Abstract
Deletions of the CDKN2A/B tumor suppressor locus and of the IKAROS and PAX5 genes that promote B-lineage development occur frequently in lymphoid, but not myeloid leukemias initiated by the BCR-ABL tyrosine kinase. Why is this the case, and how do these genetic lesions contribute to an aggressive disease that fails to durably respond to targeted kinase inhibitors?
Keywords: SNP genomic microarrays, acute lymphoblastic leukemia (ALL), chronic myeloid leukemia (CML), imatinib (Gleevec), dasatinib (Sprycel), targeted therapy
The Philadelphia chromosome (Ph+) (Nowell and Hungerford 1960), a balanced translocation arising from chromosomes 9 and 22 (Rowley 1973), was the first defined cytogenetic anomaly linked to a human cancer and is the founding genetic lesion of both chronic myeloid leukemia (CML) and Ph+ acute lymphoblastic leukemia (ALL). These translocations fuse the ABL1 oncogene on chromosome 9 to a breakpoint cluster region (BCR) from chromosome 22 to generate the constitutively activated BCR-ABL tyrosine kinase, the initiator of both diseases (Groffen et al. 1984; Chan et al. 1987; Clark et al. 1987). In CML, a p210BCR-ABL isoform (hereafter p210) is initially expressed in hematopoietic stem cells (HSCs) capable of giving rise to both differentiated myeloid and lymphoid progeny, whereas in de novo Ph+ ALL, the expression of either of two alternative p185 and p210 isoforms is restricted to the B-cell lineage (Wong and Witte 2004). CML typically presents as an indolent myeloproliferative disorder in which patients are said to be in chronic phase (CML-CP). Untreated, the disease ultimately accelerates to blast crisis (CML-BC), in which poorly differentiated malignant myeloid or lymphoid cells flood the circulation and hematopoietic organs, eventually resulting in death of the patient. BCR-ABL expression increases during disease progression, and this may help to promote the acquisition of additional genetic changes that are essential for the expansion of clones with greater malignant potential (Calabretta and Perrotti 2004). From a clinical perspective, de novo Ph+ ALL resembles CML lymphoid blast crisis, but without a preceding chronic phase. Although they are triggered by the same “runaway” tyrosine kinase, CML-CP and Ph+ ALL differ markedly in their aggressiveness and response to therapy, with Ph+ ALL portending a decidedly poorer outcome, regardless of the therapeutic modalities used in treating these patients. Ph+ ALL is rare in children (<5% of pediatric ALL) but common in adults (∼35% of adult ALL) and carries an equally poor prognosis in both age groups (Arico et al. 2000; Gleissner et al. 2002).
Drugs that target and inhibit the BCR-ABL kinase have revolutionized the treatment of CML. Imatinib (Gleevec) was the first such FDA-approved drug and has been hailed as “the poster child” for rational targeted therapy in cancer, since long-term remissions are achieved in virtually all CML-CP patients who are continuously treated (Wong and Witte 2004; Deininger et al. 2005). However, these patients still harbor leukemic stem cells, since those who terminate therapy almost invariably redevelop disease. Moreover, a small percentage of treated patients relapse (∼5% in the first year and fewer thereafter) (Druker et al. 2006) and, in general, most harbor leukemic clones that express mutant forms of BCR-ABL to which imatinib no longer binds (Shah et al. 2002, 2004). The advent of broader spectrum and more potent kinase inhibitors such as dasatinib (Sprycel) or nilotinib (Tasigna) “cover” most mutant forms of BCR-ABL and can reinduce remissions in many CML patients who fail imatinib therapy (Kantarjian et al. 2006; Talpaz et al. 2006; Hochhaus et al. 2007). In contrast, treatment of Ph+ ALL patients with these same drugs typically induces significant hematological and cytogenetic responses that are rapidly lost despite persistent treatment, suggesting that additional mutations “downstream” from the kinase contribute to more aggressive disease and to the reduced therapeutic response. What might these additional mutations be, and how might they contribute to disease?
