Abstract
WD40 repeat proteins have been shown to bind the histone H3 tail at the center of their β-propeller structure. In contrast, in this issue of Genes & Development, Song and colleagues (pp. 1313–1318) demonstrate that the WD40 repeat protein p55 binds a structured region of H4 through a novel binding pocket on the side of β-propeller, illustrating a diversity of histone recognition by WD40 repeat proteins.
Keywords: p55, Nurf55, RbAp48, histone, chromatin, WD40
The association of WD40 repeat proteins with histones and nucleosomes is of increasing interest because of their functions in a variety of histone/chromatin-modifying complexes. In this issue of Genes & Development, Song et al. (2008) solve the crystal structure of p55, a Drosophila seven-WD40-repeat protein. Binding of p55 is shown to be incompatible with the histone fold of H4 and results in a change in H4 structure. The interaction of p55 with H4 is distinct from that of previously characterized WD40 repeat proteins and utilizes a novel binding pocket on the side of the β-propeller instead of the center of the propeller. This novel p55–H4 interaction extends the known contacts by which WD40 repeat proteins recognize histones and reveals diversity in the manner in which this important recognition domain functions.
WD40 repeat proteins are important for a variety of cellular functions
WD40 repeats were first noted in the β-subunit of the heterotrimeric GTP-binding protein (G protein) (Fong et al. 1986). The G protein structure contained seven WD40 repeats. Each repeating unit formed one of seven β-propeller blades with four small anti-parallel β-sheets built into a toroidal structure with a tapered end and central canal (Wall et al. 1995; Neer and Smith 2000). The conserved core of repeating units contained 44–60 residues that ended with tryptophan (W) and aspartate (D). Since the initial identification of the WD40 repeats, >160 WD40 repeat proteins have been identified, with the majority found in higher eukaryotes (Smith et al. 1999). Each family member has between four and 10 copies of the WD40 repeats. In general, WD40 repeat proteins have been associated with a diverse range of cellular processes, including RNA processing, transcriptional regulation (Williams et al. 1991; Hoey et al. 1993), mitotic spindle formation (de Hostos et al. 1991; Vaisman et al. 1995), regulation of vesicle formation and vesicular trafficking (Pryer et al. 1993), and control of cell division (Feldman et al. 1997).
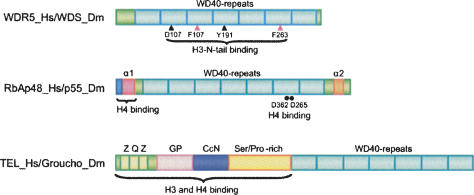
Four WD40 repeat proteins—WDR5, RbBP5, RbAp48/46 (each with seven repeats)—and the Drosophila homolog of RbAp48/46, p55, are components of histone-modifying complexes (Fig. 1). WDR5 is associated with SET domain-containing methyltransferases, such as Set1, MLL, MLL2, and MLL3/4 (Wysocka et al. 2005; Dou et al. 2006; Mendjan et al. 2006; Cho et al. 2007). In addition to its role in histone methylation, WDR5 and WDS, the Drosophila homolog of WDR5, are associated with several histone acetyltransferase (HAT) complexes, including the MSL (male-specific lethal) complex (Mendjan et al. 2006); the ATAC (Ada Two A-containing) complex, which functions as both an H3 and H4 HAT (Suganuma et al. 2008); and MOF, a H4K16 acetyltransferase complex (Dou et al. 2005). Thus, WDR5 plays important roles in multiple complexes that display distinct catalytic activities on histone substrates.
Figure 1.
Schematic domain structure of seven-WD40-repeat-containing proteins. (Black triangles) Generally accepted histoneH3 recognition sites; (red triangles) structurally rearranged phenylalanine by binding histone H3; (α1 and α2) α-helix1 and α-helix2; (black filled circles) backbone of the binding loop; (Z) putative leucine zipper domain; (Q) glutamine-rich domain; (GP) glycine/proline-rich domain; (CcN) containing nuclear localization and phosphorylation sites; (Ser/Pro-rich) serine/threonine/proline-rich domain.
