Abstract
Rho GTPases are believed to make important contributions to the development and progression of human cancer, but direct evidence in the form of somatic mutations analogous to those affecting Ras has been lacking. A recent study in Genes & Development by Xue and colleagues (1439–1444) now provides in vivo evidence that DLC1, a negative regulator of Rho, is a tumor suppressor gene deleted almost as frequently as p53 in common cancers such as breast, colon, and lung.
Keywords: DLC1, HCC, RNAi, RhoA, mouse model
Cancer is a complex set of diseases arising from combinations of genetic and epigenetic events, including base mutations, chromosomal rearrangements, DNA methylation, and chromatin modification. Genetic changes were first seen cytologically and revealed gross chromosomal abnormalities, such as translocations, deletions, amplifications (of entire chromosomes or parts of chromosomes), and inversions. Subsequently, DNA sequencing of candidate genes and then whole genomes has uncovered large numbers of more subtle genetic alterations. The recent and continuing successes of sequencing and other nonfunctional based genomic approaches have raised new problems in how to determine which changes have significance for tumor development. This is not a trivial problem and will require combinations of cell-based assays, in vivo animal models, and ultimately clinical intervention.
The identification of the Ras oncogene was the first major triumph of the early application of molecular biology to the cancer problem (Malumbres and Barbacid 2003). Although originally identified as a viral oncogene in a rodent sarcoma-inducing retrovirus, it was the seminal work of the Weinberg and Cooper laboratories in 1981 (Krontiris and Cooper 1981; Shih et al. 1981), using DNA transfection assays of human tumor DNA into immortalized mouse fibroblasts, that led to the identification of Ras as a true human oncogene. Several groups went on to show that any one of the three Ras genes (HRAS, KRAS, and NRAS) could be converted into a human oncogene by a single base mutation leading to a single amino acid substitution in the encoded Ras protein. Ras mutations are found in ∼30% of most, though not all, cancer types and it remains the most frequently mutated dominant oncogene so far identified (Bos 1989). We now know much about the consequences of those amino acid substitutions and the cellular and physiological importance of Ras in controlling proliferation and differentiation. Ras is an example of a regulatory GTPase that cycles between active (GTP-bound) and inactive (GDP-bound) conformations to control biochemical pathways and processes. These molecular switches are activated by guanine nucleotide exchange factors (GEFs), which catalyze exchange of GDP for GTP, and are inactivated by GTPase-activating proteins (GAPs), which promote the otherwise slow, intrinsic GTPase activity of the proteins (Fig. 1). The amino acid substitutions identified in Ras in human cancers are found at codons 12, 61, and to a lesser extent 13, and the common consequence of these changes is to prevent GAP-mediated stimulation of GTP hydrolysis leading to permanent activation of the switch (Trahey and McCormick 1987). Inspection of Figure 1 suggests possible alternative ways in which this molecular switch could be inappropriately activated. For example, activating mutations in one of the nine RasGEF genes or inactivation of one of the eight RasGAP genes could lead to hyperactivation of the switch. To date, no such mutations have been reported in GEF genes in human cancers, but one of the GAPs, neurofibromin, is encoded by the NF1 tumor suppressor gene. Patients with neurofibromatosis type I inherit only one functional NF1 gene and are then predisposed to cancer through complete loss of NF1. In addition, mutational activation of components of downstream signaling pathways (Fig. 1) could bypass the need for Ras and this is clearly the case with somatic mutations in BRAF (which encodes a Ras effector), found most frequently in malignant melanomas (>50%), but also in thyroid, colorectal, and ovarian cancer (Davies et al. 2002; Wellbrock et al. 2004).
Figure 1.
The Ras GTP/GDP cycle. Ras GTPases are molecular switches and the GDP/GTP cycle is controlled by GEFs and GAPs. The output of the switch is through the interaction of Ras.GTP with effector proteins.
Interestingly, in addition to the somatic mutations found in Ras pathways in human cancer, heritable germline mutations occur in several developmental disorders, though these rarely lead to cancer (Bentires-Alj et al. 2006). The disorders share phenotypic features and are referred to as “Neuro-Cardio-Facial-Cutaneous” syndromes and include Noonan syndrome, Leopard syndrome, Cardio-facio-cutaneous syndrome (CFC), and Costello syndrome. In addition to Ki-Ras (Noonan and CFC), Ha-Ras (Costello), and B-Raf (CFC) mutations, Sos1 (a Ras GEF) and MEK (downstream from B-Raf in MAP kinase cascade) mutations are found in Noonan and CFC, respectively, but these have not been found in cancer (Rajalingam et al. 2007; Schubbert et al. 2007).