Frequent deletions of IKFZ1 (IKAROS), PAX5, and CDKN2A/B (INK4A/B-ARF) in Ph+ ALL
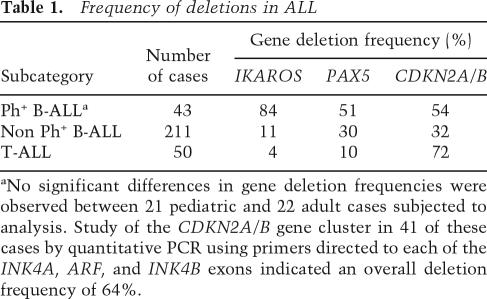
A recent genome-wide single nucleotide polymorphism (SNP) array analysis of leukemic blasts taken from 43 pediatric and adult Ph+ ALL patients, revealed the presence of three common genetic deletions involving IKZF1, PAX5, and the CDKN2A/B locus (Mullighan et al. 2008). IKZF1 (IKAROS) and PAX5 encode transcription factors required for normal lymphoid development (Nutt and Kee 2007), whereas the CDKN2A/B locus encodes three closely linked tumor suppressor genes that are widely inactivated in many forms of human cancer (Sherr 2001). These findings were confirmed by fluorescence in situ hybridization and quantitative genomic PCR analyses and, where appropriate, by nucleotide sequencing. In samples taken at the time patients first presented clinically with Ph+ ALL, 84% had already sustained IKFZ1 deletions, whereas >50% exhibited PAX5 or predominantly biallelic losses encompassing the entire CDKN2A (INK4A–ARF) and CDKN2B (INK4B) gene cluster (Table 1). Quantitative genomic PCR analysis of 41 of these cases using primers directed to each of the coding exons in the CDKN2A/B locus have revealed a higher deletion frequency (64%), reflecting better coverage of the region than that facilitated by SNP array alone; further observations indicate that CDKN2A/B inactivation is even more common in patients who fail therapy (C.G. Mullighan, unpubl.). IKFZ1 deletions were limited to the gene in 25 cases, were primarily monoallelic, and included cases that expressed a dominant-negative form of the protein, as well as others in which protein expression was absent. The fact that deletion of PAX5 and CDKN2A/B generally occurred together with IKFZ1 loss implies that disruption of each of these genes contributes independently to Ph+ B-cell ALL. In T-cell ALL, the frequencies of IKFZ1 and PAX5 deletions were much lower (Table 1), consistent with findings that these two genes play key roles in regulating B-cell lineage commitment and differentiation (Nutt and Kee 2007). Although differentiation arrest is a distinctive feature of ALL, the manner by which IKFZ1 and PAX5 inactivation collaborate with BCR-ABL to induce lymphoblastic leukemia is not yet understood.
Table 1.
Frequency of deletions in ALL
aNo significant differences in gene deletion frequencies were observed between 21 pediatric and 22 adult cases subjected to analysis. Study of the CDKN2A/B gene cluster in 41 of these cases by quantitative PCR using primers directed to each of the INK4A, ARF, and INK4B exons indicated an overall deletion frequency of 64%.
CDKN2A/B deletions occur in all lymphoid malignancies (Table 1), pointing to their general role in tumor suppression in both T- and B-cell ALL, as well as in many other tumor types. In contrast, quantitative genomic and methylation-specific PCR analysis previously undertaken with 32 CML-CP samples failed to provide evidence for inactivation of CDKN2A (K. Tago and C.J. Sherr, unpubl.). SNP array analysis performed on 23 additional CML-CP cases not only extended these results but, more strikingly, revealed no recurrent genomic lesions of any kind, suggesting that BCR-ABL alone might be sufficient to drive CML-CP. However, a small survey again identified IKFZ1, PAX5, and CDKN2A deletions in lymphoid (three cases) but not myeloid (12 cases) blast crisis, underscoring their lineage-specific occurrence in aggressive BCR-ABL-induced lymphoblastic disease (Mullighan et al. 2008).
How does BCR-ABL Select for CDKN2A/B deletions in B-cells?
