Abstract
The long-term health of the cell is inextricably linked to protein quality control. Under optimal conditions this is accomplished by protein homeostasis, a highly complex network of molecular interactions that balances protein biosynthesis, folding, translocation, assembly/disassembly, and clearance. This review will examine the consequences of an imbalance in homeostasis on the flux of misfolded proteins that, if unattended, can result in severe molecular damage to the cell. Adaptation and survival requires the ability to sense damaged proteins and to coordinate the activities of protective stress response pathways and chaperone networks. Yet, despite the abundance and apparent capacity of chaperones and other components of homeostasis to restore folding equilibrium, the cell appears poorly adapted for chronic proteotoxic stress when conformationally challenged aggregation-prone proteins are expressed in cancer, metabolic disease, and neurodegenerative disease. The decline in biosynthetic and repair activities that compromises the integrity of the proteome is influenced strongly by genes that control aging, thus linking stress and protein homeostasis with the health and life span of the organism.
Keywords: Stress responses, molecular chaperones, protein misfolding
Protein homeostasis is the cellular process that governs the “life of proteins.” More than protein quality control alone, protein homeostasis encompasses RNA metabolism and processing, protein synthesis, folding, translocation, assembly/disassembly, and clearance (Balch et al. 2008). These processes and their roles in protein homeostasis can be viewed as a systems network in which each process, or hub, is organized as mininetworks interconnected to other hubs. An implicit assumption of protein homeostasis is that each hub should be dynamic, and therefore have sufficient “buffering” capacity to respond to various imbalances in network components and flux of misfolded species and damaged proteins. Tissue-specific differences in expression of components of the homeostasis network such as chaperones and components of the proteasome, autophagy, or transport machinery could be a basis for the sensitivity of neuronal cells during aging and disease.
Two prominent modulators of protein homeostasis are molecular chaperones and stress-inducible responses. Chaperones are abundantly expressed in multiple compartments of the cell and are thought to comprise a significant fraction of the cellular machinery (Bukau et al. 2006; Ron and Walter 2007). Only limited information is available on the concentration of chaperones in a eukaryotic cell and the fraction that is functionally engaged with substrates or freely available in reserve. Nevertheless, this has led to a commonly held expectation that within the cell, chaperones have excess folding capacity and can exist in a free state to buffer unexpected folding requirements. This would require the cell to maintain a stockpile of chaperones for protein folding emergencies. A counter view is that the cell has little excess chaperone capacity, and that the concentration of chaperones is titrated closely to the folding requirements for each client protein within a specific cell type. This would imply that the folding environment in the cell is delicate, which would seem to be a risky proposition. For this to be compatible with the complex and varying cellular environment, the delicate nature of protein homeostasis would need to be paired with a highly robust stress response that is very sensitive and responds rapidly to any flux in protein biogenesis. Moreover, because chaperones are growth-regulated and stress-responsive, they afford the cell with a stress sensor to link stress signaling processes with protein homeostasis (Nollen and Morimoto 2002).
Chaperone networks
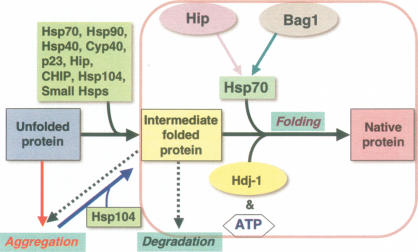
Molecular chaperones have diverse roles to regulate protein conformation, and are essential to protect nascent polypeptides from misfolding, to facilitate co- and post-translational folding, to assist in assembly and disassembly of macromolecular complexes, and to regulate translocation (Hartl and Hayer-Hartl 2002; Deuerling and Bukau 2004; Bukau et al. 2006; Ron and Walter 2007). Chaperone terminology has its origins as heat-shock proteins (Hsps) annotated by molecular size; Hsp 100, Hsp90, Hsp70, Hsp60, Hsp40, and small heat-shock protein (sHSP) families (Chang et al. 2007; Tang et al. 2007). Much of our understanding of chaperone function comes from biochemical studies of assisted protein folding using protein substrates and purified prokaryotic and eukaryotic chaperones from Escherichia coli, Saccharomyces cerevisiae, and mammalian species. A defining general characteristic of chaperones and their regulatory cochaperones is the ability to suppress misfolding and aggregation of unfolded polypeptides and to “hold” nonnative species in intermediate “on-pathway” states that remain competent for subsequent folding to the native state (Fig. 1). “Holding” activity is a property common to many chaperones as exemplified by Hsp104, Hsp90, Hsp70, the sHSPs, immunophilins/cyclophilins (FKPB52 and CyP40), the steroid aporeceptor complex protein p23, and Hip (Hsp70- and Hsp90-interacting protein) (Parsell et al. 1994; Freeman and Morimoto 1996; Freeman et al. 1996; Obermann et al. 1998). An important functional distinction among chaperones is that “holding” of intermediates is a common property, whereas folding to the native state is highly regulated by Hsp70 and ATP hydrolysis (Langer et al. 1992; McCarty et al. 1995; Freeman et al. 1996). Likewise, the Hsp60/CCT/Clp/Hsp104 chaperone machines also have a central role in both folding and disaggregation (Fig. 1; Martin et al. 1993; Weissman et al. 1995; Weibezahn et al. 2004; Doyle et al. 2007). Cycles of Hsp70 nucleotide binding and hydrolysis coupled to the release of the folded protein are regulated by cochaperones including dnaJ/Hsp40, Hip, and Bag proteins (Fig. 1; Bukau et al. 2006). Unlike the Hsp90, Hsp70, or Hsp60/CCT chaperones that have limited substrate selectivity in eukaryotes, cochaperones provide specificity for interaction with chaperones and specific client proteins (Pratt and Toft 2003). Consequently, the association of chaperones such as Hsp70 or Hsp90 with different partners including J-domain proteins, p23, cyclophilins, and immunophilins can influence the conformation and activities of a wide range of substrates such as p53 (Blagosklonny et al. 1996), HSF1 (Shi et al. 1998), and other transcription factors including steroid aporeceptors, in addition to kinases and phosphatases (Pratt and Toft 2003; Whitesell and Lindquist 2005). The assembly of macromolecular chaperone complexes containing these specific “client” proteins is a common structural motif in signal transduction to ensure that proteins whose activities must be precisely regulated are in a “hair-trigger” conformational state. For example, steroid aporeceptors are assembled in a partially folded state in macromolecular complexes with the Hsp70 and Hsp90 and their specificity cochaperones; upon hormone binding a precise sequence of events is initiated leading to chaperone dissociation and folding to a native state as a nuclear localized transcription factor (Picard et al. 1990; Pratt and Toft 2003). The folding and assembly of the large and diverse class of steroid aporeceptors emphasizes the importance of intermediate species and conformational choice as a potent step in protein biogenesis.
Figure 1.
Chaperone networks and the regulation of protein conformation. The unfolded protein transitions to the intermediate and native state assisted by molecular chaperones. The “holding” activity of chaperones is represented by formation of transient and stable intermediates that continue on-pathway to the native state upon interaction with an ATP-dependent chaperone machine such as Hsp70 (together with ATP and the cochaperones Bag1, Hip, and Hdj1). Unfolded proteins and intermediates can form aggregates that can be rescued by the Hsp104 class of chaperones. Likewise, intermediates can form aggregates, become degraded, or fold to the native state assisted by chaperones.
