Abstract
Three N-linked glycosylation sites were removed from the envelope glycoproteins of Friend, Moloney, and AKV mouse ecotropic gammaretroviruses: gs1 and gs2, in the receptor binding domain; and gs8, in a region implicated in post-binding cell fusion. Mutants were tested for their ability to infect rodent cells expressing 4 CAT-1 receptor variants. Three mutants (Mo-gs1, Mo-gs2, Fr-gs1) infect NIH 3T3 and rat XC cells, but are severely restricted in M. dunni cells and Lec8, a Chinese hamster cell line susceptible to ecotropic virus. This restriction is reproduced in ferret cells expressing M. dunni dCAT-1, but not in cells expressing NIH 3T3 mCAT-1. Virus binding assays, pseudotype assays, and use of glycosylation inhibitors further suggest that restriction is primarily due to receptor polymorphism and, in M. dunni cells, to glycosylation of cellular proteins. Virus envelope glycan size or type does not affect infectivity. Thus, host range variation due to N-glycan deletion is receptor variant-specific, cell specific, virus type-specific, and glycan site-specific.
Keywords: mouse ecotropic gammaretrovirus, CAT-1 retrovirus receptor, gammaretrovirus envelope glycosylation sites, retrovirus entry
INTRODUCTION
The initial event in the process of gammaretrovirus entry is the interaction between the viral envelope (Env) glycoprotein and its cellular receptor. For the mouse leukemia virus (MLV) gammaretroviruses, there are multiple host range variants that are determined by sequence variation in the receptor binding domain (RBD) of the Env SU (surface) component. Each host range group utilizes a different receptor, all of which are transporter proteins with multiple transmembrane domains (Tailor et al., 2003; Stocking and Kozak, 2008). Receptor binding of the RBD triggers a conformational change in Env that results in exposure of the fusion domain of the Env TM (transmembrane) component that then initiates the fusion of viral and cellular membranes.
The viral Envs as well as their cell surface receptors are modified post-translationally by glycosylation. These glycoproteins acquire N-linked oligosaccharides that are initially transferred to nascent polypeptide chains in the ER from a dolichol precursor. These glycans are subsequently modified by glycosidases and glycosyl transferases as the proteins are transported through the Golgi.
The CAT-1 receptor for the ecotropic host range subgroup of MLVs (E-MLVs) carries two consensus recognition sites for N-linked glycosylation (N-X-S/T, where X is any amino acid except P). These glycosylation sites are both occupied by N-glycans (Kim et al., 1993). Both sites are found in the 3rd extracellular loop which also carries the amino acid residues critical for receptor function (Albritton et al., 1993; Yoshimoto et al., 1993). CAT-1 receptors from which N-glycans have been removed by mutagenesis continue to mediate virus entry, and there is some indication that deglycosylated CAT-1 is a more efficient receptor (Wang et al., 1996). Glycosylation is also associated with a number of receptor-mediated resistance phenotypes that have been identified for specific virus-host combinations. These restrictions include the restriction of MoMLV by M. dunni cells (Eiden et al., 1993; Eiden et al., 1994), restriction of E-MLVs by Chinese hamster and rat cells (Miller and Miller, 1992;1993; Tavoloni and Rudenholz, 1997), and restriction of the MoMLV variant Spl574 by laboratory mouse cells (Yan et al., 2008). All of these restrictions are relieved by inhibitors of glycosylation.
The viral Env glycoprotein is also glycosylated. E-MLV Env has a small number of glycans compared to more complex retroviruses like HIV-1; MLVs have 7–9 N-glycans, and one site for O-linked glycosylation (Pinter and Honnen, 1988). Four N-glycan sites, gs3, 5, 7, 8, are variably present. One N-glycan site, gs2, uniquely carries an endoglycosidase H-sensitive high-mannose glycan; the others carry complex types (Kayman et al., 1991). Use of inhibitors to block N-linked glycosylation during virus replication has demonstrated that glycans are critical for the maturation and transport of Env. Inhibitors that specifically block the earliest steps in glycosylation can prevent cleavage of the Env precursor into SU and TM resulting in virions with no Env glycoprotein (Schultz and Oroszlan, 1979). There is limited data on the possible functional roles of the individual Env N-glycans. Felkner and Roth (1992) showed that loss of MoMLV gs2 resulted in temperature sensitive production of virus in Rat2 cells, and loss of gs4 from MoMLV and FrMLV produced noninfectious virus lacking SU protein; the gs4 mutation inhibited processing and prevented incorporation of Env into virus, although it was also shown that a glycan at this site is not required for Env function (Felkner and Roth, 1992; Kayman et al., 1991; Li et al., 1997).
In this study we assess the entry properties of 3 different E-MLV isolates mutagenized to eliminate one of three N-glycan sites: gs1 and gs2, the two glycan sites in the RBD, and gs8, located in a region of the C-terminal domain of SU known to mediate post-receptor binding fusion (Burkhart et al., 2005). We characterize these viruses for their ability to mediate binding and entry with 4 naturally occurring CAT-1 variants found in different rodent species.
RESULTS
Altered host range of viruses lacking specific N-glycan sites
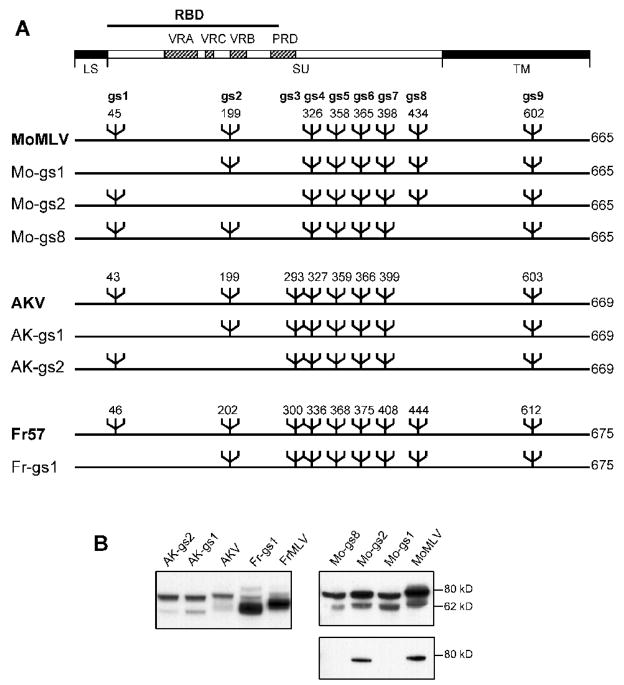
We used site-specific mutagenesis to remove individually one of three sites for N-linked glycosylation from three E-MLVs: MoMLV, FrMLV, and AKV MLV (Fig. 1A). Two sites (gs1, gs2) are located in the 236 amino acid receptor binding domain (RBD) for all three viruses, whereas a third site, gs8, is found in MoMLV and FrMLV but not AKV MLV. This third site maps to a region that has been implicated in post-binding cell fusion (Burkhart et al., 2005), and provides the epitope for the GIX cell surface marker (Rosner et al., 1980). Single glycosylation site mutants were generated for all 3 sites in MoMLV, for the two RBD sites in AKV MLV, and gs1 was removed from FrMLV.