The mechanisms that enable the continuous self-renewal of HSCs must prevent the execution of senescence programs regulated by the retinoblastoma protein (RB) and p53 tumor suppressors. The p16INK4A and p15INK4B products of the CDKN2A and CDKN2B genes are inhibitors of cyclin D-dependent kinases that help to maintain the retinoblastoma protein (RB) in its inhibitory, growth-suppressive mode. In contrast, the alternative reading frame (p14ARF) protein of the CDKN2A locus (p19Arf in the mouse) antagonizes the p53 E3 ubiquitin ligase HDM2 (Mdm2 in the mouse) to activate p53 (Sherr 2001). The Cdkn2a/b gene cluster is epigenetically silenced by polycomb complexes in mouse HSCs (Jacobs et al. 1999; Park et al. 2003; Iwama et al. 2004; Valk-Lingbeek et al. 2004), but it is remodeled and becomes poised to respond to oncogenic stress signals as the cells differentiate, thereby limiting continuous self-renewal in nonstem cells (Lowe and Sherr 2003). Hence, disruption of the Rb and p53 signaling network through deletion, mutation, aberrant repression or epigenetic silencing of Cdkn2a/b, or by mutations directly affecting Rb and p53, result in the acquisition of abnormal self-renewing capabilities that typify cancer cells.
Each of the INK4 and ARF genes is individually regulated and can separately contribute to tumor suppression, but among strains of mice lacking one or more of these genes, those that are most prone to tumor development lack the entire Cdkn2a/b cluster (Krimpenfort et al. 2007), mimicking the situation in Ph+ ALL. Expression of the CDKN2A/B locus can be triggered by chronic stress signals that accompany cellular senescence and organismal aging (Gil and Peters 2006; Kim and Sharpless 2006; Collado et al. 2007). The slow rate of induction of INK4A/B and ARF in cultured cells suggests that their three promoters integrate insidious signals that reflect a perturbed metabolic state of cells, such as a progressive accumulation of reactive oxygen species. In young mice, the two Ink4 and Arf gene products are not expressed in most normal tissues, but their induction is accelerated in response to increased and sustained signaling thresholds conveyed by activated oncogenes. For example, BCR-ABL efficiently induces Arf expression, thereby triggering a compensatory p53 response that eliminates incipient tumor cells through apoptosis. Although this provides a highly efficient tumor-suppressive mechanism, rare BCR-ABL-stimulated cells that lose Arf function can continue to expand and form tumors (Williams et al. 2006). Therefore, deletion of CDKN2A/B in a cancer cell implies that the checkpoint had been activated at some earlier stage of tumor progression.
Lineage-specific determinants of BCR-ABL-induced leukemias
CML-CP conforms well to the “cancer stem cell” model (Clarke et al. 2006), in that it arises from HSC-like progenitors that already have intrinsic self-renewal capacity and that differentiate to mature, nontumorigenic blood cells. As in normal HSCs, the CDKN2A/B gene cluster in CML-CP progenitors should be silenced, so there would be no selective pressure for its inactivation. However, if CDKN2A/B is epigenetically remodeled to respond to BCR-ABL-induced oncogenic signals during differentiation, the transition to CML lymphoid blast crisis would require that abnormal progenitors acquire intrinsic self-renewing capacity in order to be able to propagate the disease. Hence, the cells that sustain deletions of the CDKN2A/B locus will contribute selectively to the emerging malignancy. In turn, monoallelic deletions affecting IKFZ1 and PAX5 would retard B-lymphoid maturation, further expanding a proliferating pool of poorly differentiated blast cells.
The mechanism responsible for numerous IKFZ1 deletions appears to involve aberrant RAG-mediated recombination, as heptamer signal sequences were identified internal to the deletion breakpoints (Mullighan et al. 2008). Whereas CDKN2A deletion endpoints in many cancers generally tend to occur in clusters of repetitive elements that yield junctions indicative of nonhomologous end joining, the breakpoints in T-ALL also occur selectively near RAG recombinase recognition sequences (Kohno and Yokota 2006). Thus, although HSC-like cells in CML-CP can give rise to both myeloid and lymphoid lineages, RAG-mediated deletions likely drive lymphoid blast crisis. De novo Ph+ ALL follows a similar pattern, except that the founding translocations are initiated in committed lymphoid progenitors (Castor et al. 2005).
