Abstract
In this issue of Genes & Development, Mordes and colleagues (pp. 1478–1489) reveal intriguing mechanistic insights into activation of the ATR (ATM and Rad3-related) kinase critical for DNA damage resistance. They identify conserved regulatory domains within ATR and its binding partner ATRIP (ATR-interacting protein), which are contacted by the ATR activator TopBP1. These discoveries expand on our understanding of the regulation of other PIKK family members, which also contain these domains, and illustrate how functional diversity has been achieved among these kinases.
Keywords: ATR, ATRIP, TopBP1, checkpoint, PIKK, Ddc2
Every cell cycle poses a challenge to the maintenance of genomic stability. Two members of the PIKK family, ATM (ataxia-telangiectasia mutated) and ATR (ATM and Rad3-related), govern signaling cascades that meet this challenge by detecting and responding to DNA damage to elicit crucial responses such as cell cycle arrest, transcriptional changes, and DNA repair (for review, see Harper and Elledge 2007). This program can contribute directly to tumor suppression not only via maintenance of genome integrity, but also via its engagement of such programs as senescence and apoptosis, which prevent damaged cells from undergoing further divisions. Although ATM and ATR share common downstream substrates such as p53 and BRCA1, they primarily respond to different stimuli. ATM responds to double-strand breaks while ATR responds primarily to replication stress and ssDNA gaps. ATR can also respond to double-strand breaks but with much slower kinetics than ATM. As part of the PIKK family, ATM and ATR share a highly conserved C-terminal kinase domain. However, wide functional diversity has been achieved among family members, for while ATM, ATR, and DNA-PKcs are all involved in DNA damage signaling or repair, mTOR regulates protein biosynthesis, and SMG1 functions in nonsense-mediated decay. Several recent studies have highlighted how functional specificity has been achieved among these family members through their use of unique binding partners, post-translation modifications, and regulatory domains. However, unlike other serine/threonine kinases, how PIKK kinases are activated mechanistically is poorly understood. In this issue of Genes & Development, Cortez and colleagues (Mordes et al. 2008) have uncovered conserved regulatory domains in PIKKs that illuminate how activation of ATR occurs, and how specialization of ATR among PIKK family members has been achieved in the cell. These discoveries provide an important inroad into dissecting the mechanism of PIKK activation, and thus have the potential to inform a large swath of biology.
Activation of the ATR Pathway via TopBP1
ATR is recruited to ssDNA via its stable partner ATRIP (ATR-interacting protein), which binds to Replication Protein A (RPA)-coated ssDNA (Cortez et al. 2001; Zou and Elledge 2003). A second complex, the Rad17/Rfc2-5 complex, is independently recruited to ssDNA where it is loaded in an RPA-dependent manner. The presence of a dsDNA–ssDNA junction, as might be found at a stalled replication fork, activates this complex to load a second complex, the PCNA-related 9–1–1 clamp (Rad9–Rad1–Hus1). Once both complexes have been recruited to the damage site, ATR is activated and phosphorylates its effector kinase, Chk1, in addition to other substrates. How this activation is mediated was unclear, until it was recently shown that TopBP1, a mediator protein containing eight BRCT phospho-recognition motifs, binds and activates ATR/ATRIP complexes in a manner distinct from the role of TopBP1 in initiation of DNA replication (Kumagai et al. 2006). TopBP1 was first implicated in checkpoint signaling by work on the fission yeast ortholog Cut5/rad4 where it was shown to be required for prevention of mitotic entry in response to DNA replication blocks. Similar results were obtained with Dpb11 in budding yeast (Saka and Yanagida 1993; Araki et al. 1995). Furthermore, a direct connection was found between Dpb11 and Cut5 with the respective 9–1–1 complexes in yeast (Wang and Elledge 2002; Furuya et al. 2004), thereby linking it to known signaling molecules. In mammals, reduction of TopBP1 levels results in a defect in phosphorylation of Chk1 and other ATR substrates, although ATR localization to damaged DNA is unaffected. Experiments in Xenopus laevis extracts demonstrated that addition of exogenous TopBP1 could elicit ectopic ATR activity toward Chk1, as could overexpression in mammalian cells. Furthermore, TopBP1 physically interacts with ATR, and this interaction is required for stimulation of ATR in response to damage. Expression of a series of truncations revealed that TopBP1 interacts with ATR through a domain termed the ATR activation domain (AAD), between the sixth and seventh BRCT repeat (Kumagai et al. 2006).