Fig. 1.
N-linked glycosylation in Env of 3 ecotropic gammaretroviruses A) Location of N-glycan sites in the Env genes of MoMLV, FrMLV57 and AKV MLV. Six mutants were generated that lack gs1, gs2 or gs8 as indicated. The domain structure of Env is indicated at the top. RBD: receptor binding domain; VRA, VRC and VRB: variable regions A, C and B; PRD: proline-rich domain; LS: leader sequence. B) Western immunoblot analysis of protein lysates from virus infected cells. NIH 3T3 cells were infected with the indicated viruses. Panels at the left and top right were reacted with goat anti-gp70; the panel in the lower right was reacted with MoMLV specific MAb538.
Western analysis of virus infected NIH 3T3 cells with anti-gp70 shows that, as expected, removal of these sites reduces the size of the Env precursor, gPr80Env, as well as the size of the processed SU for all 6 mutants (Fig. 1B). We also used the MoMLV Env-specific antibody MAb538 to analyze the three MoMLV mutants; MAb538 reacts with gPr80Env but not SU on western blots. MAb538 detects MoMLV and Mo-gs2, but not Mo-gs1 or Mo-gs8 (Fig. 1B), indicating that removal of individual glycosylation sites alters the immunogenicity of this glycoprotein.
We tested the mutant viruses for infectivity on cells of 2 Mus species and 2 non-Mus rodent species with functional, but variant CAT-1 receptors. These 4 receptors differ in their third extracellular loop that carries critical residues for entry (Fig. 2A). dCAT-1 from the Asian M. dunni mouse differs from laboratory mouse mCAT-1 at two sites in this loop. The hamster choCAT-1 and rat xcCAT-1 genes have several substitutions relative to mCAT-1 and have additional residues flanking one of the two glycosylation sites in this loop; rat xcCAT-1 also lacks one of the 2 glycosylation sites (Kubo et al., 2002). Chinese hamster cells are normally resistant to infection by ecotropic MLVs, but this restriction is mediated by glycosylation (Miller and Miller, 1992) and is relieved in Chinese hamster Lec8 cells (Miller and Miller, 1993), a line that lacks GlcNAc-transferase I (Gottlieb et al., 1975), an enzyme involved in N-glycan processing.
Figure 2.
Infectivity of wild type ecotropic MLVs and mutated viruses that lack specific sites for N-glycans A) Amino acid sequence comparisons of rodent variants of the third extracellular loop of the CAT-1 receptor. mCAT-1 is found in laboratory mouse strains and dCAT-1 is found in the Asian wild mouse species M. terricolor (M. dunni). choCAT-1 and xcCAT-1 are the Chinese hamster and rat XC cell variants. Sites for N-glycosylation are shaded. Glycosylation sites were removed from dCAT-1 to produce dCAT-1-g as described previously (Yan et al., 2008). References or GenBank accession Nos. are provided for each sequence. B) Viruses were tested on 5 cell lines carrying 4 naturally occurring variants of the CAT-1 receptor. Virus titers were determined by the XC overlay test in which the indicated cells were infected with virus dilutions, irradiated and overlaid with XC cells to identify virus infected cells (Rowe et al., 1970; Yan et al., 2008). Virus titers are given as the log10 number of XC PFU in 0.2 ml. Chinese hamster E36 cells were resistant to infection by all viruses (not shown). Results for each set of viruses are from one representative experiment; each set was tested at least 3 times. SD of the data are also shown. Arrow: in 8 trials, XC plaques were produced once for the two viruses, with log10 virus titers of 0.2 for Mo-gs1 and 0.3 for Mo-gs2. X: not done.
Susceptibility to 3 ecotropic MLVs and glycosylation mutants was assessed by the XC overlay test (Fig. 2B). MoMLV is restricted in both of the M. dunni-derived cell lines used here; these lines were derived from tail cells cultured from a single mouse (Lander and Chattopadhyay, 1984), but cells were subsequently distributed to different laboratories and now differ in the extent of their resistance to MoMLV. Removal of the gs8 site from MoMLV had no clear effect on the ability of this MoMLV derivative to infect cells carrying mCAT-1, dCAT-1 or choCAT-1. Removal of gs1 and gs2, however, produced viruses that efficiently infected NIH 3T3 cells and XC cells, but were substantially less infectious for M. dunni, MDTF and Lec8 cells than wild type MoMLV. On M. dunni cells, no XC plaques were produced by either Mo-gs1 or Mo-gs2 in 7 of 8 tests, a million-fold reduction compared to the infectivity of these viruses on NIH 3T3, and a reduction of over 100-fold relative to the infectivity of MoMLV in these same cells. We also tested these viruses on MDTF cells, which are more sensitive than M. dunni cells to MoMLV (Fig 2B); replication of the MoMLV mutants was detected, but was reduced at least 1000-fold compared to wild type MoMLV.
AKV MLV and FrMLV57 show small differences in infectivity for NIH 3T3 and M. dunni mouse cells (Fig. 2B). Removal of either gs1 or gs2 from AKV MLV produced viruses showing no differences in infectivity on these cells. However, as was seen for Mo-gs1, Fr-gs1 was substantially less infectious than wild type FrMLV57 for M. dunni cells; this is also consistent with a previous observation that removal of both RBD glycan sites from FrMLV results in reduced infectivity on M. dunni cells (Battini et al., 1994). These results taken together indicate that removal of specific N-glycan sites from the MLV envelope can produce detectable changes in immunogenicity, and that glycans in the RBDs of MoMLV and FrMLV, but not AKV MLV, are needed for efficient entry into cells with 2 of the receptor variants tested here, dCAT-1 and choCAT-1.