What factors might further limit the frequency of CDKN2A/B deletions in CML myeloid blast crisis? In children, the lineage potential of HSCs is skewed to allow development of the adaptive immune system, but as we age, myeloid progenitors expand as lymphopoiesis declines, and this mirrors the progressively increasing incidence of myeloid versus lymphoid leukemia throughout our lifetime (Rossi et al. 2008). Aging HSCs accumulate DNA damage (Rossi et al. 2007), which can activate p53 through ARF-independent pathways and diminish stem cell function under conditions of stress. In turn, mutations of p53 occur in a significant fraction of cases of CML myeloid blast crisis, thereby inactivating the ARF–HDM2–p53 pathway and abrogating selection for ARF deletion; in contrast, p53 mutations have not been widely reported in cases of CML lymphoid blast crisis in which ARF loss predominates (Calabretta and Perrotti 2004). Moreover, the potential of BCR-ABL to drive the proliferation of myeloid blasts is enhanced by additional lineage-specific mechanisms, which include the ability of aberrant β-catenin signaling to confer self-renewal to committed granulocyte–macrophage progenitors (Jamieson et al. 2004). Although β-catenin signaling is required for myeloid disease in mouse models of CML, it is dispensable for BCR-ABL-induced ALL (Zhao et al. 2007). Conceivably, WNT/β-catenin signaling might help to maintain polycomb-mediated silencing of CDKN2A/B in the myeloid lineage.
How might CDKN2A deletions contribute to resistance to targeted therapy?
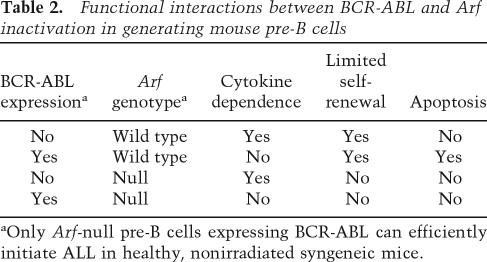
In mice, the combination of BCR-ABL expression and Arf loss are sufficient to induce aggressive B-cell ALL, and the manner by which these two genes functionally interact in B-cell progenitors is well understood (Table 2). First, constitutive BCR-ABL kinase activity triggers Arf expression and induces p53-dependent apoptosis, thereby countering BCR-ABL’s proproliferative effects. However, Arf inactivation cancels BCR-ABL-induced apoptosis and endows the cells with increased self-renewal capacity. In turn, the BCR-ABL kinase reduces the dependence of B-cell progenitors on extracellular cytokines and, in the absence of Arf-imposed restraint, drives B-cell expansion. This can even occur in p185+, Arf-null bone-marrow progenitors derived from mice that also lack the cytokine receptor common γ chain and exhibit severe combined immunodeficiency (SCID) (Williams et al. 2007). Hence, Arf inactivation enables the BCR-ABL kinase to enforce unfettered B-cell proliferation, differentiation to the pre-B-cell stage, and guarantees acquisition of full leukemic potential.
Table 2.
Functional interactions between BCR-ABL and Arf inactivation in generating mouse pre-B cells
aOnly Arf-null pre-B cells expressing BCR-ABL can efficiently initiate ALL in healthy, nonirradiated syngeneic mice.
Healthy mice infused with as few as 20 p185+, Arf-null pre-B cells die of lymphoblastic leukemia in less than a month’s time and respond poorly to imatinib therapy. However, if cytokine signaling is ablated, their response to the drug is greatly enhanced (Williams et al. 2007). This provides some evidence that p185+, Arf-null pre-B cells can be nurtured by salutary cytokines in the hematopoietic microenvironment, providing them with a measure of drug resistance. Using the more potent kinase inhibitor, dasatinib, mice with a significant tumor load can be brought into remission, but as in human Ph+ ALL, treatment failures in these mice are further undermined by clinically relevant mutations in the BCR-ABL kinase that diminish drug binding (R.T. Williams and C.J. Sherr, unpubl.). Therefore, we propose that CDKN2A/B deletions in human Ph+ ALL contribute to drug resistance by providing cells with greater fitness and facilitating the more rapid emergence of leukemic clones expressing mutant BCR-ABL isoforms.