The role of ATRIP in TopBP1-mediated ATR Activation
While it was known that TopBP1 bound to ATR/ATRIP, what surfaces were used and how ATR activation was accomplished was not known. To address this, Mordes et al. (2008) took advantage of an ATRIP mutant that lacks the C-terminal ATR-interacting domain (Ball and Cortez 2005; Falck et al. 2005), and found reduced TopBP1 binding compared with wild-type ATR–ATRIP complexes. Possible interpretations of their data are that binding of TopBP1 to ATR–ATRIP may be accomplished via binding sites on both ATR and ATRIP or by an ATR-induced conformational shift in ATRIP to generate a stronger binding site.
To begin to distinguish between these possibilities, the investigators sought to determine precisely which surface on ATRIP was bound by TopBP1. To do this, the investigators developed a clever sheared yeast two-hybrid system in which they sheared the ATRIP cDNA and made a library of ATRIP fragments with which they screened for binding to the AAD of TopBP1. Interacting ATRIP fragments corresponded to residues 203–348, which are adjacent to the ATRIP coiled-coiled domain required for ATRIP homo-oligomerization (Ball and Cortez 2005). Within these 146 amino acids, a span of 30 residues was found to be highly conserved among ATRIP homologs. A substitution mutation in this region of ATRIP, LLSS332AAAA, was able to retain ATR binding and proper localization to sites of DNA damage, while the interaction with TopBP1 was specifically lost. The substitution mutant, termed ATRIP-top, also showed decreased binding of ATR/ATRIP complexes to TopBP1, further supporting the hypothesis that ATRIP binding is partially responsible for this interaction. In vivo, the ATRIP-top mutant showed severely reduced ATR activity toward a substrate, Mcm2, compared with wild-type ATR–ATRIP complexes. Thus, the interaction of ATRIP with TopBP1 is required for TopBP1 to both bind and activate ATR kinase activity. The investigators went on to examine the replication stress response of this ATRIP-top mutant in response to hydroxyurea (HU), which causes replication fork stalling. Cells expressing ATRIP-top showed an impaired ability to complete S phase and also reduced viability, consistent with reduced signaling. These experiments demonstrate that this interaction between ATRIP and TopBP1 is required for ATR to appropriately respond to DNA damage.
Conservation of checkpoint mechanisms among eukaryotes
Most activities of the DNA damage response are conserved among eukaryotes. In Saccharomyces cerevisiae Mec1 and Ddc2 are the orthologs of ATR and ATRIP. To determine whether this ATRIP-dependent mechanism is conserved in budding yeast for Mec1 activation, corresponding mutations were made in Ddc2. Unfortunately, Ddc2 and ATRIP lack sequence homology in this region. However, the investigators reasoned that the secondary structure might be conserved, and therefore chose the location of their corresponding mutations using common secondary structure elements. Their intuition paid off when they discovered that expression of the corresponding Ddc2 substitution mutations showed increased sensitivity to both HU and methanesulfonate (MMS). While interaction with Mec1 remained intact, phosphorylation of its substrate Rad53 was decreased. This provocative experiment indicates that Ddc2 behaves similarly to ATRIP in promoting activation of Mec1 in response to DNA damage and replication stress, and suggests that while Dpb11 lacks obvious homology with the AAD of TopBP1, it likely plays a conserved role in activating Mec1.
Identification of the ATR PIKK Regulatory Domain (PRD)
Having identified the binding surface on ATRIP, the investigators next focused on determining whether the enhanced binding TopBP1 displayed to the ATR/ATRIP complexes might be explained by a second interaction with ATR. To do this, they once again turned to their shearing two-hybrid method, this time preparing a library of ATR fragments to screen against the AAD region of TopBP1. ATR contains several N-terminal HEAT repeats that participate in ATRIP binding, and a C-terminal kinase domain (Ball et al. 2005). Adjacent to this kinase domain are FAT and FATC domains, the functions of which are not thoroughly understood. The interacting residues identified in ATR mapped to a region containing the C-terminal portion of the kinase domain adjacent to the FATC domain as shown in Figure 1A. Intriguingly, sequence analysis of this uncharacterized region lying between the kinase and FATC domains showed little homology between PIKK family members, but high conservation among ATR orthologs. Scanning mutagenesis of this region produced two mutants, K2589E and HVL2591AAA, which showed markedly decreased TopBP1 activation of ATR, while both basal activity of ATR (in the absence of TopBP1) and its interaction with ATRIP were left intact. Pull-down experiments comparing wild-type ATR or ATR K2589E–ATRIP complexes showed the expected decreased TopBP1 binding in the mutant, and it is expected that the residual TopBP1 binding is due to the second binding surface on ATRIP. These experiments support the idea that TopBP1-mediated ATR activation via this domain, which they dubbed the PRD, and is required for ATR activation but not for basal activity of the kinase as observed in these assays. These ATR mutants provide new tools to separate TopBP1-mediated ATR activity from other ATR functions.