Following the discovery of Ras, ∼150 Ras-like proteins were identified in mammals, many of which are highly conserved throughout eukaryotes, and these constitute the Ras superfamily of small GTPases (Colicelli 2004). Members constituting one group within this superfamily are very closely related to Ras itself, but there has been little evidence that genetic or epigenetic changes leading to their disregulated activity have been selected for in human cancers. That is not to say that their activities may not be important—it is likely, for example, that the Ras pathway is important even in cancers that do not harbor mutations in Ras itself. Rho GTPases form a second group within the superfamily and the best studied of these, Rho, Rac, and Cdc42, not only play key roles in cell proliferation, but also other aspects of cell biology relevant to the disease, such as morphogenesis and migration. There has, therefore, been much effort to examine their contribution to human cancer, but until now no clear-cut case has been identified where a genetic alteration has been selected for in a Rho pathway component. However, work that began some 10 years ago culminated in a study published previouslyin Genes & Development that provides convincing evidence that DLC1, a gene encoding a Rho GAP, is a true tumor suppressor that is inactivated in a large number of human cancers (Xue et al. 2008).
Rho GTPases and tumorigenesis
Rho proteins are, like Ras, controlled by GEFs and GAPs, though an additional class of player, Rho GDI (GDP dissociation inhibitor), is used to regulate nucleotide-dependent membrane association (Sahai and Marshall 2002). Twenty-two Rho family GTPases have been uncovered in the human genome, though there is much additional complexity to these switches mediated through 82 GEFs, 67 GAPs, and three GDIs. Yet more striking has been the (biochemical) identification of ∼100 putative targets for Rho, Rac, and Cdc42 (Etienne-Manneville and Hall 2002; Jaffe and Hall 2005). Clearly each Rho family GTPase can be regulated in multiple ways and once activated can potentially engage multiple biochemical pathways. The best understood of these are linked to the assembly and organization of the actin cytoskeleton, but others range from kinases acting on phospholipids to oxidases generating reactive oxygen. How specificity and selectivity are achieved both upstream of and downstream from these switches is not well understood.
Given the large number and functional diversity of target proteins, it is not surprising that Rho family members are capable of influencing numerous aspects of cell biology. These have been reviewed elsewhere, but encompass many that are pertinent to cancer, such as the cell cycle (both G1 progression and mitosis), adhesion (cell–cell and cell–matrix), morphogenesis, polarity, and migration (Etienne-Manneville and Hall 2002; Jaffe and Hall 2005). Driven by this knowledge, there has been much effort to assess their importance in cancer progression. So far there are no examples of amino acid substitutions in members of the Rho family similar to those found in Ras, but there are many reports of GTPase overexpression in tumors, including the three isoforms of Rho (A, B, C), the three isoforms of Rac (1, 2, 3), RhoG, Rho H, Cdc42, and Rnd3/RhoE (Preudhomme et al. 2000; Pasqualucci et al. 2001; Ellenbroek and Collard 2007). RhoC has been singled out as playing a significant role in invasion, beginning with a paper showing that overexpression of RhoC induces metastatic activity in nonmetastatic tumor cells, through to a more recent report of microRNA-10b-dependent up-regulation of RhoC specifically in metastatic breast cancer cells (Clark et al. 2000; Ma et al. 2007). Why RhoC, and not RhoA, should be specifically linked to metastasis is totally unclear; the two proteins are almost identical in sequence and they interact with the same proteins.
Just like Ras, alternative mechanisms for activating Rho GTPases can be envisaged, such as constitutive activation of a GEF or a deletion of a GAP. Many members of the Rho GEF family have been shown to be capable of acting as oncogenes in experimental assays when rendered constitutively active, often through deletion of N-terminal regulatory sequences. Furthermore, mice lacking Tiam-1 (a Rac GEF) are significantly more resistant to skin cancer induced by chemical carcinogens—a tumor model that involves mutational activation of Ras (Malliri et al. 2002). However, with the possible exception of a translocation involving LARG (a Rho GEF) found in acute myeloid leukaemia, genetic activation of a GEF does not appear to occur in human cancer. Some GEFs, such as β-PIX and Vav1 are overexpressed in cancer, which might lead to increased activation of Rho GTPases, but these data remain circumstantial (Ahn et al. 2003; Fernandez-Zapico et al. 2005). However, the best evidence to support a role for Rho pathways in human cancer has come from the analysis of one particular GAP, DLC1.
DLC1—a bona fide tumor suppressor?
