M.W., Verdel A., Huarte M., Shi Y., Gygi S.P., Moazed D., Martienssen R.A. S. pombe LSD1 homologs regulate heterochromatin propagation and euchromatic gene transcription. Mol. Cell. 2007c;26:89–101. doi: 10.1016/j.molcel.2007.02.023. [DOI] [PubMed] [Google Scholar]
- Lando D., Peet D.J., Gorman J.J., Whelan D.A., Whitelaw M.L., Bruick R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes & Dev. 2002a;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D., Peet D.J., Whelan D.A., Gorman J.J., Whitelaw M.L. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002b;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Laumonnier F., Holbert S., Ronce N., Faravelli F., Lenzner S., Schwartz C.E., Lespinasse J., Van Esch H., Lacombe D., Goizet C., et al. Mutations in PHF8 are associated with X linked mental retardation and cleft lip/cleft palate. J. Med. Genet. 2005;42:780–786. doi: 10.1136/jmg.2004.029439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Denchi E., Attwooll C., Pasini D., Helin K. Deregulated E2F activity induces hyperplasia and senescence-like features in the mouse pituitary gland. Mol. Cell. Biol. 2005;25:2660–2672. doi: 10.1128/MCB.25.7.2660-2672.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 2007;618:130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- Lee Y., Song A.J., Baker R., Micales B., Conway S.J., Lyons G.E. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 2000;86:932–938. doi: 10.1161/01.res.86.9.932. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Wynder C., Cooch N., Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Wynder C., Schmidt D.M., McCafferty D.G., Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.G., Norman J., Shilatifard A., Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007a;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., Croce L.D., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007b;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Lee J., Thompson J.R., Botuyan M.V., Mer G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008;15:109–111. doi: 10.1038/nsmb1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Gogol M., Carey M., Pattenden S.G., Seidel C., Workman J.L. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes & Dev. 2007;21:1422–1430. doi: 10.1101/gad.1539307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G., Lin J.C., Wei V., Yoo C., Cheng J.C., Nguyen C.T., Weisenberger D.J., Egger G., Takai D., Gonzales F.A., et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Gilmont R.R., Benndorf R., Welsh M.J. Identification and characterization of a novel protein from Sertoli cells, PASS1, that associates with mammalian small stress protein hsp27. J. Biol. Chem. 2000;275:18724–18731. doi: 10.1074/jbc.M001981200. [DOI] [PubMed] [Google Scholar]
- Lockman K., Taylor J.M., Mack C.P. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ. Res. 2007;101:e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- Loh Y.H., Zhang W., Chen X., George J., Ng H.H. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes & Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P.J., Sundquist K., Baeckstrom D., Poulsom R., Hanby A., Meier-Ewert S., Jones T., Mitchell M., Pitha-Rowe P., Freemont P., et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J. Biol. Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- MacLellan W.R., Garcia A., Oh H., Frenkel P., Jordan M.C., Roos K.P., Schneider M.D. Overlapping roles of pocket proteins in the myocardium are unmasked by germ line deletion of p130 plus heart-specific deletion of Rb. Mol. Cell. Biol. 2005;25:2486–2497. doi: 10.1128/MCB.25.6.2486-2497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R., Penot G., Hubner M.R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Metzger E., Wissmann M., Yin N., Muller J.M., Schneider R., Peters A.H., Gunther T., Buettner R., Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Michaloglou C., Vredeveld L.C., Soengas M.S., Denoyelle C., Kuilman T., van der Horst C.M., Majoor D.M., Shay J.W., Mooi W.J., Peeper D.S. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–724. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Hagiwara T., Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J. Biol. Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Ng S.S., Kavanagh K.L., McDonough M.A., Butler D., Pilka E.S., Lienard B.M., Bray J.E., Savitsky P., Gileadi O., von Delft F., et al. Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity. Nature. 2007;448:87–91. doi: 10.1038/nature05971. [DOI] [PubMed] [Google Scholar]
- Nicholson G.A., Dawkins J.L., Blair I.P., Kennerson M.L., Gordon M.J., Cherryson A.K., Nash J., Bananis T. The gene for hereditary sensory neuropathy type I (HSN-I) maps to chromosome 9q22.1-q22.3. Nat. Genet. 1996;13:101–104. doi: 10.1038/ng0596-101. [DOI] [PubMed] [Google Scholar]
- Nicolas E., Lee M.G., Hakimi M.A., Cam H.P., Grewal S.I., Shiekhattar R. Fission yeast homologs of human histone H3 lysine 4 demethylase regulate a common set of genes with diverse functions. J. Biol. Chem. 2006;281:35983–35988. doi: 10.1074/jbc.M606349200. [DOI] [PubMed] [Google Scholar]