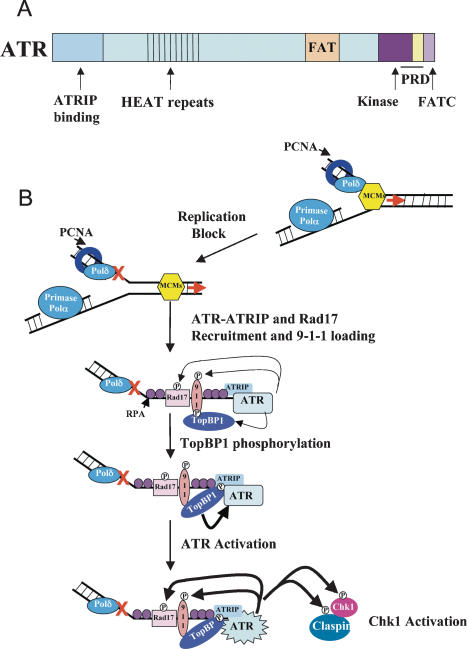
Figure 1.
How ATR is activated at stalled replication forks. (A) A schematic of ATR and its domains. (B) A model for ATR activation at a stalled replication fork. Upon encountering a replication-blocking lesion, denoted by the red X, loss of coordination between DNA polymerase and the MCM helicase generates ssDNA, which is coated by RPA. ATR–ATRIP and the RAD17/RFC2–5 complexes are independently recruited to ssDNA–RPA stretches and RAD17/RFC2–5 loads the RAD9–HUS1–RAD1 (9–1–1) clamp complex. TopBP1 binds RAD9 through a constitutively phosphorylated site on its C terminus. Several ATR-dependent phosphorylation events occur, including sites on RAD17, 9–1–1 clamp components, and TopBP1. Phosphorylated TopBP1 can bind surfaces on ATR and ATRIP, causing ATR to become activated and to phosphorylate its downstream effectors including Chk1.
Essentiality of TopBP1-mediated ATR Activation
ATR and TopBP1 are both essential genes, and the extent to which TopBP1-mediated ATR activation may be required for cell viability distinct from its role in checkpoint response had not been addressed (Brown and Baltimore 2000; Yamane et al. 2002). To determine the role of the PRD in ATR function, the investigators created a cell line containing an ATR-/flox allele and either a tetracycline-inducible wild-type or mutant K2589E ATR cDNA. While both cell lines displayed normal Chk1 phosphorylation in response to HU prior to excision of the floxed allele, the cell line expressing K2589E ATR showed marked reduction in Chk1 phosphorylation following Cre recombinase-mediated excision, indicating an intact ATR PRD is required for proper replication stress responses. Upon further examination of the viability of these different cell lines after Cre-mediated deletion of ATR, they discovered that the only colonies from the K2589E cell line that survived Cre treatment were found to have retained the endogenous wild-type ATR allele. In contrast, many clones were isolated expressing the wild-type cDNA following successful Cre-mediated excision. This experiment suggests that the ATR PRD is not only involved in replication stress signaling, but it is also required for cell viability illustrating the crucial role of ATR activation in cellular physiology.
Conservation of PRD-mediated regulation of PIKK family members
Interestingly, the region analogous to the PRD in ATR has recently been highlighted in studies of other PIKK family members. Tip60-mediated acetylation of ATM, which results in increased ATM activity in response to DNA damage, has been shown to occur on a lysine residue within the corresponding ATM PRD (Sun et al. 2005). In order to examine whether such post-translation modification may be occurring on ATR, Mordes et al. (2008) made a series of lysine-to-arginine mutations within the ATR PRD. None of these mutations appeared to have an effect on TopBP1-mediated ATR activation, and siRNA knockdown of Tip60 did not alter cell recovery from HU, suggesting that PRD acetylation is specific to ATM and does not participate in ATR regulation, although the PRD itself is important in both cases. Thus, the PRD may have a conserved regulatory function in the PIKK family, although disparate sequences correspond to its unique usage for each family member. Thus, the PRD represents a point of divergence allowing for specialization among these kinases.