The story starts in 1998 when DLC1 (Deleted in Liver Cancer 1) was identified in the Popescu laboratory during a representative difference analysis (RDA) screen, a PCR-based subtractive hybridization technique, using samples taken from a primary hepatocellular carcinoma (HCC) and adjacent noncancerous tissue (Yuan et al. 1998). It was noted then that the gene is expressed ubiquitously in normal tissue, yet is absent in ∼30% of HCC cell lines. DLC1 is localized at chromosome 8p21.3-22, a region that shows loss of hererozygosity (LOH) or heterozygous deletions in a large number and a wide variety of human cancers, including prostate, colon, breast, ovarian, liver, lung, bladder, and head and neck (Yuan et al. 1998; Arbieva et al. 2000). Furthermore, chromosome transfer experiments had provided functional evidence for a tumor suppressor gene on the short arm of chromosome 8 (Gustafson et al. 1996; Tanaka et al. 1996). Studies performed over the last 10 years have yielded persuasive evidence that DLC1 could be that gene. Thus, DLC1 mRNA is down-regulated in many human cancers and this can be accounted for either by DLC1 LOH or, more frequently, by epigenetic silencing of the gene through hypermethylation or possibly histone hypoacetylation, (Durkin et al. 2007). A recent study by Xue et al. (2008) in a previous issue of Genes & Development describes an extensive representative oligonucleotide microarray analysis (ROMA) showing heterozygous deletion of DLC1 in ∼50% of liver, 50% of breast, 70% of colon, and 50% of lung cancers. Since mutations in the coding sequence of DLC1 appear to be very rare, the suggestion is that the other allele could be silenced epigenetically. This frequency of allele loss is astonishingly high and almost as common as p53 deletions in these cancers.
Direct functional evidence for DLC1 tumor suppressor activity has been more difficult to obtain. Numerous studies involving reintroduction of DLC1 into cancer cell lines that do not express DLC1 protein have been undertaken. Expression of DLC1 in HCC or lung cancer cells, for example, inhibits anchorage-independent growth in soft agar and prevents tumor growth in mouse xenografts (Yuan et al. 2004; Zhou et al. 2004). While these experiments are consistent with tumor suppressor activity, they do not prove that loss of DLC1 is a causative step in the development of the original tumor. DLC1 expression could simply be inhibiting an essential pathway required for cell proliferation. So far mouse knockouts have not been informative—DLC1-null mice are embryonic lethal, while heterozygotes have no phenotype—and tissue-specific knockouts will be necessary to make progress in this area (Durkin et al. 2005).
The study by Xue et al. (2008) has now provided direct evidence that DLC1 can act as a tumor suppressor gene in vivo. The authors make use of a novel mouse model of liver cancer that involves the isolation and ex vivo genetic manipulation of mouse embryonic liver progenitor cells, followed by transplantation into the livers of recipient mice (Zender et al. 2006). This approach allows the use of shRNAs to attenuate protein expression and thereby study the consequences of tumor suppressor gene loss on liver carcinogenesis (Xue et al. 2007). The authors had reported previously that overexpression of c-Myc, or oncogenic Ras in liver progenitors obtained from p53-null mice induce tumors when cells are reintroduced into the livers of wild-type mice, though in the case of c-Myc this is a relatively weak effect (Zender et al. 2006). To examine DLC1 in this context, p53-deficient liver progenitor cells were cotransduced with two retroviruses, one expressing c-Myc and the other expressing a DLC1-directed shRNA. When these genetically modified liver progenitors were transplanted into the livers of syngeneic recipient mice, DLCl loss was found to cooperate with c-Myc to promote the formation of liver tumors. The effect of DLC1 suppression was not as dramatic as expression of oncogenic Ras, which produced tumors more rapidly and in the absence of c-Myc. It would have been interesting to know whether c-Myc expression is necessary to observe a DLC1 shRNA-induced phenotype, but this was not reported. Nevertheless, the experiment leaves little doubt that DLC1 can act as a tumor suppressor gene and taken together with the genomic analyses, the case becomes very strong that this is a human tumor suppressor that is not only Deleted in Liver Cancer, but also Deleted in Lots of Cancers.