Stress and other physiological demands on protein homeostasis can result in the sporadic and unpredicted flux of conformationally challenged proteins that can alter the chaperone-substrate balance. Yet, at the same time, the pool of chaperones is essential to protect nascent chains and other metastable proteins from misfolding under normal conditions and when cells are exposed to acute or chronic stress (Gidalevitz et al. 2006). Stress-induced changes in the absolute and relative levels of chaperones and cochaperones could lead to novel chaperone networks that, in turn, would redirect information flow through alternate folding pathways, with effects on translocation and subcellular localization. Consequently, some signaling pathways may be enhanced or dampened because of changes in the levels of a particular cochaperone or chaperone that affects the stability of the heteromeric complex. For example, altering the levels of Aha1, a cochaperone of Hsp90, corrects the folding and trafficking of mutant CFTR (Wang et al. 2006). Changes in the levels of Hsp90, achieved by genetic approaches or exposure to small molecules like geldanamycin, has potent pleiotropic biological consequences (Rutherford and Lindquist 1998; Bagatell and Whitesell 2004). Likewise, changes in the activity of Hsp70, by modulating the levels of the cochaperone Bag1, has consequences on the activity of the Ras/Raf-1 with downstream effects on signaling and cell growth (Song et al. 2001). In Drosophila, overexpression of Hsp70 is deleterious for growth, and expression of a dominant-negative Hsp70 causes developmental defects (Feder et al. 1992; Elefant and Palter 1999). Long-term chaperone imbalance, therefore, is likely to have dramatic consequence on diverse cellular processes.
What remains less clear is whether this is a general strategy of the cell to link stress to specific signaling pathways. Cells that have lost their ability to properly regulate proper growth control, such as tumor cells, often express higher levels of chaperones (Jaattela 1999). Depletion of Hsp90 by geldanamycin treatment or Hsp70 by antisense treatment arrests cell growth and spurs progression to cell death (Whitesell et al. 1994, 2003; Nylandsted et al. 2000). Tumor cells are dependent on elevated levels of Hsps, and tumorigenesis requires expression of Hsf1 (Dai et al. 2007). This suggests that enhanced levels of chaperones could suppress missense mutations that accumulate during transformation to promote cell growth and oncogenesis. This is exemplified by the relationship between p53 and Hsp90, where mutant forms of p53 require Hsp90 for function (Blagosklonny et al. 1996). Given these diverse activities of Hsp90, it remains a puzzle to understand how chaperone networks can simultaneously touch on multiple stages of protein biogenesis and yet regulate the functionality of critical client proteins involved in key regulatory steps, while maintaining the ability to respond to diverse cellular stresses.
Together, Hsps and chaperones serve the cell as a “master concierge,” touching on diverse cellular activities, while constantly integrating signals from the environment with protein homeostasis and metabolism. The involvement of chaperones in a multitude of signaling and regulatory pathways offers a challenge to systems biology in which networks are used to organize specific pathways. Because proteins such as Hsp90 and Hsp70 touch on hundreds if not thousands of events, an imbalance in protein homeostasis networks may have profound long-term consequences.
Chaperone regulation—the heat-shock response
Genes encoding chaperone machines are of three regulatory classes: (1) constitutively expressed and regulated during growth and development, (2) constitutive and inducibly regulated, and (3) strictly inducible. The expression of chaperones is essential for normal growth and development and to survive stress. The heat-shock response enables the cell to elevate the expression of genes that function to protect against proteotoxic stress and to initiate a regulatory cascade for recovery and adaptation (Wu 1995; Morimoto 1998; Anckar and Sistonen 2007). This occurs by a nearly instantaneous induction of heat-shock genes (Hs genes) proportional to the intensity, duration, and type of stress (Abravaya et al. 1991; Gasch et al. 2000; Hahn et al. 2004). While the primary form of regulation is at the level of transcription, the heat-shock response is also post-transcriptionally regulated by stress-induced mRNA stability (Theodorakis and Morimoto 1987), by translational control (Banerji et al. 1984), and by stress-induced effects on the activity and subcellular localization of chaperones (Welch and Suhan 1986).
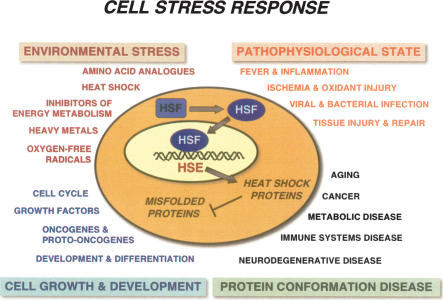
The heat-shock response is mediated by a family of heat-shock transcription factors (HSFs) that are expressed and maintained in a non-DNA-binding state under control (nonstress) conditions primed for activation to a trimeric DNA-binding state. Multiple HSF genes (HSF1, HSF2, and HSF4) are expressed in mammals, avians express HSF1–4, while Drosophila, Caenorhabditis elegans, and yeast express only HSF1 (Wu 1995; Morimoto 1998; Pirkkala et al. 2001; Anckar and Sistonen 2007). In mammals, HSF1 corresponds to the ubiquitous stress-responsive activator (McMillan et al. 1998), whereas HSF2, although widely expressed, is developmentally regulated and important for neuronal specification (Sistonen et al. 1992; Kallio et al. 2002), and HSF4 is highly cell type-specific associated with expression of crystallines (Bu et al. 2002; Fujimoto et al. 2004). The activity of HSF1 is induced by a variety of stress signals, including a wide range of acute and chronic perturbations of physiological states and disease (Fig. 2; Morimoto 1998).
Figure 2.
Cell stress response. The expression of HS genes including chaperones and components of the clearance machinery is induced in response to three classes of physiological and environmental stress conditions including environmental stress, pathophysiological stress, and protein conformational disease, and by a fourth class of cell growth and development. Indicated below each major class are representative conditions known to involve the expression of heat-shock proteins and chaperones.
Induction of the heat-shock response corresponds to a stepwise process that involves activation of HSF1 monomers to nuclear-localized trimers, binding to DNA, and attenuation of transcription with subsequent conversion back to the monomer (Wu 1995; Morimoto 1998; Anckar and Sistonen 2007). The principal targets for HSF1 are heat-shock elements (HSEs) within promoter regions of HS genes (Williams and Morimoto 1990; Xiao et al. 1991). Microarray experiments and chromatin immunoprecipitation assays have also identified genes that respond to heat shock with promoters that bind to HSF1 but lack consensus HSEs, suggesting that HSFs could exert their activity via interactions with other DNA-binding proteins (Trinklein et al. 2004). HSF1 in unstressed metazoan cells is maintained predominantly in a repressed non-DNA-binding monomeric state by transient interactions with chaperones including Hsp90, Hsp70, and Hsp40 (Abravaya et al. 1992; Shi et al. 1998; Zou et al. 1998). Activation to a stable DNA-binding trimer is associated with the stress signal. The signals that induce the heat-shock response and HSF1 have not been thoroughly elucidated. Much of the data is consistent with the primary signal being associated with a flux of intermediates that are detected as misfolded and damaged proteins (Ananthan et al. 1986). The appearance of excess nonnative protein shifts the chaperone equilibrium, thus derepressing HSF1 to undergo a conformational transformation to an active state. This involves the formation of HSF1 homotrimers via an extended heptad repeat (HR-A/B) located between the DNA-binding domain and the transcription activation domain, which releases the DNA-binding domain for binding to HSEs (Sorger and Nelson 1989). Association of HSF1 at the promoters of HS genes, in turn, releases a preinitiated paused RNA polymerase II complex upon recruitment of elongation factors including pTEFb (Lis et al. 2000; Boehm et al. 2003; Ni et al. 2004). Chromatin at HS gene loci is further regulated by recruitment of the Mediator complex that transduces signals to the transcriptional machinery (Park et al. 2001) and the FACT remodeling factor (Saunders et al. 2003). High rates of transcription are maintained only when HSF1 trimers remain bound to the HSEs; when either the stress signal is removed or damaged proteins are no longer generated, the heat-shock response attenuates rapidly (Abravaya et al. 1991; Yao et al. 2006). Association of chaperones with HSF1 suppresses transcription (Abravaya et al. 1992; Shi et al. 1998). Stress-activated HSF1 is further modified post-translationally by phosphorylation (Sorger and Pelham 1988; Knauf et al. 1996; Kline and Morimoto 1997; Holmberg et al. 2001; Guettouche et al. 2005), sumoylation (Hietakangas et al. 2003; Anckar et al. 2006), and acetylation (S. Westerheide, J. Anckar, L. Sistonen, and R. Morimoto, pers. comm.). Modification of HSF1 at conserved residues has multiple regulatory consequences, to maintain HSF1 in a repressed state, to enhance transcriptional activity, or to signal attenuation. The combination of these post-translational modifications and chaperone interactions thus affords HSF1 with multiple forms and levels of control and feedback loops to precisely regulate chaperone levels in the cell.