RbAp48 is another seven-WD40-repeat protein that was initially identified as a retinoblastoma protein (Rb)-binding protein (Qian et al. 1993). RbAp48/46 and its Drosophila homolog p55 are associated with a variety of chromatin-regulating complexes that include chromatin assembly factor CAF-1, (Tyler et al. 1996; Verreault et al. 1996), Hat1 (Parthun et al. 1996; Verreault et al. 1998), histone deacetylase HDAC1, the NuRD ATP-dependent chromatin remodeling complex (Hassig et al. 1997; Xue et al. 1998; Zhang et al. 1998, 1999; Kuzmichev et al. 2002), and the Extra Sex Combs (ESC) and Enhancer of Zeste [E(Z)], ESC–E(Z) complex, which is a type of Polycomb group (PcG) histone methyltransferase (HMT) complex (Czermin et al. 2002; Muller et al. 2002). The p55 protein has been shown to associate with newly synthesized H4 (Verreault et al. 1996), and the RbAp48/46 family to which it belongs is thought to play an important role in histone metabolism.
WD40 repeat protein structure
WD40 repeat proteins play a vital role in a variety of chromatin-modifying complexes. The structure of WDR5, a seven-WD40-repeat protein that is a component of the SET1 HMT complex family, has been well characterized. Initial peptide-binding studies with WDR5 showed that it bound preferentially to dimethylated Lys 4 on histone H3 (H3K4me2) but was also required for K4 trimethylation (Fig. 1; Wysocka et al. 2005). Several crystal structures of WDR5 associated with peptides representing various states of H3K4 methylation have clarified this issue (Couture et al. 2006; Han et al. 2006; Ruthenburg et al. 2006; Schuetz et al. 2006). While the exact type of bonding between H3 and WDR5 remains controversial, it is generally accepted that H3R2, located on the exposed tail of histone H3, contacts the center of the WDR5 β-propeller structure (Fig. 1; Couture et al. 2006; Han et al. 2006; Ruthenburg et al. 2006; Schuetz et al. 2006). Indeed, recent studies revealed that methylation of H3R2 by the PRMT6 methyltransferase impedes both H3K4 trimethylation and recruitment of WDR5 (Guccione et al. 2007; Hyllus et al. 2007). R2 is also important for the recognition of methylated K4 by CHD1 (Flanagan et al. 2005). Analysis of WDR5 crystals have shown that while this protein can interact with all forms of methylated K4, it has additional contacts with H3K4me2 via a pair of conventional and unconventional hydrogen bonds. Thus, it would appear that WDR5 binds to H3K4me2 and presents it to a SET1 family methyltransferase for subsequent trimethylation (Ruthenburg et al. 2006; Schuetz et al. 2006). Indeed, one biochemical study demonstrated that WDR5 mediated an interaction between the catalytic subunit of the MLL HMT complex and the histone substrate (Dou et al. 2006).
In contrast to the H3 tail binding the center of the WD40 repeat β-propeller in WDR5, Song et al. (2008) have discovered a very different interaction of a WD40 repeat protein with histone H4 that includes contacts with normally folded portions of the histone. The p55 protein was initially identified as a subunit of the Drosophila CAF-1 (dCAF1). Its human homolog, RbAp48/46, has been shown to bind H4 in vitro (Verreault et al. 1998). The current structural study shows p55 binding to the first helix (helix1-H4, 31–41 amino acids) of an H4 peptide (15–41 amino acids) via a binding pocket formed by its N-terminal α-helix (α1) and a binding loop protruding from the seventh blade of the β-propeller structure (Fig. 1). Thus, unlike WDR5, which binds the central canal of the WD40 repeat β-propeller, p55 interacts with H4 via a novel pocket on the side of the propeller of the structure. The p55/RbAp48 α1 helix contains a unique sequence that is not present in either WDR5 or its Drosophila homolog, WDS, which contributes to this binding site (Fig. 1). This binding pocket is not altered by interaction with H4, but the histone fold domain of H4 must be distorted in order to bind p55. Thus, the interaction of p55 with a structured domain of H4 is quite distinct from the interaction of WDR5 with the protruding H3 tail.
While the structure presented demonstrates the mode of p55 binding to histone H4, p55 also appears to bind to histone H3. Surface plasmon resonance (SPR) experiments demonstrate that an H4R39A mutation disrupts p55 binding to histone H4 alone (Song et al. 2008). However, in the context of an H32–H4R39A2 mutant histone tetramer, p55 can still bind. In addition, the p55 D362A and D365A mutants, which were mutated in the H4-binding loop of p55, were still able to bind H3, although to a lesser degree than the wild-type protein. Song et al. (2008) suggest that p55 binds different surfaces on H3 and H4 and that the interaction with H4 influences binding of p55 to H3. Through these interactions, p55 may serve to present the H3 tail for modification by various p55-containing complexes.