DLC1 is a Rho GAP
When DLC1 cDNA was first isolated, it was clear that the encoded 1091-amino-acid protein harbors a Rho GAP domain (Yuan et al. 1998). We now know that it, together with its close family relatives DLC2 and DLC3, is part of a family of 67 Rho GAPs encoded in the human genome (Tcherkezian and Lamarche-Vane 2007). Rho GAPs stimulate the otherwise low intrinsic GTPase activity of Rho family members and thus negatively regulate their activity (Diekmann et al. 1991). Full-length DLC1 and the isolated GAP domain show strong activity toward the three isoforms of Rho (i.e., RhoA, RhoB, and RhoC), weak activity on Cdc42, and no activity on Rac in in vitro assays (Durkin et al. 2007; Healy et al. 2008). The structure and mechanism of action of GAP domains are well understood and catalysis is mediated by an “arginine finger” (residue 718 in DLC1) donated by GAP, promoting hydrolysis of the γ-phosphate of GTP bound to the GTPase (Bos et al. 2007). Mutational analysis of DLC1 suggests that GAP activity is required for tumor suppressor activity, though additional regions of DLC1 likely play a role, including a START domain (located at the extreme C terminus of the protein and thought to interact with lipid) and a region located between residues 292–648 (Wong et al. 2005; Liao et al. 2007; Healy et al. 2008). One study has suggested that DLC1 has residual tumor suppressor activity in the absence of functional GAP activity, though it is hard to know what to make of this observation, since it seems to be at odds with most other studies (Healy et al. 2008). Overall, these experiments strongly suggest that hyperactivation of Rho is the key feature of DLC1’s role as a tumor suppressor. The fact that DLC1 is one of 67 Rho GAPs, many of which can act on Rho and are ubiquitously expressed, suggests that each GAP may make a unique contribution to regulating Rho activity. This could reflect their different spatial locations or, given the large number of potential downstream targets of each GTPase, their recruitment to specific signaling outputs, or both.
Proving that DLC1 tumor suppressor activity is mediated by Rho activation is not as straightforward as it sounds; Rho regulates many basic aspects of cell biology—for example, cytokinesis—and simply eliminating Rho activity would likely render cells inviable (Narumiya and Yasuda 2006). In their recent study published in Genes & Development, Xue et al. (2008) provide several lines of evidence to support the conclusion that Rho activation is key. First, constitutively active Rho (RhoAV14) promotes tumor formation with a similar efficiency and pathology to DLC1 shRNA when expressed together with c-Myc in p53−/− liver progenitor cells. Second, attenuation of RhoA expression (using shRNA) in two independent murine hematomas lacking DLC1 expression, suppresses their in vivo growth. While there are clear caveats to interpreting these experiments (RhoA depletion also had a significant, though admittedly lower, inhibitory effect on murine hepatoma cell lines that express DLC1), the overwhelming evidence points to the activation of Rho as being the key consequence of loss of DLC1’s tumor suppressor activity.
Rho GTPases can trigger numerous downstream signaling pathways by interacting with distinct effectors—to date, ∼20 such target proteins have been reported that specifically interact with Rho (Etienne-Manneville and Hall 2002). One of the best-characterized is Rho kinase (ROCK), which regulates myosin II and actin filament contractility, through its ability to phosphorylate and inactivate myosin light chain phosphatase (Fukata et al. 2001). Rho kinase is involved in many aspects of normal cell biology, such as cell cycle, morphogenesis, and migration, and in addition has been shown to participate in the proliferation, invasion, and metastasis of cancer cells (Etienne-Manneville and Hall 2002; Sahai and Marshall 2002; Narumiya and Yasuda 2006). In the final part of their study, Xue et al. (2008) show that two small molecule Rho kinase inhibitors, Y-27632 and to a lesser extent Fasudil, inhibit in vitro colony formation of p53−/− liver progenitor cells expressing c-Myc and DCL1 shRNA. It should be noted, however, that both Y-27632 and Fasudil inhibit PRK/PKN and citron kinase, two other kinases activated by Rho, so the result is not entirely conclusive (Ishizaki et al. 2000).
DLC1 function
The cumulating data point toward loss of DLC1 causing disregulation of a Rho pathway that can promote some aspect of cancer progression. But what is special about DLC1, and why aren’t other Rho GAPs also deleted in cancer? One possibility is that they are, and in fact this has been suggested for DLC2, DLC3, p190RhoGAP, and GRAF (interestingly all GAPs for Rho), although the evidence is still relatively indirect (Borkhardt et al. 2000; Wolf et al. 2003; Durkin et al. 2007). A more intriguing, though not mutually exclusive, explanation is that DLC1 is associated with a particular Rho pathway and it is the disregulation of that specific pathway that contributes to cancer. Work in S. cerevisiae has provided perhaps the clearest paradigm for this idea, with three GAPs, SAC7, BEM2, and BAG7, differentially regulating at least two different RHO1-regulated biological pathways (Schmidt et al. 2002). In flies, worms, and mammalian cells, one GAP, MgcRacGAP (a GAP for Rho despite its name), is specifically linked to Rho’s role in the assembly of the contractile ring during cytokinesis, while p190-B-RhoGAP regulates a Rho/Rho kinase pathway in mouse mesenchymal progenitor cells that is specifically linked to the inhibition of adipogenesis and the promotion of myogenesis (Sordella et al. 2003; Narumiya and Yasuda 2006). It is of great interest, therefore, to identify the normal physiological function of DLC1.