Infectivity of viruses with altered N-glycans
To determine if the viruses that showed altered infectivity after removal of specific N-glycan sites would also show altered infectivity if Env N-oligosaccharides were modified by type or size, we used two methods to generate viruses with altered glycans. First, we collected virus grown in Lec8 cells. Because of the absence of GlcNAc-transferase I, Lec8 cell glycoproteins carry smaller Man5(GlcNAc)2-oligosaccharides and the SU glycoproteins produced in these Lec8 cells are markedly reduced in size (Fig. 3A). Second, we treated MLV infected cells with 4 inhibitors of glycosylation to produce viruses with other types of altered N-glycans (Fig. 3B). The inhibitors specifically block different enzymes involved in N-glycosylation: CST blocks glucosidase I and generates immature Glc3Man7–9 oligosaccharides; DNM blocks glucosidase II and produces Glc1–2Man7–9 structures; DMM blocks mannosidase I and produces Man7–9(GlcNAc)2 structures that lack glucose. All three of these inhibitors thus produce high mannose structures. Sw blocks mannosidase II and inhibits formation of N-linked complex oligosaccharides, so hybrid structures are produced (Man4GlcNAc2, Man5GlcNAc2, hybrid chains). Western analysis indicates that the early glycosylation blocks by CST or DNM treatment increase the size of gPr80env and reduce processing into SU. SU produced in the presence of DMM, DNM and Sw is decreased in size (Fig. 3B). gPr80Envs produced in the presence of the various inhibitors are all identified by MAb538 by Western immunoblotting, except for the CST-modified glycoprotein (Fig. 3B).
Figure 3.
Western immunoblot analysis of lysates from MoMLV-infected cells. A) NIH 3T3 (N) or hamster Lec8 (L) cells infected with MoMLV or FrMLV and probed with goat anti-gp70. B) Cell lysates from NIH 3T3 cells treated with the indicated inhibitors. After the reaction with MAb538 (on the bottom), the filter was stripped and probed with anti-gp70. Lanes separated by a space were from the same exposure of a single filter.
FrMLV and MoMLV stocks produced in Lec8 cells or in the presence of DMM, DNM and Sw were tested for relative infectivity on M. dunni and NIH 3T3 cells. All stocks tested contained infectious virus. All of the FrMLVs showed comparable infectivity on the two cells by the XC overlay test (data not shown). All of the MoMLVs showed greater infectivity on NIH 3T3 cells compared to M. dunni, within the range typically observed for MoMLV (data not shown). This suggests that Env glycan type or size does not influence receptor usage of E-MLVs.
Infectivity on stable transfectants expressing different Mus CAT-1 receptor genes
To determine if the observed differences in infectivity with the deglycosylated viruses could be attributed to the CAT-1 receptor variants in these cells, the viruses were used to infect stable transfectants expressing dCAT-1 or mCAT-1 (Fig. 4). M. dunni cells expressing mCAT-1 in addition to endogenous dCAT-1 showed increased infectibility with MoMLV but infectibility of these cells with FrMLV was not altered, as previously shown (Yan et al., 2008). These cells also showed increased infectibility with Mo-gs1, Mo-gs2, and Fr-gs1. Thus, the expression of mCAT-1 in M. dunni cells resulted in virus-specific increased susceptibility to Fr-gs1 and to the 3 MoMLV variants.
Figure 4.
Virus titers of ecotropic MLVs on mouse cells and on mouse and ferret transfectants expressing specific CAT-1 receptors. Virus titers were measured as the number of XC PFU in 0.2 ml using the XC overlay test. Results are from a representative experiment. SD of the data are also shown. Arrow: no XC plaques detected in 5 trials. X, not done.
Expression of mCAT-1 or dCAT-1 in MA139 ferret cells renders them susceptible to E-MLV infection (Yan et al., 2008) (Fig. 4). As previously noted, ferret cells expressing mCAT-1 (FerrM) and dCAT-1 (FerrD) are equally infectible by MoMLV indicating that the sequence difference between mCAT-1 and dCAT-1 does not account for the MoMLV susceptibility difference between NIH 3T3 and M. dunni cells (Yan et al., 2008). However, while FerrM was equivalently susceptible to MoMLV, Mo-gs1 and Mo-gs2, FerrD was nearly 1000-fold less susceptible to Mo-gs1 and Mo-gs2. We also infected a ferret transfectant expressing dCAT-1 from which the two N-glycan sites had been deleted by mutagenesis (Fig. 2A) (Yan et al., 2008). The cells expressing deglycosylated dCAT-1-g showed the same pattern of virus susceptibility as FerrD (data not shown).
These same ferret transfectants were infected with FrMLV and Fr-gs1. Fr-gs1, like Mo-gs1, showed reduced infectivity in the transfectants expressing dCAT-1 (Fig. 4) and dCAT-1-g (data not shown) compared to the mCAT-1 transfectant. These results indicate that while the CAT-1 sequence difference does not account for the resistance of M. dunni to MoMLV, this sequence difference is an important factor that influences susceptibility to the deglycosylated viruses. Removal of gs1 from FrMLV and either RBD glycan from MoMLV compromises the ability of these MLVs to interact with the dCAT-1 receptor, but not with the mCAT-1 receptor whether the receptor is expressed in mouse cells, or cells of heterologous species.
Effect of glycosylation inhibitors on virus infectivity
The reduction in susceptibility to deglycosylated MoMLV and FrMLV in dCAT-1 versus mCAT-1 expressing ferret cells is not as dramatic as the susceptibility differences between M. dunni and NIH 3T3. This suggests that other factors may contribute to the resistance of M. dunni cells. Because glycosylation contributes to MoMLV resistance in dunni cells (Eiden et al., 1994; Yan et al., 2008), we tested the effects of inhibitors of glycosylation on infectivity of deglycosylated viruses in M. dunni cells (Table 1). All inhibitors tested increased susceptibility to MoMLV as well as to Mo-gs1, Mo-gs2, and Fr-gs1, but not to FrMLV57. Thus, glycosylation is a contributing factor to restriction of Fr-gs1 and all of the MoMLV variants in M. dunni cells.
Table 1.