The heat-shock response has often been portrayed as a universal molecular response to various stress stimuli (Fig. 2). While this is generally correct, the exceptions are instructive. There are numerous observations in the literature in which the heat-shock response is poorly or incompletely activated. These include early development or exposure of intact organisms to whole-body stress (Bienz 1984). Of particular interest have been studies on the heat-shock response in the brain and during aging (Sprang and Brown 1987; Shamovsky and Gershon 2004). Achieving a stress condition in whole mammals is challenging and has required anesthetics to prevent temperature-induced seizures. Restricted expression of HS genes has been observed in different regions of the brain, consistent with the selective expression of HS genes in cultured neuronal cells. Human neuroblastoma Y79 cells, for example, respond to heat shock by activation of HSF1 and expression of Hsp90 but not Hsp70 (Mathur et al. 1994). In intact primary hippocampal neurons from neonatal rat embryos, only HSF2 but not HSF1 is expressed until later stages of development (Marcuccilli et al. 1996). Consequently, the heat-shock response of primary hippocampal neurons is deficient, whereas astrocytes that express both HSF1 and HSF2 have a robust stress response. Similar observations have been made in primary motor neurons that exhibit a deficient heat-shock response thought to be due to a defect in activation of HSF1 (Batulan et al. 2003).
The regulation of the heat-shock response and HSF1 activity by small molecules has provided valuable tools to elucidate the mechanism of HSF1 regulation. These compounds include proteasome inhibitors, serine protease inhibitors, Hsp90 inhibitors, nonsteroidal anti-inflammatory drugs (NSAIDS), triterpenoids such as celastrol, and inhibitors of HSF1 including triptolide (Westerheide and Morimoto 2005; Westerheide et al. 2006). Protease inhibitors (DCIC, TPCK, and TLCK) and proteasome inhibitors (MG132, lactacystin) induce the heat-shock response by elevating the concentration of proteins targeted to the degradative machineries (Mathew et al. 1998; Rossi et al. 1998). In contrast, inhibitors of Hsp90 including the fungal antibiotic radicicol, the benzoquinone ansamycins geldanamycin, and 17AAG activate HSF1, in part because Hsp90 is a negative regulator of HSF1 (Bagatell et al. 2000; Whitesell et al. 2003; Bagatell and Whitesell 2004; Whitesell and Lindquist 2005; Sharp and Workman 2006). NSAIDS including sodium salicylate have multiple properties; at higher concentrations they partially activate HSF1 and at lower concentrations they synergize with other stress conditions to induce the heat-shock response (Jurivich et al. 1992). Exposure of human tissue culture cells to sodium salicylate results in activation of HSF1 trimers that bind in vivo to the HSEs of the Hsp70 gene without inducing transcription. Salicylate-treated cells, however, are sensitized to stress and readily activate HS genes upon exposure to other mild stress conditions. In a similar manner, indomethacin induces HSF1 DNA binding with full Hsp70 transcription upon exposure to a secondary stress (Lee et al. 1995). Of the inflammatory modulators, arachidonic acid, and the cyclopentenone prostaglandins, including PGA1, PGA2, and PGJ2, all induce the full complement of HSF1 activities (Amici et al. 1992; Jurivich et al. 1994). The triterpenoid celastrol isolated from the Chinese plant Triptergium wilfordii represents a herbal medicine class of bioactive molecules that induces two protective stress responses, HSF1 and the anti-oxidant response (Westerheide et al. 2004; Trott et al. 2008). The effects of celastrol are rapid like heat shock; however, unlike the heat-shock response that self-attenuates, the celastrol induction of HS genes persists for an extended period (Westerheide et al. 2004).
‘Mis’folding—challenges to the proteome and chaperone networks in disease and aging
The functional redundancy and capacity of molecular chaperones and clearance machines would suggest that protein quality control should be an efficient process. Chaperones, together with the autophagy and ubiquitin proteasome system (UPS), are abundantly expressed, yet there is evidence to suggest that limitations and malfunction of the clearance machinery are risk factors in diseases of protein conformation (Bence et al. 2001; Holmberg et al. 2004; Venkatraman et al. 2004). At the root of the “protein misfolding” problem could be the long-term consequences of homeostasis imbalance on protein stability, folding, and clearance. Over time, this could compromise innumerable other proteins that harbor mild folding defects with consequences on multiple cellular processes. Efforts by the protein homeostasis machinery to adapt to this chronic challenge could result in further decline as even more proteotoxic species accumulate in a self-amplifying vortex (Stadtman 1992).
The misfolding problem
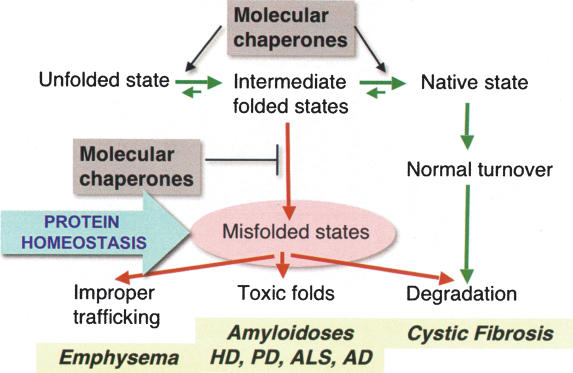
To understand the cellular context in which protein homeostasis functions will require an understanding of translational efficiency and fidelity, and beyond this to obtain estimates on the subsequent events of folding, assembly, and clearance in the cell. Even if each step in the process is relatively efficient, the overall yield is likely to be low. In cells with active protein synthesis or, for example, in early development when there is rapid cell division, it seems reasonable to suggest that the levels of chaperones and clearance machineries must be in abundance and the process highly efficient (Fig. 3). Stress responses such as the heat-shock response and the unfolded protein response (UPR) then serve to protect the cell against extreme flux as can occur when cytoplasmic or lumen-localized proteins misfold. A provocative question is whether all misfolded and damaged proteins are recognized, refolded, or cleared with equal efficiency, or if certain proteins such as those implicated in neurodegenerative diseases and other protein conformational diseases are particularly difficult for the quality control machineries.
Figure 3.
Interplay between protein quality control (transition of unfolded to intermediates to native states) and clearance mechanisms in protein conformation disease. Chaperones have a critical role to suppress the appearance of misfolded species and to enhance protein folding. The imbalance of misfolded species is associated in human disease with premature clearance of CFTR as occurs in cystic fibrosis, to prevent improper trafficking of α-1-anti-trypsin as occurs in emphysema, and to prevent proteins from adopting toxic folds as in amyloidoses including huntingtin in Huntington’s disease, α-synuclein and parkin in Parkinson’s disease, mutant SOD1 in familial ALS, and Aβ in Alzheimer’s disease.