Electron microscopy studies of another PIKK family members, DNA-PKcs, revealed that the its binding partner, the Ku70/80 heterodimer, may bind to a region adjacent to the kinase domain, which corresponds to the ATR PRD (Spagnolo et al. 2006). In order to expand on these results, Mordes et al. (2008) examined whether the PRD is also involved in regulation of the PIKK family member DNA-PKcs. A series of mutations were made within this region, and DNA-PKcs autophosphorylation was used as a read-out for DNA-PKcs activity in response to ionizing radiation. Two of these mutants exhibited defects in DNA-PKcs autophosphorylation following damage, suggesting that the PRD may have a conserved regulatory role in DNA-PKcs activation as well. It will be interesting to determine whether the PRD mediates Ku70/80 binding.
Requirement of the FATC domain for basal ATR Activity
It is clear that activation of ATR is a key aspect of its function, and a detailed understanding of its activation mechanism is likely to have important ramifications for understanding the activation of other PIKKs. It is likely that other domains conserved among PIKKs are involved in kinase activity as well. Therefore, Mordes et al. (2008) turned their attention toward the FATC domain, which is conserved but whose function is not known. A deletion of the PRD-adjacent FATC domain of ATR was made, and resulted in total loss of ATR basal activity in their assays. While it has been shown that the ATM FATC can be successfully replaced by the ATR FATC, (Jiang et al. 2006) Cortez and colleagues found that basal ATR kinase activity could not be restored by replacement with the ATM FATC. While it is difficult to draw conclusions from negative results, it suggests that specific features of the FATC domain may need to be matched with particular aspects of the resident PIKK framework. Furthermore, it is curious that ATM can function with the ATR FATC domain, since the acetylation by TIP60 that is thought to be required for ATM activity resides in this domain. It would be interesting to know if ATM activity in the swap mutant is still Tip60-dependent. It is possible that the FATC domain of ATR has a constitutive function while the FATC domain of ATM requires activation that cannot be incorporated into the ATR molecule, perhaps because additional sequences outside of FATC are required to recruit Tip60. Furthermore, while no combination of lysine-to-arginine mutations in the ATR FATC appeared to affect ATR activity, it will be interesting to learn whether perhaps other modifications are required in the ATR FATC domain. Regardless, it is clear that the ATR FATC domain is strictly required for basal ATR activity and is likely to play a similar role in all PIKKs.
Specificity in PIKK activation
Specificity among PIKK family members appears to be mediated in part by unique binding partners, such as ATRIP, Nbs1, Ku70/80, and raptor-rictor in the cases of ATR, ATM, DNA-PKCs, and mTOR, respectively. Parallels can be made between the emerging role of TopBP1 and that of GβL, which binds to the mTOR kinase domain and increases its activity and also stabilizes interaction with its binding partner raptor (Kim et al. 2003). This study by Mordes et al. (2008) has also reiterated the role of ATRIP in stabilizing the interaction between TopBP1 and ATR for ATR activation, illustrating the involvement of ATRIP in multiple steps of ATR regulation. However, some common regulation may exist among PIKKs, as suggested by the finding that the Tel2 protein can regulate the stability of all PIKK family members via interaction with conserved N-terminal HEAT repeats (Shikata et al. 2007; Takai et al. 2007). In some cases, these regulatory mechanisms can even functionally connect family members. It has recently been shown that TopBP1 can be phosphorylated by ATM following irradiation, and that this modification may assist in ATR activation by ATM in response to double-strand breaks (Yoo et al. 2007).