Conclusions about function that rely on the reintroduction of DLC1 into cells suffer from the problem that overexpression of any Rho GAP could potentially lead to down-regulation of Rho activity, particularly if the GAP domain alone is introduced (Tcherkezian and Lamarche-Vane 2007). Nevertheless, full-length DLC1 has been reported to lead to disassembly of the actin cytoskeleton and integrin adhesions complexes when expressed in NIH 3T3 fibroblasts, as well as in cancer cell lines that lack endogenous DLC1 leading to the suggestion that this GAP is specifically involved in actin cytoskeletal regulation (Yuan et al. 2004; Kim et al. 2007; Guan et al. 2008; Liao and Lo 2008). The localization of DLC1 varies between cell lines; in some it appears as diffusely cytoplasmic, while in others it has been reported at focal adhesions (Durkin et al. 2007; Liao et al. 2007). The N-terminal region of DLC1 contains a putative focal adhesion targeting sequence that binds to the SH2 domain of focal adhesion proteins, notably tensin (Durkin et al. 2007). The significance of this interaction is not clear; however, since a mutant version of DLC1 that no longer binds to tensin is unable to suppress cancer cell proliferation, it is argued that it must be important (Lo 2004; Liao et al. 2007; Qian et al. 2007).
Embryonic fibroblasts can be obtained from DLC1−/− mice and these display alterations in the organization of actin filaments and focal adhesion (Durkin et al. 2005). Confusingly, however, these knockout cells have fewer stress fibers and focal adhesions—the opposite of what would have been predicted for the loss of a GAP that regulates Rho. In fact the cytoskeletal and adhesion complex changes seen in DLC−/− fibroblasts appear to be more in keeping with Rac activation. Unfortunately the authors did not examine the levels of either Rho.GTP or Rac.GTP in these cells, which might have provided some insight into this unexpected result. In the absence of tissue-specific mouse knockouts, we must look to work in Drosophila on RhoGAP88C, the fly ortholog of DCL1, to provide some in vivo physiological data. Mutations in RhoGAP88C were first identified as crossveinless-c and result in defects in tissue morphogenesis during development (Denholm et al. 2005). Closer examination suggests that this GAP regulates tubulogenesis and convergent extension, two processes driven by reorganization of the actin cytoskeleton. An additional and provocative observation to emerge from this study is that RhoGAP88C acts through Rho in some tissues, but it acts through Rac and not Rho in others. The in vitro biochemical activity of this GAP has not been determined and so it is possible that it shows a different specificity from its mammalian counterpart. Otherwise, tissue-specific modification of its catalytic activity would need to be invoked, rendering the in vitro assays essentially useless for predicting specificity. Two subsequent studies have concluded that RhoGAP88C is localized basolaterally in epithelial cells and serves to restrict Rho activity to the apical surface and thereby generate morphogenetic tissue remodeling through polarized activation of myosin II (Brodu and Casanova 2006; Simoes et al. 2006).
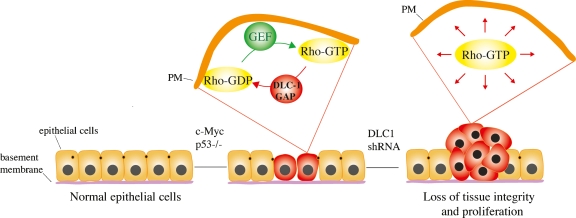
Taken together, a picture emerges of spatially localized DLC1 acting to control Rho activity so as to promote changes in the actin cytoskeleton during cell morphogenesis. The disruption of this pathway might be expected to lead to tissue disorganization during differentiation programs, which could promote inappropriate cell proliferation (Fig. 2).
Figure 2.
DLC1 is a tumor suppressor. Loss of DLC1 leads to deregulated and/or delocalized activation of Rho. This may disrupt tissue morphogenesis leading to inappropriate proliferation. (PM) Plasma membrane.
Conclusion—DLC1, can we do anything about it?