Interaction of the p55-binding pocket with H4 appears to be context-dependent. Within the Hat1 HAT complex, Song et al. (2008) showed that the interaction of p55 with H4-α1 is important for H4 acetylation. By contrast, mutations in the H4-binding pocket disrupted the ability of p55 to assemble into the PRC2 (Polycomb Repressor Complex 2) H3K27 methyltransferase complex. These data suggest that in the context of the PRC2 complex the p55-binding pocket may interact with other PRC2 subunits and not with H4. Thus, the p55 WD40 repeat domain appears to be multifunctional.
The idea that WD40 repeat proteins can serve as adaptors or presentation molecules (Ruthenburg et al. 2007) is an area of interest. Remarkably, the human ISWI-containing ATP-dependent chromatin remodeling complex NURF (nucleosome remodeling factor) contains both WDR5 and RbAp48 (the human p55 homolog). This complex also contains BPTF (bromodomain and PHD finger transcription factor), which has a PHD finger domain that was shown to mediate a direct preferential association with the H3K4me3 histone tail (Wysocka et al. 2006). siRNA knockdown of WDR5 leads to reduction of H3K4me3 and a partial disruption of the association of BPTF with chromatin (Wysocka et al. 2006). Thus, factors other that WDR5 alone may contribute to the association of NURF with chromatin. The possibility that one of these factors is RbAp48/p55 would be interesting to address.
WD40 repeat protein interactions with histones are not limited to the WD40 repeat domains; other, less conserved domains can play an important role. In some cases, it is not the WD40 repeat domain that is involved in histone recognition. The Drosophila WD40 repeat protein Groucho and its mammalian homolog TLE1 (Transducin-like Enhancer of split 1) are transcriptional corepressors involved in various signaling pathways. These proteins interact with H3 and H4 through an N-terminal conserved glutamine-rich (Q) domain and a glycine/proline-rich (GP) domain (Fig. 1). The seven-repeat WD40 domain mediates interactions with DNA-binding transcription factors such as Engrailed and Dorsal (Pickles et al. 2002). Pickles et al. (2002) compared the phenotypes resulting from missense mutation of three different positions of the WD40 repeat domain within the Caenorhabditis elegans Groucho/TLE homolog UNC-37 (Fig. 1). A mutation that perturbed the WD40 β-propeller structure disrupted UNC-37 binding to the UNC4 homeodomain transcription factor (Pickles et al. 2002). UNC-37 contains additional GP and Q domains that are required for interaction with H3 and HDACs (Chen et al. 1999). Thus, the binding of transcription factors by the WD40 repeat domain might facilitate or target HDAC activity. An analogous function of WD40 repeats is found in the yeast Tup1 protein. Tup1 is a global transcriptional repressor with a structure similar to Groucho/TLE (Sprague et al. 2000). Tup1 directly interacts with the N termini of H3 and H4 through its repression domain, which is on the N-terminal side of the WD40 repeats (Edmondson et al. 1996). In Tup1, however, the WD40 repeats are required for promoter targeting through association with DNA-binding proteins such as α2 (Fig. 1; Edmondson et al. 1996).
Conclusion
Whether it is WDR5 presentation of H3K4me for subsequent methylation, p55 modification of H4 structure, or interaction with transcription factors by Groucho/TLE1/Tup1, WD40 repeat proteins play important roles in chromatin function and gene regulation. A priori, one might have expected proteins with related domains to follow some paradigm for chromatin interactions. WD40 repeat proteins, however, are revealing a great deal of functional diversity in how they establish their interactions with histones and chromatin.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1676208.