Effect of various glycosylation inhibitors on the ability of MoMLV and FrMLV wild type virus and glycosylation mutants to replicate in M. dunni cells.
| Log10 Virus Titer with Glycosylation Inhibitorsa |
||||||
|---|---|---|---|---|---|---|
| Virus | - | Tu | 2DG | DMM | CST | Sw |
| MoMLV | 2.4 | 5.0 | 4.8 | 4.4 | 4.1 | 4.8 |
| Mo-gs1 | --- | 1.0 | 2.1 | 0.6 | 0.9 | 0.9 |
| Mo-gs2 | --- | 1.3 | 2.9 | 2.1 | 1.5 | 1.5 |
| FrMLV57 | 4.6 | ND | 4.5 | 4.4 | 4.4 | 4.6 |
| Fr-gs1 | 1.6 | ND | 3.3 | 2.9 | 2.6 | 2.6 |
Virus was quantitated using the XC overlay assay and numbers represent XC PFU in 0.2 ml. Glycosylation inhibitors were added the day before virus infection. Each virus-inhibitor combination was tested three times; the numbers represent a representative experiment. ---, no positive cells in cultures infected with 0.1 ml of undiluted virus stock. ND, not done.
Pseudotype infections
To determine if removal of envelope glycans affects later stages in the viral life cycle, we infected cells with pseudotypes carrying the LacZ reporter gene. Pseudotypes were generated for MoMLV, FrMLV57, and their respective gs1 mutants.
As observed previously, the MoMLV pseudotype is restricted in M. dunni cells although these cells are susceptible to infection with the FrMLV57 pseudotype (Table 2) (Yan et al., 2008). M. dunni cells showed no detectable infection with pseudotypes of either Mo-gs1 or Fr-gs1, consistent with the XC overlay test results (Fig. 2B).
Table 2.
Titers of ecotropic LacZ virus pseudotypes in mouse cells and MA139 cells transfected with mCAT-1 (FerrM) or dCAT-1 (FerrD)
| Log10 Titer of LacZ Pseudotypesa |
||||
|---|---|---|---|---|
| Cellsb | MoMLV | Mo-gs1 | FrMLV57 | Fr-gs1 |
| NIH 3T3 | 4.3 | 4.4 | 4.1 | 4.7 |
| M. dunni | --- | --- | 3.4 | --- |
| FerrM (mCAT-1) | 3.5 | 3.6 | 3.5 | 4.0 |
| FerrD (dCAT-1) | 2.9 | 1.5 | 3.5 | 2.3 |
Measured as the number of cells positive for β-galactosidase activity in 100 ul of virus. ---, no positive cells in cultures infected with 0.1 ml of undiluted pseudotype stock. Experiment was done twice; numbers represent results from one experiment.
CAT-1 gene expressed in transfected cells is given in ().
The transfected MA139 line carrying mCAT-1, FerrM, was efficiently infected by all 4 pseudotypes as also shown by the XC test (Fig. 4). In contrast, FerrD cells expressing dCAT-1 resemble FerrM in their susceptibility to FrMLV and MoMLV pseudotypes, but show a 60–100-fold reduction in sensitivity to pseudotypes of both gs1 mutants (Table 2). These results are consistent with the XC test results and indicate that the restriction of viruses lacking specific glycans is receptor mediated.
Virus binding assays
To determine if the removal of glycans affects virus binding to the CAT-1 receptor or a later stage in the entry process, we performed virus binding assays on NIH 3T3, M. dunni and MDTF cells with MoMLV, FrMLV57, and their respective gs1 glycosylation mutants. We also used the ferret transfectants FerrM and FerrD to evaluate binding of FrMLV57 and Fr-gs1.
Binding of each virus was initially assessed on NIH 3T3 cells (Fig 5A, B). Strong virus binding was observed on NIH 3T3 cells for both wild type viruses, and removal of glycosylation sites only slightly modulated the virus-binding characteristics in a virus-specific manner. Mo-gs1 had a slightly decreased binding efficacy compared to MoMLV (Fig. 5A) whereas Fr-gs1 showed slightly increased binding (Fig. 5B), reflecting the different XC plaque titers produced by these virus stocks on NIH 3T3 (Fig. 2B).
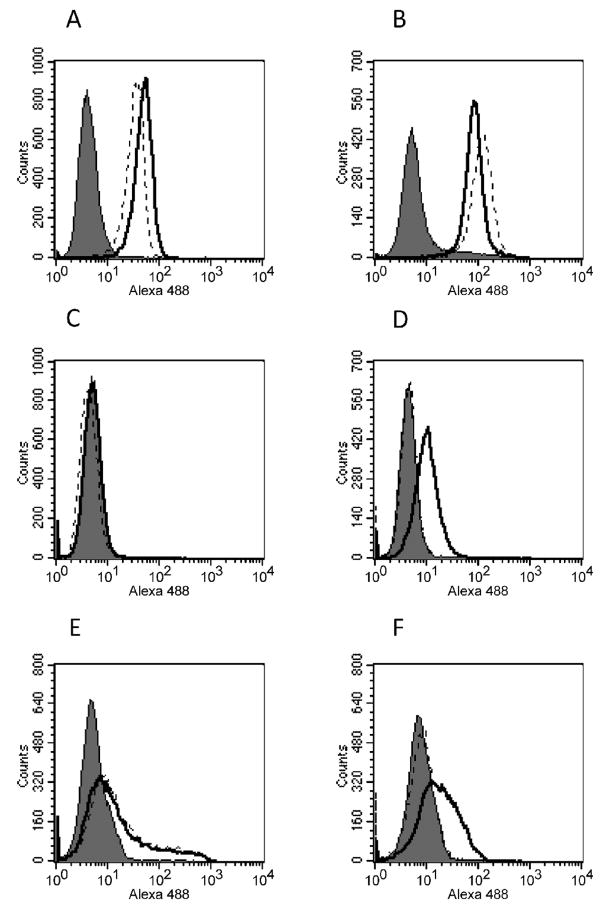
Figure 5.
Virus binding assay performed on NIH3T3 (A, B), MDTF (C, D) FerrM (E) and FerrD (F) cells. Dead cells were excluded by gating 7-AAD negative cells. Viruses used in these experiments included Mo-MuLV (black line, panel A and C) and its Mo-gs1 glycosylation mutant (broken line, panel A and C) and Friend MuLV (black line, panel B, D, E and F) and its Fr-gs1 glycosylation mutant (broken line, panel B, D, E and F). Cells treated similarly in absence of virus suspension served as negative control (grey histogram in all panels).