Protein folding has been studied extensively by biochemical, biophysical, and computational methods to observe and describe the events by which polypeptides fold from the unfolded state (U) to the native (N) state with the appearance of ensembles of intermediate species (I) (Fig. 3; Chiti and Dobson 2006). Visualized as a folding landscape, proteins enroute to the native state can enter energy minima that correspond to nonproductive intermediates with unfolded and misfolded regions that can seed aggregates (Chiti and Dobson 2006). A challenge to understanding protein folding in the cell is to know which of these intermediates and pathways are favored or disfavored, and how mutations in expressed proteins, or changes in protein homeostasis, can result in nonnative species. The appearance of aggregates must occur within the normal context of quality control. Molecular chaperones have essential roles to keep the expression of nonproductive misfolded species at a minimum and to enhance unfolding of such species to enter either on-pathway to productive folding pathways or degradation (Fig. 3).
Misfolding and aggregation are now recognized as common molecular events for a large number of human diseases (Fig. 3; Perutz 1999; Chiti and Dobson 2006). This has attracted much attention to a class of proteins that can form cross-β-sheet structures that form ordered homotypic species that can associate with other cellular proteins to form heterotypic aggregates. Conformational diseases also have in common that aggregate species cause “gain-of-function” proteotoxicity and result in cellular pathology and disease due to improper trafficking, premature degradation, or the appearance of toxic folds (Fig. 3; Kakizuka 1998; Chiti and Dobson 2006). Such diseases include CAG-repeat/polyglutamine (polyQ) expansion diseases including Huntington’s disease (HD), Kennedy’s disease, and spinocerebellar ataxias, and non-CAG diseases including Parkinson’s disease, amyotrophic lateral sclerosis (ALS), prion disease, and Alzheimer’s disease. Each of these diseases has the distinctive characteristic of selective neuronal vulnerability despite broader patterns of expression, age-dependent onset, and a progressive usually fatal clinical course. The expression of alternate self-associated folded states associated with aggregation and fibril formation, while typically associated with cellular dysfunction and pathology, need not always be toxic. For example, the biology of yeast prions exemplified by Sup35, a yeast translation termination factor, offers a valuable perspective for the role of alternate protein conformations in biological function. Sup35 can exist as a soluble translation elongation factor or in a fibrillar state that allows translational read-through and transmission as a “gain-of-function” prion state (Serio et al. 2000; True and Lindquist 2000). Biophysical studies on yeast prions have identified multiple alternative folded states, each of which can self-propagate as distinct stable prion strains (Tessier and Lindquist 2007; Toyama et al. 2007). Therefore, protein misfolding, while often deleterious, can also provide a diverse range of alternate shapes and properties to the biology of proteins.
Insights on neurodegenerative disease and protein conformational disease from nonmammalian model systems
A striking characteristic of proteins that cause neurodegenerative disease are the dissimilarities in primary sequence, function, and native structures, yet all share the ability to form various alternate unfolded and misfolded states that are highly aggregation-prone and cause toxicity. This is supported by compelling evidence from in vitro biophysical studies, observations at the cellular and molecular level in cultured cells and transgenic animals, and from studies of human tissues. Contributing to this knowledge has been the ability to replicate molecular, cellular, and behavioral phenotypes associated with neurodegenerative disease in the model systems of S. cerevisiae, C. elegans, and Drosophilia melanogaster (Warrick et al. 1998; Faber et al. 1999; Feany and Bender 2000; Krobitsch and Lindquist 2000; Satyal et al. 2000; Parker et al. 2001; Morley et al. 2002). The development of these nonmammalian models for protein conformational disease has been invaluable for the discovery of modifiers and pathways, to elucidate underlying mechanisms of toxicity, and for the testing of small molecules (Warrick et al. 1999; Kazemi-Esfarjani and Benzer 2000; Morley et al. 2002; Nollen et al. 2004; Garcia et al. 2007).
Drosophila and C. elegans exhibit an organismal complexity that is accessible at the molecular level and can be readily exploited to interrogate cellular and behavioral phenotypes during development and aging. These advantages, together with genome-wide tools for genetic screens and a short life cycle and life span, also allow rapid testing of hypotheses. Observations from many laboratories have shown that polyQ or Aβ proteins are toxic in multiple cells and tissues of Drosophila and C. elegans including retinal neurons, chemosensory, mechanosensory, motor neurons, and body wall muscle cells (Link 1995; Warrick et al. 1998; Faber et al. 1999; Marsh et al. 2000; Parker et al. 2001; Morley et al. 2002; Takeyama et al. 2002; Brignull et al. 2006). While no single model recapitulates all features of a disease it is evident that each system has contributed unique insights that have validated the general approach.
The expression of proteins with expanded polyQ underlies a larger class of neurodegenerative diseases known as CAG or triplet-repeat disease including HD, spinocerebellar ataxias, and Kennedy’s disease (Perutz 1999; Zoghbi and Orr 2000; Ross 2002). Expression of polyQ alone, in the context of flanking coding sequences, or when inserted into unrelated proteins, is sufficient to recapitulate many of the defining characteristics of aggregation toxicity. A unique characteristic of the polyQ diseases is the relationship between the polyQ repeat length polymorphism with a pathogenic threshold of 35–40 glutamine residues. Genetic studies have established that Huntingtin alleles from normal chromosomes contain <30–34 CAG repeats, whereas those from affected chromosomes have >37 repeats with a strong inverse correlation between repeat length and age of onset (Zoghbi and Orr 2000; Ross 2002). Among the most striking features of polyQ length-dependent phenotypes is the consistency of observations whether from in vitro studies with polyQ peptides or from in vivo studies in C. elegans neurons and nonneuronal cells compared with human disease and neuropathology. The inverse correlation between polyQ length and age at onset observed in the human diseases suggests that the intrinsic biophysical properties of polyQ motifs must have a dominant role in pathology and toxicity.
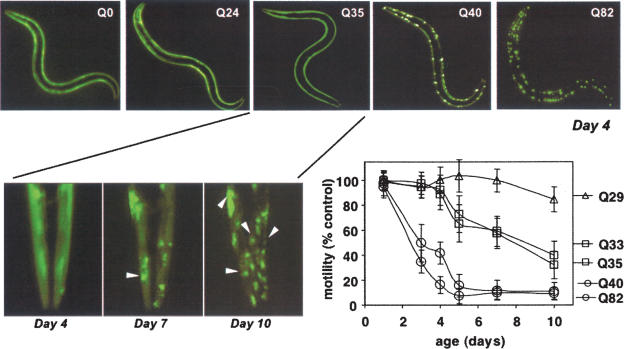
In transgenic C. elegans expressing polyQ repeats of different lengths up to Q86 as YFP fusions, there is a clear relationship between polyQ length and aggregation toxicity in neurons and muscle cells. In young adult animals expressing polyQ in body wall muscle cells, <Q35 is diffuse and soluble whereas >Q40 form discrete aggregates visualized by the chimeric fluorescent YFP tag (Fig. 4; Satyal et al. 2000; Morley et al. 2002). When expressed throughout the C. elegans nervous system, Q19 is diffuse and soluble in the cell bodies of commissural neurons and the dorsal nerve cord whereas Q86 forms aggregates in the early larval stage neurons that persist in adults (Brignull et al. 2006). The appearance of polyQ aggregates in neurons and muscle cells is associated with toxicity and has dramatic effects on motility (Fig. 4; Morley et al. 2002; Brignull et al. 2006). Neurotoxicity is observed in C. elegans expressing intermediate polyQ lengths (Q35 and Q40) even though polyQ aggregates are detected only in specific neurons (Brignull et al. 2006). Another dimension is the transparency of C. elegans, which has allowed live cell imaging methods. FRAP (fluorescence recovery after photobleaching) and FLIP (fluorescence lifetime in photobleaching) methods provide direct measures of diffusion rates of protein species that can be used to compare different cell types during development and aging, whereas FRET (fluorescence resonance energy transfer) efficiencies measures molecular proximities in protein–protein interactions. Neuronal Q40 exhibits heterogeneous properties with some ventral nerve cord neurons expressing both soluble and aggregate species, whereas other neurons (head or tail ganglia) expressing only soluble Q40 (Brignull et al. 2006). This cell-to-cell variation in the biophysical properties of threshold lengths of polyQ in neurons could have many explanations, including subtle differences in expression and differences in chaperones and clearance machineries or other cell type-specific modifiers.