Take home lessons
Overall, these new findings emphasize specific genetic differences between Ph+ ALL and CML and suggest that recurrent gene copy number losses affecting B-cell differentiation are universal in Ph+ ALL. While CDKN2A/B deletions occur in 50%–60% of Ph+ ALL cases at diagnosis, their frequency is increased in relapsed patients, consistent with data in a mouse model that their inactivation diminishes the effectiveness of therapy targeting the BCR-ABL kinase. Further studies incorporating higher resolution interrogation of genomic copy number alterations, deeper sequencing, and analysis of epigenetic changes might well reveal that the CDKN2A/B gene cluster is compromised in an even greater number of Ph+ ALL and lymphoid CML-BC cases. The aggressive nature of these BCR-ABL-induced lymphoid malignancies calls for treatment by potent second generation kinase inhibitors that are anticipated to more efficaciously prevent the emergence of mutant clones, either in conjunction with combination chemotherapy (Schultz et al. 2007) or potentially with other agents that target signaling between leukemia-initiating cells and their hematopoietic microenvironment.
Acknowledgments
The underlying work was supported in part by ALSAC of St. Jude Children’s Research Hospital and Cancer Center Core Grant CA-21765 from the National Cancer Institute, NIH. C.J.S. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1673908.
References
- Arico M., Valsecchi M.G., Camitta B., Schrappe M., Chessells J., Baruchel A., Gaynon P., Silverman L., Janka-Schaub G., Kamps W., et al. Outcome of treatment in children with Philadelphia chromosome-positive acute lymphoblastic leukemia. N. Engl. J. Med. 2000;342:998–1006. doi: 10.1056/NEJM200004063421402. [DOI] [PubMed] [Google Scholar]
- Calabretta B., Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- Castor A., Nilsson L., Astrand-Grundstrom I., Buitenhuis M., Ramirez C., Anderson K., Strombeck B., Garwicz S., Bekassy A.N., Schmiegelow K., et al. Distinct patterns of hematopoietic stem cell involvement in acute lymphoblastic leukemia. Nat. Med. 2005;11:630–637. doi: 10.1038/nm1253. [DOI] [PubMed] [Google Scholar]
- Chan L.C., Karhi K.K., Rayter S.I., Heisterkamp N., Eridani S., Powles R., Lawler S.D., Groffen J., Faulkes J.G., Greaves M.F., et al. A novel abl protein expressed in Philadelphia chromosome positive acute lymphoblastic leukemia. Nature. 1987;325:635–637. doi: 10.1038/325635a0. [DOI] [PubMed] [Google Scholar]
- Clark S.S., McLaughlin J., Crist W.M., Champlin R., Witte O.N. Unique forms of the abl tyrosine kinase distinguish Ph1-positive CML from Ph1-positive ALL. Science. 1987;235:85–88. doi: 10.1126/science.3541203. [DOI] [PubMed] [Google Scholar]
- Clarke M.F., Dick J.E., Dirks P.B., Eaves C.J., Jamieson C.H.M., Jones D.L., Visvader J., Weissman I.L., Wahl G.M. Cancer stem cells—Perspectives on current status and future directions: AACR workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Collado M., Blasco M.A., Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–233. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Deininger M., Buchdunger E., Druker B.J. The development of imatinib as a therapeutic agent for chronic myelogenous leukemia. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- Druker B.J., Guilhot F., O’Brien S.G., Gathmann I., Kantarjian H., Gattermann N., Deininger M.W., Silver R.T., Goldman J.M., Stone R.M., et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N. Engl. J. Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- Gil J., Peters G. Regulation of the INK4b–ARF–INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Gleissner B., Gokbuget N., Bartram C.R., Janssen B., Rieder H., Janssen J.W., Fonatsch C., Heyll A., Voliotis D., Beck J., et al. Leading prognostic relevance of the BCR-ABL translocation in adult B-lineage lymphoblastic leukemia: A prospective study of the German Multicenter Trial Group and confirmed polymerase chain reaction analysis. Blood. 2002;99:1536–1543. doi: 10.1182/blood.v99.5.1536. [DOI] [PubMed] [Google Scholar]
- Groffen J., Heisterkamp N., de Klein A., Bartram C.R., Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Hochhaus A., Kantarjian H.M., Baccarani M., Lipton J.H., Apperley J.F., Druker B.J., Facon T., Goldberg S.L., Cervantes F., Niederwieser D., et al. Dasatinib induces notable hematologic and cytogenetic responses in chronic-phase chronic myeloid leukemia after failure of imatinib therapy. Blood. 2007;109:2303–2309. doi: 10.1182/blood-2006-09-047266. [DOI] [PubMed] [Google Scholar]