Virus binding properties were drastically different on M. dunni (data not shown) and MDTF cells (Fig 5C, D), which showed nearly identical properties. Binding of MoMLV and Mo-gs1 were undetectable on M. dunni and MDTF cells (Fig. 5C), consistent with the poor infectivity of both viruses on these cells. Binding of FrMLV57 was detected on both dunni cell lines but was reduced compared to NIH 3T3 cells. This difference did not correlate with the XC plaque titer of this virus which was 5–10 fold higher on M. dunni than on NIH 3T3 cells suggesting that other cell-specific factors may influence virus replication. In contrast to wild type FrMLV57, Fr-gs1 had no detectable binding ability on either M. dunni or MDTF cells (Fig 5D). These data suggest that removal of gs1 from FrMLV57, a virus capable of infecting M. dunni cells, drastically reduces its ability to recognize the dCAT-1 receptor, preventing efficient replication on these cells.
Virus binding of FrMLV57 and Fr-gs1 was also assessed on FerrM and FerrB cells. Both viruses showed reduced but equivalent binding to FerrM cells (Fig. 5E), consistent with the observation that these cells are less susceptible to MLVs than mouse cells (Fig. 4). FerrD resembled FerrM in its ability to bind FrMLV57, but showed very reduced binding with Fr-gs1 (Fig. 5F). These data suggest that receptor polymorphism is responsible for reduced infectivity of the gs1 mutants.
DISCUSSION
Among the various factors responsible for intrinsic resistance to retroviruses are several that inhibit virus entry. Such factors include ERV produced Env glycoproteins that interfere with virus-receptor binding, and mutations in receptor genes that alter their interactions with viruses (Stocking and Kozak, 2008). For the MLVs, receptor polymorphisms that recognize different Env variants have been identified for the XPR1 receptor that mediates infection of xenotropic and polytropic MLVs (XMVs and PMVs) (Marin et al., 1999; Yan et al., 2007), viruses originally described as different host range groups (Hartley et al., 1977). For the E-MLVs, there are fewer examples of receptor mediated host range variations, in part because, unlike the X/PMVs that can infect all mammals tested, E-MLV infection is limited to rodents. In addition, most known examples of E-MLV entry restrictions are due, at least in part, to glycosylation of cellular proteins. Because glycosylation of the viral Env has also been associated with altered infectivity of multiple viruses including MLVs (Battini et al., 1994) and other retroviruses (Ogert et al., 2001), we examined the need for specific N-glycans in Env regions important for entry. Here we show that deletion of specific N-glycan sites from ecotropic MLV env genes produces viruses with altered host range. This effect is receptor variant specific, virus type specific, and glycan site specific. Deletion of glycans gs1 and gs2 alters the host range of MoMLV and FrMLV but not AKV MLV, and this restriction is observed only in cells expressing two of the four naturally occurring CAT-1 variants.
Virus infectivity was shown to be affected by glycan site deletion but not by the alteration of glycan type or size. This suggests that the role of glycans in host range may be a consequence of their cotranslational placement on the Env protein, a process that may result in conformational changes in the Env glycoprotein that differentially alters recognition by the 4 receptor variants. Glycan additions generally ensure proper folding of proteins, help maintain their stability, and are needed for intracellular routing and further processing. For MLV Env, blockage of glycosylation by tunicamycin halts processing and translocation to the plasma membrane (Schultz and Oroszlan, 1975). That glycan removal affects infectivity due to altered folding rather than a specific role for glycans in entry is consistent with the crystal structure of FrMLV (Fass et al., 1997) which shows that neither gs1 nor gs2 is positioned near the residues in VRA that form the pocket for virus binding, namely residues S84, D86 and W102 (Davey et al., 1999).
Deletion of either of the 2 RBD glycan sites of MoMLV and gs1 of FrMLV produces viruses that identify a functional distinction among the 4 naturally occurring CAT-1 variants. These mutated viruses are much less able to infect dunni and hamster cells, while infectivity on rat XC cells and NIH 3T3 cells is not altered. These infectivity differences are also seen in MA139 ferret cells expressing the dCAT-1 and mCAT-1 receptor variants indicating that receptor polymorphism is the major responsible host cell factor. Virus binding assays further demonstrate there is an association between infectivity and the binding ability of Fr-gs1 compared to the wild type FrMLV in M. dunni cells as well as in the transfected ferret cells. Infectivity can be, however, affected by other, secondary cellular factors as shown by the fact that glycosylation inhibitors can reduce this entry restriction in M. dunni cells.
The three viruses used here have ecotropic host range, but show significant sequence variation in their RDBs and also differ in their patterns of glycosylation (Fig. 1A). FrMLV and MoMLV are laboratory isolates that, unlike AKV, are oncogenic in mice. Both viruses were isolated after serial passage of tumors in mice and neither of these viruses have counterparts that are found naturally in laboratory mice or wild mouse species. In contrast, AKV MLVs are naturally occurring MLVs carried as endogenous retroviruses by many inbred laboratory strains and are also found in Asian species of Mus (Kozak and O’Neill, 1987). Sequence comparisons of their Env genes indicate closer relationship between FrMLV and MoMLV (88% amino acid identity in RBD) than between either of these viruses and AKV (78% identify). These similarities are reflected in our results; removal of RBD glycan sites from FrMLV and MoMLV similarly compromises infectivity in cells expressing dCAT-1, whereas AKV mutants show no changes in infectivity. Examination of other MLV isolates indicates there is naturally occurring polymorphism at 4 glycan sites (gs3,5,7,8) among ecotropic viruses suggesting that these sites are not critical for producing infectious virus as shown here for gs8.
Mutational changes in the retroviral RBD can lead to the generation of viruses that can use alternative receptors, multiple receptors or even multiple determinants on the same receptor protein (Stocking and Kozak, 2008). The coevolution of Env and receptor has resulted in their sequence diversity and functional plasticity, and previous studies have identified sequence variations in E-MLVs responsible for variations in receptor usage (Yan et al., 2008; Masuda et al, 1996). Such variants show altered infectivity into different mouse or hamster cells, and differential ability to induce multinucleated syncytia. The present study indicates that RBD glycans also serve to control virus host range variation, a control that likely depends on protein folding rather than a direct role in receptor interaction.
METHODS
Viruses and Cells
Two ecotropic MLV isolates were obtained from J. W. Hartley (NIAID, Bethesda, MD): AKV MLV and Moloney MLV (MoMLV). We also produced stocks of Friend and MoMLV from molecular clones of FrMLV57 and MoMLV (pNCA-HA) obtained from S. Ruscetti (NCI, Frederick, MD) (Oliff et al., 1980) and J. Silver (NIAID, Bethesda, MD) (Ou et al., 2006).