Figure 4.
PolyQ length and age-dependent aggregation toxicity in C. elegans. Animals expressing different lengths of polyQ-YFP (Q0, Q24, Q35, Q40, and Q82) exhibit a length-dependent aggregation phenotype in day 4 young adult animals. Intermediate lengths of polyQ (i.e., Q35) show an age-dependent aggregation phenotype of soluble protein at day 4 and increasing numbers of aggregates in day 7 and day 10 of adulthood. The graph shows that there is a corresponding age-dependent increase in cellular toxicity (loss of motility) with polyQ expansion. Figure adapted from Morley et al. (2002).
The aggregation and toxicity phenotypes in C. elegans, together with genome-wide tools for genetic screens, have been used to identify modifiers of polyQ. For example, genes that prevent polyQ aggregation should exhibit enhanced aggregation when knocked down by RNAi. Using C. elegans strains expressing Q33 and Q35, 186 modifier genes that caused premature onset of aggregation were identified (Nollen et al. 2004). These modifiers sort into five major classes: genes involved in RNA metabolism, protein synthesis, protein folding, protein trafficking, and protein degradation. Examples of some of these genes are: RNA helicases, splicing factors, and transcription factors for RNA metabolism; initiation and elongation factors and ribosomal subunits for protein synthesis; chaperonins and Hsp70 family members for protein folding; nuclear import and cytoskeletal genes for protein trafficking; and proteasomal genes for protein degradation. These five classes can be further sorted into two categories: genes that regulate the expression of a damaged protein, and genes involved in folding, transport, and clearance. For example, perturbation of the RNA-processing machinery leads to accelerated aggregation of polyQ perhaps due to an increased burden of abnormal proteins requiring the activity of the protein folding buffer. These results reveal that the transition between soluble and aggregated states of polyQ is regulated by a much more complex network that extends well beyond protein quality control. It is interesting to note that a genetic screen for modifier genes that are osmoprotective in C. elegans overlaps substantially with these polyQ modifier genes revealing common targets for regulators of protein homeostasis (Lamitina et al. 2006).
The results of a forward genetic screen performed in C. elegans expressing expanded polyQ in muscle cells identified a loss-of-function mutation in unc-30 expressed only in the GABAergic neurons that results in cholinergic overstimulation and imbalance in protein homeostasis (Garcia et al. 2007). Similar results were observed with small molecule agonists and antagonists that work at the synaptic junction to effect overstimulation. These results suggest that protein homeostasis is also regulated in a cell-nonautonomous manner, and reveal an important and unexpected role for the nervous system on the health of the post-synaptic cell.
Aging, a potent modifier of protein homeostasis
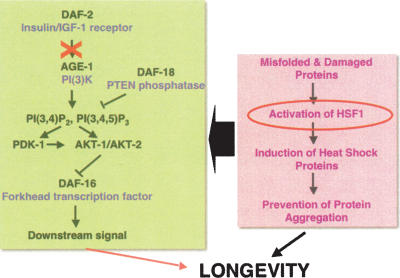
The association with aging is among the most characteristic aspects of protein conformational disease. For neurodegenerative disease, the age at which neurological symptoms appear varies, with Alzheimer’s disease and Parkinson’s disease being late-onset, ALS occurring most frequently in early to mid-life, and HD age-associated but more closely linked with polyQ length polymorphism. Aging and protein homeostasis are strongly intertwined. Genes in the insulin-like signaling (ILS) pathway that regulate aging and prolong life span have been shown to be potent modulators of aggregation toxicity (Figs. 4, 5; Morley et al. 2002). In wild-type C. elegans, the kinase activity of AGE-1 is required for a signaling cascade that results in constitutive repression of the forkhead transcription factor DAF-16 (Morris et al. 1996; Lin et al. 1997). Derepression of DAF-16 in age-1 animals extends life span and daf-16 mutations suppress longevity. The age-1 effects on longevity and polyQ aggregation toxicity require the DAF-16 pathway, revealing a common genetic pathway (Fig. 5; Morley et al. 2002; Hsu et al. 2003). The modulation of protein misfolding and aggregation by the ILS pathway appears to be a general feature, and has also been observed in C. elegans expressing Aβ as a model for Alzheimer’s disease (Cohen et al. 2006).
Figure 5.
Roles of the ILS pathway and HSF1 in aggregation toxicity and longevity. Both HSF1 and DAF-16 are essential components and function in concert to promote longevity and to maintain protein homeostasis. Reduction of the ILS signal (depicted by the X) leads to the activation of DAF-16 and enhancement of life span.
The role of genetic pathways that regulate life span and suppress proteotoxicity associated with multiple aggregation-prone proteins suggests an important relationship between the genetics of aging and disease. The molecular interactions between these pathways are subserved, in part, by factors that detect and respond to misfolded proteins; namely, DAF-16, HSF1, and molecular chaperones. Inhibition of HSF1 suppresses the ILS protection against misfolded proteins. Moreover, HSF1 down-regulation leads to decreased life span and an accelerated aging phenotype in C. elegans, while conversely, overexpression of HSF1 extends life span (Fig. 5; Morley et al. 2002; Hsu et al. 2003). Inactivation of daf-16, hsf-1, and sHSPs in C. elegans accelerated the aggregation of polyQ supporting the proposition that the ILS could coordinately influence aging and protein aggregation through the action of DAF-16 and HSF1 (Hsu et al. 2003; Morley and Morimoto 2004). The ILS pathway and HSF1 function via stress responses and chaperone networks, while potent modifiers of aggregation toxicity must function in the context of multiple networks that regulate different aspects of protein homeostasis (Hsu et al. 2003; Morley and Morimoto 2004; Cohen et al. 2006).
The challenge to the cell and organism, in the face of chronic proteotoxic stress associated with disease, is the global, system-wide decline in cellular function with deleterious consequences on viability. In turn, this causes an acceleration in the levels of damaged proteins that leads to the decline in multiple biosynthetic and repair activities, which, over time, has deleterious consequences on the health and aging of the organism. The idea that the molecular determinants of longevity influence proteotoxicity is supported by observations that the time until pathology develops—days in C. elegans, weeks in Drosophila, months in mice, and years in humans—correlates approximately with the life span of the organism.