- Nigro J.M., Baker S.J., Preisinger A.C., Jessup J.M., Hostetter R., Cleary K., Bigner S.H., Davidson N., Baylin S., Devilee P., et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–708. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- Noma K., Allis C.D., Grewal S.I. Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro K., Gaubatz S., Livingston D.M., Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Okada Y., Scott G., Ray M.K., Mishina Y., Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Panteleyev A.A., Botchkareva N.V., Sundberg J.P., Christiano A.M., Paus R. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am. J. Pathol. 1999;155:159–171. doi: 10.1016/S0002-9440(10)65110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Christensen J., Agger K., Cloos P.A.C., and Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes & Dev. 2008 doi: 10.1101/gad.470008. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.R., Kim D., Levitan I., Dressler G.R. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavri R., Zhu B., Li G., Trojer P., Mandal S., Shilatifard A., Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Pedrosa E., Ye K., Nolan K.A., Morrell L., Okun J.M., Persky A.D., Saito T., Lachman H.M. Positive association of schizophrenia to JARID2 gene. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007;144:45–51. doi: 10.1002/ajmg.b.30386. [DOI] [PubMed] [Google Scholar]
- Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pothof J., van Haaften G., Thijssen K., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H., Tijsterman M. Identification of genes that protect the C. elegansgenome against mutations by genome-wide RNAi. Genes & Dev. 2003;17:443–448. doi: 10.1101/gad.1060703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G.B., Beaudoin G.M., DeRenzo C.L., Zarach J.M., Chen S.H., Thompson C.C. The hairless gene mutated in congenital hair loss disorders encodes a novel nuclear receptor corepressor. Genes & Dev. 2001;15:2687–2701. doi: 10.1101/gad.916701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter G.B., Zarach J.M., Sisk J.M., Thompson C.C. The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain. Mol. Endocrinol. 2002;16:2547–2560. doi: 10.1210/me.2002-0115. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M.G., Lunyak V.V., Glass C.K. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes & Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Rudolph T., Yonezawa M., Lein S., Heidrich K., Kubicek S., Schafer C., Phalke S., Walther M., Schmidt A., Jenuwein T., et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol. Cell. 2007;26:103–115. doi: 10.1016/j.molcel.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Sanchez C., Sanchez I., Demmers J.A., Rodriguez P., Strouboulis J., Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell. Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Santos C., Rodriguez-Revenga L., Madrigal I., Badenas C., Pineda M., Mila M. A novel mutation in JARID1C gene associated with mental retardation. Eur. J. Hum. Genet. 2006;14:583–586. doi: 10.1038/sj.ejhg.5201608. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Sarmento O.F., Digilio L.C., Wang Y., Perlin J., Herr J.C., Allis C.D., Coonrod S.A. Dynamic alterations of specific histone modifications during early murine development. J. Cell Sci. 2004;117:4449–4459. doi: 10.1242/jcs.01328. [DOI] [PubMed] [Google Scholar]
- Schneider J.E., Bose J., Bamforth S.D., Gruber A.D., Broadbent C., Clarke K., Neubauer S., Lengeling A., Bhattacharya S. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev. Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Secombe J., Eisenman R.N. The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: The Myc connection. Cell Cycle. 2007;6:1324–1328. doi: 10.4161/cc.6.11.4269. [DOI] [PubMed] [Google Scholar]
- Secombe J., Li L., Carlos L., Eisenman R.N. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Whetstine J.R. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y.J., Matson C., Lan F., Iwase S., Baba T., Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Siderius L.E., Hamel B.C., van Bokhoven H., de Jager F., van den Helm B., Kremer H., Heineman-de Boer J.A., Ropers H.H., Mariman E.C. X-linked mental retardation associated with cleft lip/palate maps to Xp11.3-q21.3. Am. J. Med. Genet. 1999;85:216–220. [PubMed] [Google Scholar]
- Suzuki T., Minehata K., Akagi K., Jenkins N.A., Copeland N.G. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J. 2006;25:3422–3431. doi: 10.1038/sj.emboj.7601215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Takeuchi T. A gene trap approach to identify genes that control development. Dev. Growth Differ. 1997;39:127–134. doi: 10.1046/j.1440-169x.1997.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Yamazaki Y., Katoh-Fukui Y., Tsuchiya R., Kondo S., Motoyama J., Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes & Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Kojima M., Nakajima K., Kondo S. jumonji gene is essential for the neurulation and cardiac development of mouse embryos with a C3H/He background. Mech. Dev. 1999;86:29–38. doi: 10.1016/s0925-4773(99)00100-8. [DOI] [PubMed] [Google Scholar]
- Tan K., Shaw A.L., Madsen B., Jensen K., Taylor-Papadimitriou J., Freemont P.S. Human PLU-1 Has transcriptional repression properties and interacts with the developmental transcription factors BF-1 and PAX9. J. Biol. Chem. 2003;278:20507–20513. doi: 10.1074/jbc.M301994200. [DOI] [PubMed] [Google Scholar]