Virus stocks were made by collecting culture fluids from infected or transfected cells. Infectious virus in these stocks was quantitated by the XC overlay test (Rowe et al., 1970; Yan et al., 2008) following infection of NIH 3T3 and 2 lines derived from tail cells of a single M. terricolor (M. dunni) mouse; the two cell lines are here designated M. dunni and MDTF (Lander and Chattopadhyay, 1984). The XC overlay test was also used to assess virus susceptibility on Chinese hamster E36 (Gillin et al., 1972) and Lec8 cells (ATCC CRL-1737), XC rat cells, and MA139 ferret or M. dunni cells stably transfected with 3 CAT-1 receptor variants: mCAT-1 from NIH 3T3, dCAT-1 from M. dunni, and dCAT-1-g, a variant of dCAT-1 from which both glycosylation sites were removed by mutagenesis (Yan et al., 2008). Western analysis indicates equivalent expression of mCAT-1 and dCAT-1 in the ferret transfectants (Yan et al., 2008)
Subconfluent cultures were infected with appropriate dilutions of virus stocks in the presence of polybrene (4 ug/ml; Aldrich, Milwaukee, WI). The cells were irradiated 4 days after virus infection with ultraviolet light from germicidal bulbs (30 sec at 60 ergs/mm2), and were then overlaid with XC cells. XC cells produce plaques of multicleated syncytia in response to focal areas of virus-infected cells. Cultures were fixed and stained 3 days later and examined for plaques of syncytia. Virus spread was poor in infected XC and Lec8 cells making XC plaques difficult to read, so infected cells were first irradiated and overlaid with NIH 3T3 or H3T3-1, a line derived in this laboratory from tail cells of M. spicilegus; these cells do not restrict any ecotropic MLVs. After 3 days, cells were again irradiated and overlaid with XC cells.
Inhibitors of N-linked glycosylation
Cells were treated by various inhibitors of N-linked glycosylation as follows: deoxymannojirimycin (DMM, 100 ug/ml); castanospermine (CST, 100 ug/ml), tunicamycin (Tu, 0.2 ug/ml); 2-deoxy-D-glucose (2DG, 25 mM) deoxynojirimycin (DNM, 200 ug/ml); swainsonine (Sw, 200 ug/ml). All inhibitors were obtained from SIGMA (La Jolla, CA). Inhibitors were added to cultures that had been seeded the previous day and were not removed when virus and polybrene were added 18–24 hours later. Cells were lysed for Western immunoblotting 3–4 days later or were irradiated for the XC overlay test. To generate viruses with altered N-glycans, inhibitors were added to virus producing cells. Medium was replaced the next day and fresh inhibitor was added. Virus was collected 3–4 days later.
Pseudotype assay
MoMLV, FrMLV57 and their respective gs1 mutants were used to infect an NIH 3T3 derived cell line stably transfected with pCLMFG-LacZ (Imgenex Co., San Diego, Calif.) and kindly provided by Dr. J. Silver (NIAID, Bethesda, MD). Filtered media from the virus infected cultures contained a mixture of infectious virus and LacZ pseudotypes. Cells were infected with appropriate dilutions of these pseudotype virus stocks in the presence of 4–8 μg/ml polybrene. One day after infection, cells were fixed with 0.4% glutaraldehyde and assayed for β-galactosidase activity using as substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal, 2 mg/ml; ICN Biomedicals, Aurora, Ohio). Infectious titers were expressed as the number of blue cells per 100 microliters of virus supernatant.
Mutagenesis of MoMLV, AKV and FrMLV
Six mutant viruses lacking glycosylation sites gs1, gs2, or gs8 (Fig. 1A) were produced by overlapping PCR using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). MoMLV mutants Mo-gs1, Mo-gs2 and Mo-gs8 were generated using clone pNCA-HA (obtained from J. Silver, NIAID, Bethesda, MD) (Ou et al., 2006). AKV MLV mutants AK-gs1 and AK-gs2 were made using clone pGEM-AKV, containing the PstI fragment of AKV MLV from pAKV23 (obtained from J. Lenz, Albert Einstein College of Medicine, Bronx, NY). FrMLV mutant Fr-gs1 was produced using the FrMLV57 clone (obtained from S. Ruscetti, NCI, Frederick, MD) (Oliff et al., 1980).
For each mutagenesis, sense and antisense primers containing the various mutations were used in PCR reactions with flanking 5′ and 3′ primers as indicated in Table 3. The two products of these reactions were combined and used as template for a second PCR using the flanking primers. MoMLV PCR products lacking gs1 or gs2 were digested with PmlI and NsiI and ligated into pNCA-HA; the MoMLV fragment lacking gs8 was introduced as an HpaI fragment. The Fr-gs1 mutant was introduced into FrMLV57 as a SacII insertion. The AK-gs1 and AK-gs2 mutants were generated as follows: a 2.7 kb BsmI fragment was recovered from pGEM-AKV using forward primer: 5′-GCAGGAGTGAGAGTACGAGGACAT and reverse primer 5′-TGGACCAGGCGATTGAGAA and cloned it into the vector pCR2.1-TOPO. Overlapping PCR was performed using indicated primers and the BsgI/AvrII fragments of the PCR product were cloned into the AKV subclone. Two 2.7 kb BsmI fragmentsfrom these subclones containing N43Q and N199Q were ligated into pGEM-AKV. All mutations were confirmed by sequencing; all clones of AK-gs1 and AK-gs2 contained the additional mutation Q341H. Clones were transfected into NIH 3T3 cells and culture medium containing virus was collected.
Table 3.