Future outlook and perspectives
The integrity of the proteome is central to ensure the efficient functioning of the cell and to ensure that biological processes remain healthy throughout the life span of the organism. Currently, much of our knowledge on protein homeostasis is from studies in yeast and tissue culture cells under “normal” conditions of cell growth or upon challenge by acute environmental and physiological stress. Continued efforts to study the fundamentals of protein folding in the cell will be essential to understand the complexity of events that occur during proteotoxic stress in diseases of protein conformation. While much of the current attention is at the level of the cell, it will become increasingly important to understand how these events are organized and integrated system-wide at the organismal level. Among the more challenging questions are those associated with cell type specificity and cell-nonautonomous interactions. What is the basis for the selective toxicity of proteins such as huntingtin or Aβ, and can we learn from protection mechanisms of cells that are less sensitive to proteotoxicity? Are specific neuronal cells less well adapted than others to the stress of misfolded proteins because they lack protective stress responses or because they have limited capacities for clearance that, over time, lead to eventual cellular dysfunction and tissue collapse? While most studies have emphasized universal cell-autonomous considerations of folding within the cell, there are increasing numbers of observations in which cell-nonautonomous interactions between cells have significant consequence on aggregation toxicity.
The expression of disease-associated aggregation-prone proteins causes an imbalance in protein homeostasis with deleterious consequences on the folding of other metastable proteins. This reveals that the cell does not have excess capacity to buffer against protein misfolding, and suggests that the cellular machineries and quality control may be maladapted to certain types of chronic stress. Efforts to alleviate proteotoxicity by enhancing molecular chaperones or proteasome and autophagy function in numerous cell-based assays and transgenic model systems have shown promise as potential therapeutic strategies. However, beyond folding and clearance mechanisms, the molecular interactions that regulate the functional properties and health of proteins include a much larger proteostasis network with RNA biogenesis, protein synthesis, and translocation having prominent roles (Balch et al. 2008). The demonstration that the DAF-16 and HSF1 stress pathways have essential roles in the regulation of proteostasis and life span provides an important insight. Manipulation of these functional proteostasis networks could therefore restore folding, transport, and clearance machineries and thus prevent the further appearance and accumulation of damaged and misfolded proteins. The arrest of age-associated decline in proteostasis by restoring the capacity of the quality control machinery could therefore have broad-reaching consequences and benefits.
Acknowledgments
I thank members of my laboratory for their comments, critical discussions, and reading of the manuscript. R.I.M. is supported by grants from the National Institutes of Health (NIGMS, NIA, and NINDS), the Huntington Disease Society of America Coalition for the Cure, the ALS Association, and the Daniel F. and Ada L. Rice Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1657108.
References
- Abravaya K., Phillips B., Morimoto R.I. Attenuation of the heat shock response in HeLa cells is mediated by the release of bound heat shock transcription factor and is modulated by changes in growth and in heat shock temperatures. Genes & Dev. 1991;5:2117–2127. doi: 10.1101/gad.5.11.2117. [DOI] [PubMed] [Google Scholar]
- Abravaya K., Myers M.P., Murphy S.P., Morimoto R.I. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes & Dev. 1992;6:1153–1164. doi: 10.1101/gad.6.7.1153. [DOI] [PubMed] [Google Scholar]
- Amici C., Sistonen L., Santoro M.G., Morimoto R.I. Antiproliferative prostaglandins activate heat shock transcription factor. Proc. Natl. Acad. Sci. 1992;89:6227–6231. doi: 10.1073/pnas.89.14.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthan J., Goldberg A.L., Voellmy R. Abnormal proteins serve as eukaryotic stress signals and trigger the activation of heat shock genes. Science. 1986;232:522–524. doi: 10.1126/science.3083508. [DOI] [PubMed] [Google Scholar]
- Anckar J., Sistonen L. Heat shock factor 1 as a coordinator of stress and developmental pathways. Adv. Exp. Med. Biol. 2007;594:78–88. doi: 10.1007/978-0-387-39975-1_8. [DOI] [PubMed] [Google Scholar]
- Anckar J., Hietakangas V., Denessiouk K., Thiele D.J., Johnson M.S., Sistonen L. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol. Cell. Biol. 2006;26:955–964. doi: 10.1128/MCB.26.3.955-964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagatell R., Whitesell L. Altered Hsp90 function in cancer: A unique therapeutic opportunity. Mol. Cancer Ther. 2004;3:1021–1030. [PubMed] [Google Scholar]
- Bagatell R., Paine-Murrieta G.D., Taylor C.W., Pulcini E.J., Akinaga S., Benjamin I.J., Whitesell L. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents. Clin. Cancer Res. 2000;6:3312–3318. [PubMed] [Google Scholar]
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Manipulating proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Banerji S.S., Theodorakis N.G., Morimoto R.I. Heat shock-induced translational control of HSP70 and globin synthesis in chicken reticulocytes. Mol. Cell. Biol. 1984;4:2437–2448. doi: 10.1128/mcb.4.11.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batulan Z., Shinder G.A., Minotti S., He B.P., Doroudchi M.M., Nalbantoglu J., Strong M.J., Durham H.D. High threshold for induction of the stress response in motor neurons is associated with failure to activate HSF1. J. Neurosci. 2003;23:5789–5798. doi: 10.1523/JNEUROSCI.23-13-05789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bence N.F., Sampat R.M., Kopito R.R. Impairment of the ubiquitin–proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Bienz M. Developmental control of the heat shock response in Xenopus. Proc. Natl. Acad. Sci. 1984;81:3138–3142. doi: 10.1073/pnas.81.10.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagosklonny M.V., Toretsky J., Bohen S., Neckers L. Mutant conformation of p53 translated in vitro or in vivo requires functional HSP90. Proc. Natl. Acad. Sci. 1996;93:8379–8383. doi: 10.1073/pnas.93.16.8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm A.K., Saunders A., Werner J., Lis J.T. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignull H.R., Moore F.E., Tang S.J., Morimoto R.I. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J. Neurosci. 2006;26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L., Jin Y., Shi Y., Chu R., Ban A., Eiberg H., Andres L., Jiang H., Zheng G., Qian M., et al. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat. Genet. 2002;31:276–278. doi: 10.1038/ng921. [DOI] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Chang H.C., Tang Y.C., Hayer-Hartl M., Hartl F.U. SnapShot: Molecular chaperones, Part I. Cell. 2007;128:212. doi: 10.1016/j.cell.007.01.001. [DOI] [PubMed] [Google Scholar]
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- Cohen E., Bieschke J., Perciavalle R.M., Kelly J.W., Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. 2006;313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- Dai C., Whitesell L., Rogers A.B., Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130:1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E., Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit. Rev. Biochem. Mol. Biol. 2004;39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- Doyle S.M., Shorter J., Zolkiewski M., Hoskins J.R., Lindquist S., Wickner S. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat. Struct. Mol. Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefant F., Palter K.B. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol. Biol. Cell. 1999;10:2101–2117. doi: 10.1091/mbc.10.7.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber P.W., Alter J.R., MacDonald M.E., Hart A.C. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc. Natl. Acad. Sci. 1999;96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany M.B., Bender W.W. A Drosophila model of Parkinson’s disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- Feder J.H., Rossi J.M., Solomon J., Solomon N., Lindquist S. The consequences of expressing hsp70 in Drosophila cells at normal temperatures. Genes & Dev. 1992;6:1402–1413. doi: 10.1101/gad.6.8.1402. [DOI] [PubMed] [Google Scholar]