- Iwama A., Oguro H., Negishi M., Kato Y., Morita Y., Tsukui H., Ema H., Kamijo T., Katoh-Fukui Y., Koseki H., et al. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Jacobs J.J.L., Kieboom K., Marino S., DePinho R.A., van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylia S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A., et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast crisis CML. N. Engl. J. Med. 2004;371:634–636. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Kantarjian H., Giles F., Wunderle L., Bhalla K., O’Brien S., Wassmann B., Tanaka C., Manley P., Rae P., Mietlowski W., et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N. Engl. J. Med. 2006;354:2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Sharpless N.E. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Kohno T., Yokota J. Molecular processes of chromosome 9p21 deletions causing inactivation of the p16 tumor suppressor gene in human cancer: Deduction from structural analysis of breakpoints for deletions. DNA Repair (Amst.) 2006;5:1273–1281. doi: 10.1016/j.dnarep.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., Ijpenberg A., Song J.Y., van der Valk M., Nawjin M., Zevenhoven J., Berns A. p15Ink4b is a critical tumour suppressor in the absence of p16Ink4a. Nature. 2007;448:943–946. doi: 10.1038/nature06084. [DOI] [PubMed] [Google Scholar]
- Lowe S.W., Sherr C.J. Tumor suppression by Ink4a-Arf: Progress and puzzles. Curr. Opin. Genet. Dev. 2003;13:77–83. doi: 10.1016/s0959-437x(02)00013-8. [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.-H., et al. 2008BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros Nature .453110–114. [DOI] [PubMed] [Google Scholar]
- Nowell P., Hungerford D. Chromosomes of normal and leukemic human leukocytes. J. Natl. Cancer Inst. 1960;25:85. [PubMed] [Google Scholar]
- Nutt S.L., Kee B.L. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Park I.-K., Qian D., Kiel M., Becker M.W., Pihalja M., Weissman I.L., Morrison S.J., Clarke M.F. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- Rossi D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I.L. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- Rossi D.J., Jamieson C.H.M., Weissman I.L. Stem cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Rowley J.D. A new consistent chromosomal abnormality in chronic myelogenous leukemia. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Schultz K.R., Bowman W.P., Slayton W., Devidas M., Sather H., Borowitz M.J., Davies S.M., Trigg M., Pasut B., Jorstad D., et al. Blood. 2007. Improved early event free survival (EFS) in children with philadelphia chromosome-positive (Ph+) acute lymphoblastic leukemia (ALL) with intensive imatinib in combination with high dose chemotherapy: Children’s Oncology Group (COG) Study AALL0031. (ASH Annual Meeting Abstracts) 110: Abstract 4. [Google Scholar]
- Shah N.P., Nicoll J.M., Nagar B., Gorre M.E., Paquette R.L., Kuriyan J., Sawyers C.L. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- Shah N.P., Tran C., Lee F.Y., Chen P., Norris D., Sawyers C.L. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:319–321. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell Biol. 2001;2:731–737. doi: 10.1038/35096061. [DOI] [PubMed] [Google Scholar]
- Talpaz M., Shah N.P., Kantarjian H., Donato N., Nicoll J., Paquette R., Cortes J., O’Brien S., Nicaise C., Bleickardt E., et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek M.E., Bruggeman S.W., van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Williams R.T., Roussel M.F., Sherr C.J.2006Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia Proc. Natl. Acad. Sci. 103 :6688–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R.T., den Besten W., Sherr C.J. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes & Dev. 2007;21:2283–2287. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., Witte O.N. The BCR-ABL story: Bench to bedside and back. Annu. Rev. Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- Zhao C., Blum J., Chen A., Kwon H.Y., Jung S.H., Cook J.M., Lagoo A., Reya T. Loss of β-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]