Primers used to generate MLVs lacking specific glycosylation sites
| Mutant | Mutagenic Primer* | Flanking Primer* |
|---|---|---|
| Mo-gs1 (N45Q) | 5′-CAGTCCTCATCAAGTCTATcAgATCACCTGGGAGGTAACC (5890–5929) | 5′-ATCGCAGCTTGGATACACGC (5706–5725) |
| 5′-GGTTACCTCCCAGGTGATcTgATAGACTTGATGAGGACTG (5890–5929) | 5′-AGTGGTAAGGTTCAGTATGG (7068–7087) | |
|
| ||
| Mo-gs2 (N199D) | 5′-ATTTCATCACAGTAAACAACgATCTCACCTCTGACCAGGC (6351–6390) | 5′-ATCGCAGCTTGGATACACGC (5706–5725) |
| 5′-GCCTGGTCAGAGGTGAGATcGTTGTTTACTGTGATGAAAT (6351–6390) | 5′-AGTGGTAAGGTTCAGTATGG (7068–7087) | |
|
| ||
| Mo-gs8 (N434Q) | 5′-CATCTCCACCACCATACTGcAgCTTACCACTGATTATTGTG (7057–7097) | 5′-ATCGCAGCTTGGATACACGC (5706–5725) |
| 5′-CACAATAATCAGTGGTAAGcTgCAGTATGGTGGTGGAGATG (7057–7097) | 5′-TGAGTGGCCATTAGAGCAGTAG (7263–7284) | |
|
| ||
| AK-gs1 (N43Q) | 5′-GCCCCCACCAGGTTTTTcAgCTCACCTGGGAAGTGAC (5892–5928) | 5′-GACTGACTCCGTATGAAATC (5407–5426) |
| 5′-GTCACTTCCCAGGTGAGcTgAAAAACCTGGTGGGGGC (5892–5928) | 5′-TGGGGCAGAAGTATGGTT (6857–6874) | |
|
| ||
| AK-gs2 (N199Q) | 5′-CATCACAGTAAGCAACcAgCTAACCTCAGACCAGGCAACC (6361–6400) | 5′-GACTGACTCCGTATGAAATC (5407–5426) |
| 5′-GGTTGCCTGGTCTGAGGTTAGcTgGTTGCTTACTGTGATG (6361–6400) | 5′-TGGGGCAGAAGTATGGTT (6857–6874) | |
|
| ||
| Fr-gs1 (N46Q) | 5′-GCCCTCACCAGGTCTACcAgATTACCTGGGAAGTGA (5896–5931) | 5′-GCACCCCCGCCCCTTGTAA (5422–5440) |
| 5′-TCACTTCCCAGGTAATcTgGTAGACCTGGTGAGGGC (5896–5931) | 5′-CATGGAGCTGCTGGAACTGC (7316–7335) | |
Western immunoblotting
Lysates from virus infected cells were tested for viral Env by western immunoblot analysis. Cell lysates were subjected to electrophoresis on NuPAGE 4–12% Bis-Tris gels (Invitrogen, Carlsbad, CA). Western blots were probed with two antibodies against MLV Env: a goat polyclonal anti-Rauscher MLV gp70 antiserum (ViroMed Biosafety Laboratories, Camden, NJ) with peroxidase conjugated rabbit anti-goat IgG as the secondary antibody (Invitrogen, Eugene, OR), and MAb538, a monoclonal anti-MoMLV antibody (Lander et al., 1984) provided by J. Portis (NIAID, Hamilton, MT) with peroxidase-conjugated goat anti-mouse IgG (Invitrogen).
Virus binding Assay
The virus binding assay was performed essentially as described previously (Kadan et al., 1992)]. NIH 3T3, M. dunni, MDTF and tranfected ferret cells were detached from culture flasks by a short treatment at room temperature with trypsin-EDTA (Gibco, Grand Island, NY). They were washed once and resuspended at 2 million cells per ml. One million cells were incubated in presence of 100 μl of virus suspension and 8 μg/ml polybrene for 40 minutes at 37°C in a final volume of 1 ml. Cells treated similarly in the absence of virus stock served as negative control. Samples were mixed every 10 minutes. At the end of the incubation, cells were washed twice with wash buffer (PBS supplemented with 1% bovine serum albumin). Virions bound to the cells were detected by staining the cells first with the goat-anti gp70 antiserum (1:200) in 100μl for 30 minutes at 4°C. After two washes with wash buffer, cells were incubated with Alexa Fluor 488 labeled donkey anti-goat IgG (Invitrogen, Carlsbad, CA)(1:500) in 100μl for 30 minutes at 4°C. After two additional washes, cells were resuspended in 500μl PBS. To exclude dead cells from the analysis, the cell suspension was incubated at room temperature for 10 minutes with 20 μl 7-AAD solution (BD Via Probe, BD Biosciences, San Jose CA) before being analyzed by flow cytometry (FACScalibur, BD Biosciences Immunocytometry Systems). Fluorescence data of a minimum of 50,000 cells were recorded and analysis was performed on live cells (7-AAD negative cells, >90% of the population) using Cell Quest Pro (BD Biosciences).
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIAID.
We thank Qingping Liu for expert technical assistance and Alicia Buckler-White for sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albritton LM, Kim JW, Tseng L, Cunningham JM. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battini JL, Kayman SC, Pinter A, Heard JM, Danos O. Role of N-linked glycosylation in the activity of the Friend murine leukemia virus SU protein receptor-binding domain. Virology. 1994;202:496–499. doi: 10.1006/viro.1994.1369. [DOI] [PubMed] [Google Scholar]
- Burkhart MD, D’Agostino P, Kayman SC, Pinter A. Involvement of the C-terminal disulfide-bonded loop of murine leukemia virus SU protein in a postbinding step critical for viral entry. J Virol. 2005;79:7808–7876. doi: 10.1128/JVI.79.12.7868-7876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey RA, Zuo Y, Cunningham JM. Identification of a receptor-binding pocket on the envelope protein of Friend murine leukemia virus. J Virol. 1999;73:3758–3763. doi: 10.1128/jvi.73.5.3758-3763.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiden MV, Farrell K, Warsowe J, Mahan LC, Wilson CA. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J Virol. 1993;67:4056–4061. doi: 10.1128/jvi.67.7.4056-4061.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiden MV, Farrell K, Wilson CA. Glycosylation-dependent inactivation of the ecotropic murine leukemia virus receptor. J Virol. 1994;68:626–631. doi: 10.1128/jvi.68.2.626-631.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- Felkner RH, Roth MJ. Mutational analysis of the N-linked glycosylation sites of the SU envelope protein of Moloney murine leukemia virus. J Virol. 1992;66:4258–4264. doi: 10.1128/jvi.66.7.4258-4264.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillin FD, Roufa DJ, Beaudet AL, Caskey CT. 8-Azaguanine resistance in mammalian cells I. Hypoxanthine-guanine phosphoribosyltransferase. Genetics. 1972;72:239–252. doi: 10.1093/genetics/72.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb C, Baenziger J, Kornfeld S. Deficient uridine diphosphate-N-acetylglucosamine:glycoprotein N-acetylglucosaminyltransferase activity in a clone of Chinese hamster ovary cells with altered surface glycoproteins. J Biol Chem. 1975;250:3303–3309. [PubMed] [Google Scholar]