- Freeman B.C., Morimoto R.I. The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J. 1996;15:2969–2979. [PMC free article] [PubMed] [Google Scholar]
- Freeman B.C., Toft D.O., Morimoto R.I. Molecular chaperone machines: Chaperone activities of the cyclophilin Cyp-40 and the steroid aporeceptor-associated protein p23. Science. 1996;274:1718–1720. doi: 10.1126/science.274.5293.1718. [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Izu H., Seki K., Fukuda K., Nishida T., Yamada S., Kato K., Yonemura S., Inouye S., Nakai A. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J. 2004;23:4297–4306. doi: 10.1038/sj.emboj.7600435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia S.M., Casanueva M.O., Silva M.C., Amaral M.D., Morimoto R.I. Neuronal signaling modulates protein homeostasis in Caenorhabditis elegans post-synaptic muscle cells. Genes & Dev. 2007;21:3006–3016. doi: 10.1101/gad.1575307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A.P., Spellman P.T., Kao C.M., Carmel-Harel O., Eisen M.B., Storz G., Botstein D., Brown P.O. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidalevitz T., Ben-Zvi A., Ho K.H., Brignull H.R., Morimoto R.I. Progressive disruption of cellular protein folding in models of polyglutamine diseases. Science. 2006;311:1471–1474. doi: 10.1126/science.1124514. [DOI] [PubMed] [Google Scholar]
- Guettouche T., Boellmann F., Lane W.S., Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J.S., Hu Z., Thiele D.J., Iyer V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 2004;24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hietakangas V., Ahlskog J.K., Jakobsson A.M., Hellesuo M., Sahlberg N.M., Holmberg C.I., Mikhailov A., Palvimo J.J., Pirkkala L., Sistonen L. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C.I., Hietakangas V., Mikhailov A., Rantanen J.O., Kallio M., Meinander A., Hellman J., Morrice N., MacKintosh C., Morimoto R.I., et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmberg C.I., Staniszewski K.E., Mensah K.N., Matouschek A., Morimoto R.I. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–4318. doi: 10.1038/sj.emboj.7600426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A.L., Murphy C.T., Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Jaattela M. Heat shock proteins as cellular lifeguards. Ann. Med. 1999;31:261–271. doi: 10.3109/07853899908995889. [DOI] [PubMed] [Google Scholar]
- Jurivich D.A., Sistonen L., Kroes R.A., Morimoto R.I. Effect of sodium salicylate on the human heat shock response. Science. 1992;255:1243–1245. doi: 10.1126/science.1546322. [DOI] [PubMed] [Google Scholar]
- Jurivich D.A., Sistonen L., Sarge K.D., Morimoto R.I. Arachidonate is a potent modulator of human heat shock gene transcription. Proc. Natl. Acad. Sci. 1994;91:2280–2284. doi: 10.1073/pnas.91.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizuka A. Protein precipitation: A common etiology in neurodegenerative disorders? Trends Genet. 1998;14:396–402. doi: 10.1016/s0168-9525(98)01559-5. [DOI] [PubMed] [Google Scholar]
- Kallio M., Chang Y., Manuel M., Alastalo T.P., Rallu M., Gitton Y., Pirkkala L., Loones M.T., Paslaru L., Larney S., et al. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J. 2002;21:2591–2601. doi: 10.1093/emboj/21.11.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemi-Esfarjani P., Benzer S. Genetic suppression of polyglutamine toxicity in Drosophila. Science. 2000;287:1837–1840. doi: 10.1126/science.287.5459.1837. [DOI] [PubMed] [Google Scholar]
- Kline M.P., Morimoto R.I. Repression of the heat shock factor 1 transcriptional activation domain is modulated by constitutive phosphorylation. Mol. Cell. Biol. 1997;17:2107–2115. doi: 10.1128/mcb.17.4.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf U., Newton E.M., Kyriakis J., Kingston R.E. Repression of human heat shock factor 1 activity at control temperature by phosphorylation. Genes & Dev. 1996;10:2782–2793. doi: 10.1101/gad.10.21.2782. [DOI] [PubMed] [Google Scholar]
- Krobitsch S., Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. 2000;97:1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamitina T., Huang C.G., Strange K. Genome-wide RNAi screening identifies protein damage as a regulator of osmoprotective gene expression. Proc. Natl. Acad. Sci. 2006;103:12173–12178. doi: 10.1073/pnas.0602987103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer T., Lu C., Echols H., Flanagan J., Hayer M.K., Hartl F.U. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- Lee B.S., Chen J., Angelidis C., Jurivich D.A., Morimoto R.I. Pharmacological modulation of heat shock factor 1 by antiinflammatory drugs results in protection against stress-induced cellular damage. Proc. Natl. Acad. Sci. 1995;92:7207–7211. doi: 10.1073/pnas.92.16.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Dorman J.B., Rodan A., Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Link C.D. Expression of human β-amyloid peptide in transgenic Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J.T., Mason P., Peng J., Price D.H., Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes & Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Marcuccilli C.J., Mathur S.K., Morimoto R.I., Miller R.J. Regulatory differences in the stress response of hippocampal neurons and glial cells after heat shock. J. Neurosci. 1996;16:478–485. doi: 10.1523/JNEUROSCI.16-02-00478.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., Mayhew M., Langer T., Hartl F.U. The reaaction cycle of GroEL and GroES in chaperonin-assisted protein folding. Nature. 1993;366:228–233. doi: 10.1038/366228a0. [DOI] [PubMed] [Google Scholar]
- Marsh J.L., Walker H., Theisen H., Zhu Y.Z., Fielder T., Purcell J., Thompson L.M. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- Mathew A., Mathur S.K., Morimoto R.I. Heat shock response and protein degradation: Regulation of HSF2 by the ubiquitin–proteasome pathway. Mol. Cell. Biol. 1998;18:5091–5098. doi: 10.1128/mcb.18.9.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur S.K., Sistonen L., Brown I.R., Murphy S.P., Sarge K.D., Morimoto R.I. Deficient induction of human hsp70 heat shock gene transcription in Y79 retinoblastoma cells despite activation of heat shock factor 1. Proc. Natl. Acad. Sci. 1994;91:8695–8699. doi: 10.1073/pnas.91.18.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty J.S., Buchberger A., Reinstein J., Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- McMillan D.R., Xiao X., Shao L., Graves K., Benjamin I.J. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 1998;273:7523–7528. doi: 10.1074/jbc.273.13.7523. [DOI] [PubMed] [Google Scholar]
- Morimoto R.I. Regulation of the heat shock transcriptional response: Cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes & Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- Morley J.F., Morimoto R.I. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol. Biol. Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley J.F., Brignull H.R., Weyers J.J., Morimoto R.I. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2002;99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.Z., Tissenbaum H.A., Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- Ni Z., Schwartz B.E., Werner J., Suarez J.R., Lis J.T. Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol. Cell. 2004;13:55–65. doi: 10.1016/s1097-2765(03)00526-4. [DOI] [PubMed] [Google Scholar]
- Nollen E.A., Morimoto R.I. Chaperoning signaling pathways: Molecular chaperones as stress-sensing ‘heat shock’ proteins. J. Cell Sci. 2002;115:2809–2816. doi: 10.1242/jcs.115.14.2809. [DOI] [PubMed] [Google Scholar]
- Nollen E.A., Garcia S.M., van Haaften G., Kim S., Chavez A., Morimoto R.I., Plasterk R.H. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc. Natl. Acad. Sci. 2004;101:6403–6408. doi: 10.1073/pnas.0307697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nylandsted J., Rohde M., Brand K., Bastholm L., Elling F., Jaattela M. Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. 2000;97:7871–7876. doi: 10.1073/pnas.97.14.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann W.M., Sondermann H., Russo A.A., Pavletich N.P., Hartl F.U. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.M., Werner J., Kim J.M., Lis J.T., Kim Y.J. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- Parker J.A., Connolly J.B., Wellington C., Hayden M., Dausset J., Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc. Natl. Acad. Sci. 2001;98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D.A., Kowal A.S., Singer M.A., Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Perutz M.F. Glutamine repeats and neurodegenerative diseases: Molecular aspects. Trends Biochem. Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- Picard D., Khursheed B., Garabedian M.J., Fortin M.G., Lindquist S., Yamamoto K.R. Reduced levels of hsp90 compromise steroid receptor action in vivo. Nature. 1990;348:166–168. doi: 10.1038/348166a0. [DOI] [PubMed] [Google Scholar]