How ATR signaling is initiated: dual sensor engagement with a positive activation loop
The ATR pathway must be finely tuned in order to respond appropriately to a range of challenges, whether it be dynamic replication fork structures during S phase in each cell cycle, or widespread damage from an exogenous source. It is clear that early events are subject to multiple levels of regulation, in their recruitment, stabilization, post-translational modifications, and catalytic activity. An explanation for how TopBP1 gains access to ATR at sites of DNA damage was suggested by the interactions with 9–1–1 found in yeast. Mammalian TopBP1 binds the constitutively phosphorylated C-terminal tail of Rad9 on the 9–1–1 complex, and this binding occurs via its first pair of BRCT repeats on TopBP1. In chicken DT40 cells, it has been shown that signaling defects of Rad17−/− cells can be restored by expression of a histone 2B-TopBP1-AAD fusion protein. This, in addition to other experiments, suggests that a major function of the 9–1–1 complex is to recruit TopBP1 for ATR activation (Furuya et al. 2004; Delacroix et al. 2007; Lee et al. 2007). Thus, although considered a mediator protein, TopBP1 has been implicated in early events in signaling following recruitment of ATR–ATRIP and 9–1–1 to sites of DNA damage and replication stress. Furthermore, TopBP1 is a substrate for ATM and ATR, and its phosphorylation appears to be important for activation of ATR. These observations, together with the results of Cortez and colleagues, have led to the model shown in Figure 1B. In response to DNA replication blocks, coordination between the DNA polymerases and the MCM helicases are disrupted leading to the accumulation of ssDNA at the stalled replication fork. This ssDNA is rapidly bound by the free RPA complex. The presence of extensive RPA–ssDNA complexes then recruits both the ATR/ATRIP complex and the Rad17–RFC complex. The presence of a ssDNA–dsDNA junction then activates the Rad17–RFC complex to load the 9–1–1 complex onto DNA. TopBP1 is associated with the 9–1–1 complex, allowing it to now colocalize with ATR/ATRIP. The sequence of the next series of events is not yet clear, but involves phosphorylation of TopBP1, which has been shown to be required for TopBP1 activity. One appealing possibility is that the basal activity of ATR phosphorylates TopBP1 on activating residues. Previous work in vitro has shown that ATR can phosphorylate Rad17 when both are colocalized by binding RPA–ssDNA complexes (Zou et al. 2003). Phosphorylated TopBP1 then becomes competent to bind ATR/ATRIP and activates ATR kinase activity forming a positive feedback loop. This then sets in motion the events leading to Claspin and Chk1 activation and the phosphorylation of additional downstream effectors.
Future directions
The precise physical mechanisms by which the interactions between TopBP1 and ATR–ATRIP elicit increased ATR activity remain to be determined. It is not yet clear whether the initial association with ATRIP occurs first followed by docking of the TopBP1 AAD with the PRD of ATR. However, it is likely that the direct binding to the PRD of ATR results in a conformational change in the structure of ATR to generate an active kinase domain. Whether this is analogous to phosphorylation on the D loop that reorganizes the kinase domain of standard serine/threonine kinase domain or is more like the relief of an inhibitory domain as in the activation of Protein Kinase C, remains to be determined. It is clear that the domains and separation-of-function mutants discovered by Mordes et al. (2008) will be key to the elucidation of this mechanism. Preliminary kinetic studies discussed in Mordes et al. (2008) indicated that the interaction between TopBP1 and the ATR PRD may reduce the apparent Km of ATR. Structural and kinetic studies to determine the precise function of the ATR PRD on ATR activity can now be carried out. If it is a conformational change that activates ATR, it should be possible to mimic that change by mutagenesis of the PRD to generate a constitutively active ATR, which might be capable of suppressing AAD-defective mutants in TopBP1. Furthermore, this study also supports the idea that the PRD has a conserved regulatory function for other PIKKs such as DNA-PKcs, perhaps by mediating Ku70/80 binding. These possibilities remain to be further investigated to determine their generality.
The function of the highly conserved FATC domain in PIKK activation remains to be determined. Cortez and colleagues also shed light on this domain, which they found to be required for basal ATR kinase activity. It will be of interest to determine whether additional interactions or post-translational modifications may occur on the ATR FATC domain. This domain is well-positioned to affect stability or conformational changes in the kinase domain, and it is quite possible that binding to the PRD mediates its effects through changes in the FATC structure. Now that these domains and key residues have been identified within ATR, future studies can build upon these tools to further define the function and potential interplay of these domains in ATR regulation.