Directed therapeutic intervention depends on a deep understanding of the relevant signaling pathways through which DLC1 loss is manifest. It is a sobering thought that the signaling pathways downstream from Ras responsible for human cancer are still debated some 25 years after its discovery as a human oncogene and it would be optimistic to believe that identifying Rho pathways will be any easier. Inhibiting the GTPase itself, whether Ras or Rho, is challenging. One of the most promising potential targets for Ras inactivation has been farnesyltransferase (FT), the enzyme required for carboxy-terminal, post-translational modification by a farnesyl lipid (Wright and Philips 2006). FT inhibitors are currently in clinical trials, though the data reported so far are not encouraging. Inhibiting Rho using a similar strategy seems less attractive, since it uses a geranylgeranyltransferase to add a geranylgeranyl group; a much more widespread modification than farnesyl addition. Two other processing enzymes that act on both Ras and Rho, a carboxyl-protease and an isoprenylcysteine carboxyl methyltransferase, are being considered as Ras targets, but in tissue culture at least these seem not to be essential for Rho function (Michaelson et al. 2005). Another possibility that is distinctive to DLC1 might be to attack the epigenetic mechanisms that appear to be commonly used to silence this gene in human cancers. Inhibitors of DNA methyltransferase and histone deacetylase (HDAC) have already been shown to induce the restoration of DLC1 expression in cancer cells, making Zebularine, a new and highly effective DNA demethylating agent, as well as HDAC inhibitors attractive therapeutic approaches (Guan et al. 2006; Neureiter et al. 2007; Seng et al. 2007; Xu et al. 2007). Finally, if it turns out that Rho kinase mediates the key signaling pathway downstream from DLC1 loss, then there is already a huge effort underway to develop small molecule inhibitors of this protein. Rho kinase has been implicated in various forms of cardiovascular disease—such as pulmonary hypertension, myocardial hypertrophy, and atherosclerosis—and in fact one compound, Fasudil, is already being used clinically in Japan for cerebral ischemia (Rikitake and Liao 2005; Tawara and Shimokawa 2007). With over a dozen pharmaceutical companies reportedly working on this problem, and if the work from Xue et al. (2008) implicating Rho kinase downstream from DLC1 turns out to be correct, those companies may end up with a blockbuster!
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1691408.
References
- Ahn S.J., Chung K.W., Lee R.A., Park I.A., Lee S.H., Park D.E., Noh D.Y. Overexpression of βPix-a in human breast cancer tissues. Cancer Lett. 2003;193:99–107. doi: 10.1016/s0304-3835(03)00004-1. [DOI] [PubMed] [Google Scholar]
- Arbieva Z.H., Banerjee K., Kim S.Y., Edassery S.L., Maniatis V.S., Horrigan S.K., Westbrook C.A. High-resolution physical map and transcript identification of a prostate cancer deletion interval on 8p22. Genome Res. 2000;10:244–257. doi: 10.1101/gr.10.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentires-Alj M., Kontaridis M.I., Neel B.G. Stops along the RAS pathway in human genetic disease. Nat. Med. 2006;12:283–285. doi: 10.1038/nm0306-283. [DOI] [PubMed] [Google Scholar]
- Borkhardt A., Bojesen S., Haas O.A., Fuchs U., Bartelheimer D., Loncarevic I.F., Bohle R.M., Harbott J., Repp R., Jaeger U., et al. The human GRAF gene is fused to MLL in a unique t(5;11)(q31;q23) and both alleles are disrupted in three cases of myelodysplastic syndrome/acute myeloid leukemia with a deletion 5q. Proc. Natl. Acad. Sci. 2000;97:9168–9173. doi: 10.1073/pnas.150079597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos J.L. ras oncogenes in human cancer: A review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- Bos J.L., Rehmann H., Wittinghofer A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Brodu V., Casanova J. The RhoGAP crossveinless-c links trachealess and EGFR signaling to cell shape remodeling in Drosophila tracheal invagination. Genes & Dev. 2006;20:1817–1828. doi: 10.1101/gad.375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark E.A., Golub T.R., Lander E.S., Hynes R.O. Genomic analysis of metastasis reveals an essential role for RhoC. Nature. 2000;406:532–535. doi: 10.1038/35020106. [DOI] [PubMed] [Google Scholar]
- Colicelli J. Human RAS superfamily proteins and related GTPases. Sci. STKE. 2004;2004:RE13. doi: 10.1126/stke.2502004re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., Teague J., Woffendin H., Garnett M.J., Bottomley W., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- Denholm B., Brown S., Ray R.P., Ruiz-Gomez M., Skaer H., Hombria J.C. crossveinless-c is a RhoGAP required for actin reorganisation during morphogenesis. Development. 2005;132:2389–2400. doi: 10.1242/dev.01829. [DOI] [PubMed] [Google Scholar]