- Pirkkala L., Nykanen P., Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- Pratt W.B., Toft D.O. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 2003;228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ross C.A. Polyglutamine pathogenesis: Emergence of unifying mechanisms for Huntington’s disease and related disorders. Neuron. 2002;35:819–822. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- Rossi A., Elia G., Santoro M.G. Activation of the heat shock factor 1 by serine protease inhibitors. An effect associated with nuclear factor-κB inhibition. J. Biol. Chem. 1998;273:16446–16452. doi: 10.1074/jbc.273.26.16446. [DOI] [PubMed] [Google Scholar]
- Rutherford S.L., Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Satyal S.H., Schmidt E., Kitagawa K., Sondheimer N., Lindquist S., Kramer J.M., Morimoto R.I. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A., Werner J., Andrulis E.D., Nakayama T., Hirose S., Reinberg D., Lis J.T. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003;301:1094–1096. doi: 10.1126/science.1085712. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar A.G., Kowal A.S., Sawicki G.J., Moslehi J.J., Serpell L., Arnsdorf M.F., Lindquist S.L. Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- Shamovsky I., Gershon D. Novel regulatory factors of HSF1 activation: Facts and perspectives regarding their involvement in the age-associated attenuation of the heat shock response. Mech. Ageing Dev. 2004;125:767–775. doi: 10.1016/j.mad.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Sharp S., Workman P. Inhibitors of the HSP90 molecular chaperone: Current status. Adv. Cancer Res. 2006;95:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- Shi Y., Mosser D.D., Morimoto R.I. Molecular chaperones as HSF1-specific transcriptional repressors. Genes & Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L., Sarge K.D., Phillips B., Abravaya K., Morimoto R.I. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol. Cell. Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Takeda M., Morimoto R.I. Bag1-Hsp70 mediates a physiological stress signalling pathway that regulates Raf-1/ERK and cell growth. Nat. Cell Biol. 2001;3:276–282. doi: 10.1038/35060068. [DOI] [PubMed] [Google Scholar]
- Sorger P.K., Nelson H.C. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- Sorger P.K., Pelham H.R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988;54:855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Sprang G.K., Brown I.R. Selective induction of a heat shock gene in fiber tracts and cerebral neurons of the rabbit brain detected by in situ hybridization. Brain Res. 1987;427:89–93. doi: 10.1016/0169-328x(87)90049-0. [DOI] [PubMed] [Google Scholar]
- Stadtman E.R. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]
- Tang Y.C., Chang H.C., Hayer-Hartl M., Hartl F.U. SnapShot: Molecular chaperones, Part II. Cell. 2007;128:412. doi: 10.1016/j.cell.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Tessier P.M., Lindquist S. Prion recognition elements govern nucleation, strain specificity and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodorakis N.G., Morimoto R.I. Posttranscriptional regulation of hsp70 expression in human cells: Effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol. Cell. Biol. 1987;7:4357–4368. doi: 10.1128/mcb.7.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyama B.H., Kelly M.J., Gross J.D., Weissman J.S. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- Trinklein N.D., Murray J.I., Hartman S.J., Botstein D., Myers R.M. The role of heat shock transcription factor 1 in the genome-wide regulation of the mammalian heat shock response. Mol. Biol. Cell. 2004;15:1254–1261. doi: 10.1091/mbc.E03-10-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott A., West J.D., Klaic L., Westerheide S.D., Silverman R.B., Morimoto R.I., Morano K.A. Activation of heat shock and antioxidant responses by the natural product celastrol: Transcriptional signatures of a thiol-targeted molecule. Mol Biol Cell. 2008;19:1104–1112. doi: 10.1091/mbc.E07-10-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H.L., Lindquist S.L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Venkatraman P., Wetzel R., Tanaka M., Nukina N., Goldberg A.L. Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol. Cell. 2004;14:95–104. doi: 10.1016/s1097-2765(04)00151-0. [DOI] [PubMed] [Google Scholar]
- Wang X., Venable J., LaPointe P., Hutt D.M., Koulov A.V., Coppinger J., Gurkan C., Kellner W., Matteson J., Plutner H., et al. Hsp90 cochaperone Aha1 downregulation rescues misfolding of CFTR in cystic fibrosis. Cell. 2006;127:803–815. doi: 10.1016/j.cell.2006.09.043. [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Paulson H.L., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- Warrick J.M., Chan H.Y., Gray-Board G.L., Chai Y., Paulson H.L., Bonini N.M. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat. Genet. 1999;23:425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- Weibezahn J., Tessarz P., Schlieker C., Zahn R., Maglica Z., Lee S., Zentgraf H., Weber-Ban E.U., Dougan D.A., Tsai F.T., et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Weissman J.S., Hohl C.M., Kovalenko O., Kashi Y., Chen S., Braig K., Saibil H.R., Fenton W.A., Horwich A.L. Mechanism of GroEL action: Productive release of polypeptide from a sequestered position uder GroES. Cell. 1995;83:577–587. doi: 10.1016/0092-8674(95)90098-5. [DOI] [PubMed] [Google Scholar]
- Welch W.J., Suhan J.P. Cellular and biochemical events in mammalian cells during and after recovery from physiological stress. J. Cell Biol. 1986;103:2035–2052. doi: 10.1083/jcb.103.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerheide S.D., Morimoto R.I. Heat shock response modulators as therapeutic tools for diseases of protein conformation. J. Biol. Chem. 2005;280:33097–33100. doi: 10.1074/jbc.R500010200. [DOI] [PubMed] [Google Scholar]
- Westerheide S.D., Bosman J.D., Mbadugha B.N., Kawahara T.L., Matsumoto G., Kim S., Gu W., Devlin J.P., Silverman R.B., Morimoto R.I. Celastrols as inducers of the heat shock response and cytoprotection. J. Biol. Chem. 2004;279:56053–56060. doi: 10.1074/jbc.M409267200. [DOI] [PubMed] [Google Scholar]
- Westerheide S.D., Kawahara T.L., Orton K., Morimoto R.I. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J Biol Chem. 2006 doi: 10.1074/jbc.M512044200. [DOI] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., Neckers L.M. Inhibition of heat shock protein HSP90–pp60v–src heteroprotein complex formation by benzoquinone ansamycins: Essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. 1994;91:8324–8328. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Bagatell R., Falsey R. The stress response: Implications for the clinical development of hsp90 inhibitors. Curr. Cancer Drug Targets. 2003;3:349–358. doi: 10.2174/1568009033481787. [DOI] [PubMed] [Google Scholar]
- Williams G.T., Morimoto R.I. Maximal stress-induced transcription from the human HSP70 promoter requires interactions with the basal promoter elements independent of rotational alignment. Mol. Cell. Biol. 1990;10:3125–3136. doi: 10.1128/mcb.10.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Heat shock transcription factors: Structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- Xiao H., Perisic O., Lis J.T. Cooperative binding of Drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–593. doi: 10.1016/0092-8674(91)90242-q. [DOI] [PubMed] [Google Scholar]
- Yao J., Munson K.M., Webb W.W., Lis J.T. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Zoghbi H.Y., Orr H.T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- Zou J., Guo Y., Guettouche T., Smith D.F., Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94:471–480. doi: 10.1016/s0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]