The different ways through which TopBP1 can gain proximity to ATR also requires further attention. In Xenopus laevis it has been shown that ATR is capable of phosphorylating Rad1 in the absence of the Rad9 C terminus, which binds TopBP1 and is required for Chk1 phosphorylation by ATR (Lupardus and Cimprich 2006). This suggests that ATR may be differentially active toward a subset of substrates. It is possible that Rad1 phosphorylation is carried out by the basal activity of ATR upon colocalization, although Rad1 phosphorylation has been reported to be TopBP1-dependent in Xenopus extracts (Lupardus and Cimprich 2006). Alternatively, there may be a Rad9-independent way for TopBP1 to localize to ATR. It is possible that some forms of TopBP1 will have high enough affinity for ATR that it will no longer need the enhanced local concentration provided by Rad9 binding to activate ATR.
The discoveries provided here by Cortez and colleagues now sets the stage for a full-scale assault to unravel the ATR activation mechanism. Understanding this mechanism will have important implications for many different signaling pathways. Furthermore, manipulation of DNA damage signaling pathways is being explored in treatment of cancer in the cases of ATM and DNA-PKcs. Elucidation of the surfaces involved in ATR regulation could lead to the discovery of small molecules capable of inducing or inhibiting ATR kinase activity that could prove to be important in cancer treatment. Further insights into this mechanism are eagerly awaited.
Acknowledgments
This work was supported by NIH grants to S.J.E. S.J.E. is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1685108.
References
- Araki H., Leem S.H., Phongdara A., Sugino A. Dpb11, which interacts with DNA polymerase IIε in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl. Acad. Sci. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball H.L., Cortez D. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J. Biol. Chem. 2005;280:31390–31396. doi: 10.1074/jbc.M504961200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball H.L., Myers J.S., Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR–ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E.J., Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes & Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S.J. ATR and ATRIP: Partners in checkpoint signaling. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Delacroix S., Wagner J.M., Kobayashi M., Yamamoto K., Karnitz L.M. The Rad9–Hus1–Rad1 (9–1–1) clamp activates checkpoint signaling via TopBP1. Genes & Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Furuya K., Poitelea M., Guo L., Caspari T., Carr A.M. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes & Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. The DNA damage response: Ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Jiang X., Sun Y., Chen S., Roy K., Price B.D. The FATC domains of PIKK proteins are functionally equivalent and participate in the Tip60-dependent activation of DNA-PKcs and ATM. J. Biol. Chem. 2006;281:15741–15746. doi: 10.1074/jbc.M513172200. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Sarbassov D.D., Ali S.M., Latek R.R., Guntur K.V., Erdjument-Bromage H., Tempst P., Sabatini D.M. GβL, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell. 2003;11:895–904. doi: 10.1016/s1097-2765(03)00114-x. [DOI] [PubMed] [Google Scholar]
- Kumagai A., Lee J., Yoo H.Y., Dunphy W.G. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J., Kumagai A., Dunphy W.G. The Rad9–Hus1–Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Lupardus P.J., Cimprich K.A. Phosphorylation of Xenopus Rad1 and Hus1 defines a readout for ATR activation that is independent of Claspin and the Rad9 carboxy terminus. Mol. Biol. Cell. 2006;17:1559–1569. doi: 10.1091/mbc.E05-09-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes D.A., Glick G.G., Zhao R., Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes & Dev. 2008 doi: 10.1101/gad.1666208. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y., Yanagida M. Fission yeast cut5+, required for S phase onset and M phase restraint, is identical to the radiation-damage repair gene rad4+ Cell. 1993;74:383–393. doi: 10.1016/0092-8674(93)90428-s. [DOI] [PubMed] [Google Scholar]
- Shikata M., Ishikawa F., Kanoh J. Tel2 is required for activation of the Mrc1-mediated replication checkpoint. J. Biol. Chem. 2007;282:5346–5355. doi: 10.1074/jbc.M607432200. [DOI] [PubMed] [Google Scholar]
- Spagnolo L., Rivera-Calzada A., Pearl L.H., Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol. Cell. 2006;22:511–519. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Sun Y., Jiang X., Chen S., Fernandes N., Price B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Wang R.C., Takai K.K., Yang H., de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Wang H., Elledge S.J. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Wu X., Chen J. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 2002;22:555–566. doi: 10.1128/MCB.22.2.555-566.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H.Y., Kumagai A., Shevchenko A., Dunphy W.G. Ataxia-telangiectasia mutated (ATM)-dependent activation of ATR occurs through phosphorylation of TopBP1 by ATM. J. Biol. Chem. 2007;282:17501–17506. doi: 10.1074/jbc.M701770200. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L., Elledge S.J. RPA-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]