- Diekmann D., Brill S., Garrett M.D., Totty N., Hsuan J., Monfries C., Hall C., Lim L., Hall A. Bcr encodes a GTPase-activating protein for p21rac. Nature. 1991;351:400–402. doi: 10.1038/351400a0. [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Avner M.R., Huh C.G., Yuan B.Z., Thorgeirsson S.S., Popescu N.C. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 2005;579:1191–1196. doi: 10.1016/j.febslet.2004.12.090. [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Yuan B.Z., Zhou X., Zimonjic D.B., Lowy D.R., Thorgeirsson S.S., Popescu N.C. DLC-1: A Rho GTPase-activating protein and tumour suppressor. J. Cell. Mol. Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek S.I., Collard J.G. Rho GTPases: Functions and association with cancer. Clin. Exp. Metastasis. 2007;24:657–672. doi: 10.1007/s10585-007-9119-1. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Fernandez-Zapico M.E., Gonzalez-Paz N.C., Weiss E., Savoy D.N., Molina J.R., Fonseca R., Smyrk T.C., Chari S.T., Urrutia R., Billadeau D.D. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Fukata Y., Amano M., Kaibuchi K. Rho-Rho-kinase pathway in smooth muscle contraction and cytoskeletal reorganization of non-muscle cells. Trends Pharmacol. Sci. 2001;22:32–39. doi: 10.1016/s0165-6147(00)01596-0. [DOI] [PubMed] [Google Scholar]
- Guan M., Zhou X., Soulitzis N., Spandidos D.A., Popescu N.C. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: Potential clinical applications. Clin. Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- Guan M., Tripathi V., Zhou X., Popescu N.C. Adenovirus-mediated restoration of expression of the tumor suppressor gene DLC1 inhibits the proliferation and tumorigenicity of aggressive, androgen-independent human prostate cancer cell lines: Prospects for gene therapy. Cancer Gene Ther. 2008;15:371–381. doi: 10.1038/cgt.2008.13. [DOI] [PubMed] [Google Scholar]
- Gustafson C.E., Wilson P.J., Lukeis R., Baker E., Woollatt E., Annab L., Hawke L., Barrett J.C., Chenevix-Trench G. Functional evidence for a colorectal cancer tumor suppressor gene at chromosome 8p22-23 by monochromosome transfer. Cancer Res. 1996;56:5238–5245. [PubMed] [Google Scholar]
- Healy K.D., Hodgson L., Kim T.Y., Shutes A., Maddileti S., Juliano R.L., Hahn K.M., Harden T.K., Bang Y.J., Der C.J. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 2008;47:326–337. doi: 10.1002/mc.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki T., Uehata M., Tamechika I., Keel J., Nonomura K., Maekawa M., Narumiya S. Pharmacological properties of Y-27632, a specific inhibitor of rho-associated kinases. Mol. Pharmacol. 2000;57:976–983. [PubMed] [Google Scholar]
- Jaffe A.B., Hall A. Rho GTPases: Biochemistry and biology. Annu. Rev. Cell Dev. Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Kim K.Y., Cho H.S., Jung W.H., Kim S.S., Cheon H.G. Phosphatase and tensin homolog deleted on chromosome 10 suppression is an important process in peroxisome proliferator-activated receptor-γ signaling in adipocytes and myotubes. Mol. Pharmacol. 2007;71:1554–1562. doi: 10.1124/mol.106.031948. [DOI] [PubMed] [Google Scholar]
- Krontiris T.G., Cooper G.M. Transforming activity of human tumor DNAs. Proc. Natl. Acad. Sci. 1981;78:1181–1184. doi: 10.1073/pnas.78.2.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.C., Lo S.H. Deleted in liver cancer-1 (DLC-1): A tumor suppressor not just for liver. Int. J. Biochem. Cell Biol. 2008;40:843–847. doi: 10.1016/j.biocel.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.C., Si L., deVere White R.W., Lo S.H. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 2007;176:43–49. doi: 10.1083/jcb.200608015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo S.H. Tensin. Int. J. Biochem. Cell Biol. 2004;36:31–34. doi: 10.1016/s1357-2725(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Malliri A., van der Kammen R.A., Clark K., van der Valk M., Michiels F., Collard J.G. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. RAS oncogenes: The first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- Michaelson D., Ali W., Chiu V.K., Bergo M., Silletti J., Wright L., Young S.G., Philips M. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol. Biol. Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narumiya S., Yasuda S. Rho GTPases in animal cell mitosis. Curr. Opin. Cell Biol. 2006;18:199–205. doi: 10.1016/j.ceb.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Neureiter D., Zopf S., Leu T., Dietze O., Hauser-Kronberger C., Hahn E.G., Herold C., Ocker M. Apoptosis, proliferation and differentiation patterns are influenced by Zebularine and SAHA in pancreatic cancer models. Scand. J. Gastroenterol. 2007;42:103–116. doi: 10.1080/00365520600874198. [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Kuppers R., Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–346. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- Preudhomme C., Roumier C., Hildebrand M.P., Dallery-Prudhomme E., Lantoine D., Lai J.L., Daudignon A., Adenis C., Bauters F., Fenaux P., et al. Nonrandom 4p13 rearrangements of the RhoH/TTF gene, encoding a GTP-binding protein, in non-Hodgkin’s lymphoma and multiple myeloma. Oncogene. 2000;19:2023–2032. doi: 10.1038/sj.onc.1203521. [DOI] [PubMed] [Google Scholar]