- Thompson C.C., Sisk J.M., Beaudoin G.M. Hairless and Wnt signaling: Allies in epithelial stem cell differentiation. Cell Cycle. 2006;5:1913–1917. doi: 10.4161/cc.5.17.3189. [DOI] [PubMed] [Google Scholar]
- Toyoda M., Shirato H., Nakajima K., Kojima M., Takahashi M., Kubota M., Suzuki-Migishima R., Motegi Y., Yokoyama M., Takeuchi T. jumonji downregulates cardiac cell proliferation by repressing cyclin D1 expression. Dev. Cell. 2003;5:85–97. doi: 10.1016/s1534-5807(03)00189-8. [DOI] [PubMed] [Google Scholar]
- Trewick S.C., Henshaw T.F., Hausinger R.P., Lindahl T., Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature. 2002;419:174–178. doi: 10.1038/nature00908. [DOI] [PubMed] [Google Scholar]
- Trewick S.C., McLaughlin P.J., Allshire R.C. Methylation: Lost in hydroxylation? EMBO Rep. 2005;6:315–320. doi: 10.1038/sj.embor.7400379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Tzschach A., Lenzner S., Moser B., Reinhardt R., Chelly J., Fryns J.P., Kleefstra T., Raynaud M., Turner G., Ropers H.H., et al. Novel JARID1C/SMCX mutations in patients with X-linked mental retardation. Hum. Mutat. 2006;27:389. doi: 10.1002/humu.9420. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek M.E., Bruggeman S.W., van Lohuizen M. Stem cells and cancer; The polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- van Zutven L.J., Onen E., Velthuizen S.C., van Drunen E., von Bergh A.R., van den Heuvel-Eibrink M.M., Veronese A., Mecucci C., Negrini M., de Greef G.E., et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45:437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- Vermeulen M., Mulder K.W., Denissov S., Pijnappel W.W., van Schaik F.M., Varier R.A., Baltissen M.P., Stunnenberg H.G., Mann M., Timmers H.T. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Volcik K.A., Zhu H., Finnell R.H., Shaw G.M., Canfield M., Lammer E.J. Evaluation of the jumonji gene and risk for spina bifida and congenital heart defects. Am. J. Med. Genet. A. 2004;126:215–217. doi: 10.1002/ajmg.a.20574. [DOI] [PubMed] [Google Scholar]
- Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R.S., Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang Y., Wysocka J., Sayegh J., Lee Y.H., Perlin J.R., Leonelli L., Sonbuchner L.S., McDonald C.H., Cook R.G., Dou Y., et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Wang J., Malloy P.J., Feldman D. Interactions of the vitamin D receptor with the corepressor hairless: Analysis of hairless mutants in atrichia with papular lesions. J. Biol. Chem. 2007a;282:25231–25239. doi: 10.1074/jbc.M702939200. [DOI] [PubMed] [Google Scholar]
- Wang J., Scully K., Zhu X., Cai L., Zhang J., Prefontaine G.G., Krones A., Ohgi K.A., Zhu P., Garcia-Bassets I., et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007b;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wissmann M., Yin N., Muller J.M., Greschik H., Fodor B.D., Jenuwein T., Vogler C., Schneider R., Gunther T., Buettner R., et al. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 2007;9:347–353. doi: 10.1038/ncb1546. [DOI] [PubMed] [Google Scholar]
- Wolf S.S., Patchev V.K., Obendorf M. A novel variant of the putative demethylase gene, s-JMJD1C, is a coactivator of the AR. Arch. Biochem. Biophys. 2007;460:56–66. doi: 10.1016/j.abb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Wu J., Ellison J., Salido E., Yen P., Mohandas T., Shapiro L.J. Isolation and characterization of XE169, a novel human gene that escapes X-inactivation. Hum. Mol. Genet. 1994a;3:153–160. doi: 10.1093/hmg/3.1.153. [DOI] [PubMed] [Google Scholar]
- Wu J., Salido E.C., Yen P.H., Mohandas T.K., Heng H.H., Tsui L.C., Park J., Chapman V.M., Shapiro L.J. The murine Xe169 gene escapes X-inactivation like its human homologue. Nat. Genet. 1994b;7:491–496. doi: 10.1038/ng0894-491. [DOI] [PubMed] [Google Scholar]
- Xi Z.Q., Sun J.J., Wang X.F., Li M.W., Liu X.Z., Wang L.Y., Zhu X., Xiao F., Li J.M., Gong Y., et al. HSPBAP1 is found extensively in the anterior temporal neocortex of patients with intractable epilepsy. Synapse. 2007;61:741–747. doi: 10.1002/syn.20417. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C.D. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007a;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Ye X., Xu B., Peng Z., Ma Y., Yu Y., Lin H., Chen A.P., et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. 2007b;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Yamane K., Tateishi K., Klose R.J., Fang J., Fabrizio L.A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Yang Z.Q., Imoto I., Pimkhaokham A., Shimada Y., Sasaki K., Oka M., Inazawa J. A novel amplicon at 9p23-24 in squamous cell carcinoma of the esophagus that lies proximal to GASC1 and harbors NFIB. Jpn. J. Cancer Res. 2001;92:423–428. doi: 10.1111/j.1349-7006.2001.tb01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Gocke C.B., Luo X., Borek D., Tomchick D.R., Machius M., Otwinowski Z., Yu H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell. 2006;23:377–387. doi: 10.1016/j.molcel.2006.07.012. [DOI] [PubMed] [Google Scholar]
- You A., Tong J.K., Grozinger C.M., Schreiber S.L. CoREST is an integral component of the CoREST–human histone deacetylase complex. Proc. Natl. Acad. Sci. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]