- Hartley JW, Wolford NK, Old LJ, Rowe WP. A new class of murine leukemia virus associated with development of spontaneous lymphomas. Proc Natl Acad Sci USA. 1977;74:789–792. doi: 10.1073/pnas.74.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadan MJ, Sturm S, Anderson WF, Eglitis MA. Detection of receptor-specific murine leukemia virus binding to cells by immunofluorescence analysis. J Virol. 1992;66:2281–2287. doi: 10.1128/jvi.66.4.2281-2287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayman SC, Kopelman R, Projan S, Kinney DM, Pinter A. Mutational analysis of N-linked glycosylation sites of Friend murine leukemia virus envelope protein. J Virol. 1991;65:5323–5332. doi: 10.1128/jvi.65.10.5323-5332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Cunningham JM. N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. J Biol Chem. 1993;268:16316–16320. [PubMed] [Google Scholar]
- Kozak CA, O’Neill RR. Diverse wild mouse origins of xenotropic, mink cell forus-forming, and two types of ecotropic proviral genes. J Virol. 1987;61:3082–3088. doi: 10.1128/jvi.61.10.3082-3088.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y, Ono T, Ogura M, Ishimoto A, Amanuma H. A glycosylation-defective variant of the ecotropic murine retrovirus receptor is expressed in rat XC cells. Virology. 2002;303:338–344. doi: 10.1006/viro.2002.1641. [DOI] [PubMed] [Google Scholar]
- Lander JK, Chesebro B, Fan H. Appearance of mink cell focus-inducing recombinants during in vivo infection by Moloney murine leukemia virus (M-MuLV) or the Mo+PyF101 M-MuLV enhancer variant: Implications for sites of generation and roles in leukemogenesis. J Virol. 1999;73:5671–5680. doi: 10.1128/jvi.73.7.5671-5680.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander MR, Chattopadhyay SK. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Pinter A, Kayman SC. The critical N-linked glycan of murine leukemia virus envelope protein promotes both folding of the C-terminal domains of the precursor polyprotein and stability of the postcleavage envelope complex. J Virol. 1997;71:7012–7019. doi: 10.1128/jvi.71.9.7012-7019.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin M, Tailor CS, Nouri A, Kozak SL, Kabat D. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J Virol. 1999;53:100–106. doi: 10.1128/jvi.73.11.9362-9368.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Masuda M, Hanson CA, Hoffman PM, Ruscetti SK. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J Virol. 1996;70:8534–8539. doi: 10.1128/jvi.70.12.8534-8539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Miller AD. Tunicamycin treatment of CHO cells abrogates multiple blocks to retrovirus infection, one of which is due to a secreted inhibitor. J Virol. 1992;66:78–84. doi: 10.1128/jvi.66.1.78-84.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DG, Miller AD. Inhibitors of retrovirus infection are secreted by several hamster cell lines and are also present in hamster sera. J Virol. 1993;67:5346–5352. doi: 10.1128/jvi.67.9.5346-5352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogert RA, Lee MK, Ross W, Buckler-White A, Martin MA, Chom MW. N-linked glycosylation sites adjacent to and within the V1/V2 and the V3 loops of dualtropic human immunodeficiency virus type 1 isolat DH12 gp120 affect coreceptor usage and cellular tropism. J Virol. 2001;75:5998–6006. doi: 10.1128/JVI.75.13.5998-6006.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliff AI, Hager GL, Chang EH, Scolnick EM, Chan HW, Lowy DR. Transfection of molecularly cloned Friend murine leukemia virus DNA yields a high leukemogenic helper-independent type C virus. J Virol. 1980;33:475–486. doi: 10.1128/jvi.33.1.475-486.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou W, Lu N, Yu SS, Silver J. Effect of epitope position on neutralization by anti-human immunodeficiency virus monoclonal antibody 2F5. J Virol. 2006;80:2539–2547. doi: 10.1128/JVI.80.5.2539-2547.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter A, Honnen WJ. O-linked glycosylation of retroviral envelope gene products. J Virol. 1988;62:1016–1021. doi: 10.1128/jvi.62.3.1016-1021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner MR, Tung JS, Hopkins N, Robbins PW. Relationship of GIX antigen expression to the glycosylation of murine leukemia virus glycoprotein. Proc Natl Acad Sci USA. 1980;77:6420–6424. doi: 10.1073/pnas.77.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe WP, Pugh WE, Hartley JW. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Schultz AM, Oroszlan S. Tunicamycin inhibits glycosylation of precursor polyprotein encoded by env gene of Rauscher murine leukemia virus. Biochem Biophys Res Comm. 1979;86:1206–1213. doi: 10.1016/0006-291x(79)90245-6. [DOI] [PubMed] [Google Scholar]
- Stocking C, Kozak CA. Murine endogenous retroviruses. Cell Mol Life Sci. 2008;65:3383–3398. doi: 10.1007/s00018-008-8497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor CS, Lavillette D, Marin M, Kabat D. Cell surface receptors for gammaretroviruses. Curr Top Microbiol Immunol. 2003;281:29–106. doi: 10.1007/978-3-642-19012-4_2. [DOI] [PubMed] [Google Scholar]
- Tavoloni N, Rudenholz A. Variable efficiency of murine leukemia retroviral vector on mammalian cells: role of cellular glycosylation. Virology. 1997;229:49–56. doi: 10.1006/viro.1996.8412. [DOI] [PubMed] [Google Scholar]
- Yan Y, Knoper RC, Kozak CA. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruese and their XPR1 receptors elucidate receptor determinants of virus entry. J Virol. 2007;81:10550–10557. doi: 10.1128/JVI.00933-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Jung YT, Wu T, Kozak CA. Role of receptor polymorphism and glycosylation in syncytium induction and host range variation of ecotropic mouse gammaretroviruses. Retrovirology. 2008;5:2. doi: 10.1186/1742-4690-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto T, Yoshimoto E, Meruelo D. Identification of amino acid residues critical for infection with ecotropic murine leukemia retrovirus. J Virol. 1993;67:1310–1314. doi: 10.1128/jvi.67.3.1310-1314.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Klamo E, Kuhmann SE, Kozak SL, Kavanaugh MP, Kabat D. Modulation of ecotropic murine retroviruses by N-linked glycosylation of the cell surface receptor/amino acid transporter. J Virol. 1996;70:6884–6891. doi: 10.1128/jvi.70.10.6884-6891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]