References
- Chen G., Fernandez J., Mische S., Courey A.J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophiladevelopment. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y.W., Hong T., Hong S., Guo H., Yu H., Kim D., Guszczynski T., Dressler G.R., Copeland T.D., Kalkum M., et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.F., Collazo E., Trievel R.C. Molecular recognition of histone H3 by the WD40 protein WDR5. Nat. Struct. Mol. Biol. 2006;13:698–703. doi: 10.1038/nsmb1116. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi R., McCabe D., Seitz V., Imhof A., Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- de Hostos E.L., Bradtke B., Lottspeich F., Guggenheim R., Gerisch G. Coronin, an actin binding protein of Dictyostelium discoideumlocalized to cell surface projections, has sequence similarities to G protein β subunits. EMBO J. 1991;10:4097–4104. doi: 10.1002/j.1460-2075.1991.tb04986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Tackett A.J., Smith E.R., Fukuda A., Wysocka J., Allis C.D., Chait B.T., Hess J.L., Roeder R.G. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith M.M., Roth S.Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Feldman R.M., Correll C.C., Kaplan K.B., Deshaies R.J. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Flanagan J.F., Mi L.Z., Chruszcz M., Cymborowski M., Clines K.L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Fong H.K., Hurley J.B., Hopkins R.S., Miake-Lye R., Johnson M.S., Doolittle R.F., Simon M.I. Repetitive segmental structure of the transducin βsubunit: Homology with the CDC4 gene and identification of related mRNAs. Proc. Natl. Acad. Sci. 1986;83:2162–2166. doi: 10.1073/pnas.83.7.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guccione E., Bassi C., Casadio F., Martinato F., Cesaroni M., Schuchlautz H., Luscher B., Amati B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–937. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- Han Z., Guo L., Wang H., Shen Y., Deng X.W., Chai J. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol. Cell. 2006;22:137–144. doi: 10.1016/j.molcel.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Hassig C.A., Fleischer T.C., Billin A.N., Schreiber S.L., Ayer D.E. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- Hoey T., Weinzierl R.O., Gill G., Chen J.L., Dynlacht B.D., Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- Hyllus D., Stein C., Schnabel K., Schiltz E., Imhof A., Dou Y., Hsieh J., Bauer U.M. PRMT6-mediated methylation of R2 in histone H3 antagonizes H3 K4 trimethylation. Genes & Dev. 2007;21:3369–3380. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka K., Erdjument-Bromage H., Tempst P., Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Muller J., Hart C.M., Francis N.J., Vargas M.L., Sengupta A., Wild B., Miller E.L., O’Connor M.B., Kingston R.E., Simon J.A. Histone methyltransferase activity of a DrosophilaPolycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Neer E.J., Smith T.F. A groovy new structure. Proc. Natl. Acad. Sci. 2000;97:960–962. doi: 10.1073/pnas.97.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthun M.R., Widom J., Gottschling D.E. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- Pickles L.M., Roe S.M., Hemingway E.J., Stifani S., Pearl L.H. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure. 2002;10:751–761. doi: 10.1016/s0969-2126(02)00768-2. [DOI] [PubMed] [Google Scholar]
- Pryer N.K., Salama N.R., Schekman R., Kaiser C.A. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J. Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y.W., Wang Y.C., Hollingsworth R.E., Jones D., Ling N., Lee E.Y. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature. 1993;364:648–652. doi: 10.1038/364648a0. [DOI] [PubMed] [Google Scholar]
- Ruthenburg A.J., Wang W., Graybosch D.M., Li H., Allis C.D., Patel D.J., Verdine G.L. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat. Struct. Mol. Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg A.J., Allis C.D., Wysocka J. Methylation of lysine 4 on histone H3: Intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Schuetz A., Allali-Hassani A., Martin F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A.N., Arrowsmith C.H., Min J. Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 2006;25:4245–4252. doi: 10.1038/sj.emboj.7601316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T.F., Gaitatzes C., Saxena K., Neer E.J. The WD repeat: A common architecture for diverse functions. Trends Biochem. Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Song J.-J., Garlick J.D., Kingston R.E. Structural basis of histone H4 recognition by p55. Genes & Dev. 2008 doi: 10.1101/gad.1653308. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague E.R., Redd M.J., Johnson A.D., Wolberger C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 2000;19:3016–3027. doi: 10.1093/emboj/19.12.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T., Gutierrez J.L., Li B., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 2008;15:364–367. doi: 10.1038/nsmb.1397. [DOI] [PubMed] [Google Scholar]
- Tyler J.K., Bulger M., Kamakaka R.T., Kobayashi R., Kadonaga J.T. The p55 subunit of Drosophila chromatin assembly factor 1 is homologous to a histone deacetylase-associated protein. Mol. Cell. Biol. 1996;16:6149–6159. doi: 10.1128/mcb.16.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaisman N., Tsouladze A., Robzyk K., Ben-Yehuda S., Kupiec M., Kassir Y. The role of Saccharomyces cerevisiae Cdc40p in DNA replication and mitotic spindle formation and/or maintenance. Mol. Gen. Genet. 1995;247:123–136. doi: 10.1007/BF00705642. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P.D., Kobayashi R., Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Verreault A., Kaufman P.D., Kobayashi R., Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wall M.A., Coleman D.E., Lee E., Iniguez-Lluhi J.A., Posner B.A., Gilman A.G., Sprang S.R. The structure of the G protein heterotrimer Gi α 1 β 1 γ 2. Cell. 1995;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Williams F.E., Varanasi U., Trumbly R.J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol. Cell. Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G.T., Young M.K., Cote J., Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ng H.H., Erdjument-Bromage H., Tempst P., Bird A., Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes & Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]