- Qian X., Li G., Asmussen H.K., Asnaghi L., Vass W.C., Braverman R., Yamada K.M., Popescu N.C., Papageorge A.G., Lowy D.R. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. 2007;104:9012–9017. doi: 10.1073/pnas.0703033104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajalingam K., Schreck R., Rapp U.R., Albert S. Ras oncogenes and their downstream targets. Biochim. Biophys. Acta. 2007;1773:1177–1195. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Rikitake Y., Liao J.K. ROCKs as therapeutic targets in cardiovascular diseases. Expert Rev. Cardiovasc. Ther. 2005;3:441–451. doi: 10.1586/14779072.3.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E., Marshall C.J. RHO-GTPases and cancer. Nat. Rev. Cancer. 2002;2:133–142. doi: 10.1038/nrc725. [DOI] [PubMed] [Google Scholar]
- Schmidt A., Schmelzle T., Hall M.N. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 2002;45:1433–1441. doi: 10.1046/j.1365-2958.2002.03110.x. [DOI] [PubMed] [Google Scholar]
- Schubbert S., Shannon K., Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- Seng T.J., Low J.S., Li H., Cui Y., Goh H.K., Wong M.L., Srivastava G., Sidransky D., Califano J., Steenbergen R.D., et al. The major 8p22 tumor suppressor DLC1 is frequently silenced by methylation in both endemic and sporadic nasopharyngeal, esophageal, and cervical carcinomas, and inhibits tumor cell colony formation. Oncogene. 2007;26:934–944. doi: 10.1038/sj.onc.1209839. [DOI] [PubMed] [Google Scholar]
- Shih C., Padhy L.C., Murray M., Weinberg R.A. Transforming genes of carcinomas and neuroblastomas introduced into mouse fibroblasts. Nature. 1981;290:261–264. doi: 10.1038/290261a0. [DOI] [PubMed] [Google Scholar]
- Simoes S., Denholm B., Azevedo D., Sotillos S., Martin P., Skaer H., Hombria J.C., Jacinto A. Compartmentalisation of Rho regulators directs cell invagination during tissue morphogenesis. Development. 2006;133:4257–4267. doi: 10.1242/dev.02588. [DOI] [PubMed] [Google Scholar]
- Sordella R., Jiang W., Chen G.C., Curto M., Settleman J. Modulation of Rho GTPase signaling regulates a switch between adipogenesis and myogenesis. Cell. 2003;113:147–158. doi: 10.1016/s0092-8674(03)00271-x. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kikuchi-Yanoshita R., Muraoka M., Konishi M., Oshimura M., Miyaki M. Suppression of tumorigenicity and invasiveness of colon carcinoma cells by introduction of normal chromosome 8p12-pter. Oncogene. 1996;12:405–410. [PubMed] [Google Scholar]
- Tawara S., Shimokawa H. Progress of the study of rho-kinase and future perspective of the inhibitor. Yakugaku Zasshi. 2007;127:501–514. doi: 10.1248/yakushi.127.501. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J., Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol. Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Trahey M., McCormick F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 1987;238:542–545. doi: 10.1126/science.2821624. [DOI] [PubMed] [Google Scholar]
- Wellbrock C., Ogilvie L., Hedley D., Karasarides M., Martin J., Niculescu-Duvaz D., Springer C.J., Marais R. V599EB-RAF is an oncogene in melanocytes. Cancer Res. 2004;64:2338–2342. doi: 10.1158/0008-5472.can-03-3433. [DOI] [PubMed] [Google Scholar]
- Wolf R.M., Draghi N., Liang X., Dai C., Uhrbom L., Eklof C., Westermark B., Holland E.C., Resh M.D. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: A putative tumor suppressor encoded on human chromosome 19q13.3. Genes & Dev. 2003;17:476–487. doi: 10.1101/gad.1040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C.M., Yam J.W., Ching Y.P., Yau T.O., Leung T.H., Jin D.Y., Ng I.O. Rho GTPase-activating protein deleted in liver cancer suppresses cell proliferation and invasion in hepatocellular carcinoma. Cancer Res. 2005;65:8861–8868. doi: 10.1158/0008-5472.CAN-05-1318. [DOI] [PubMed] [Google Scholar]
- Wright L.P., Philips M.R. Thematic review series: Lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- Xu W.S., Parmigiani R.B., Marks P.A. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Krasnitz A., Lucito R., Sordella R., VanAelst L., Cordon-Cardo C., Singer S., Kuehnel F., Wigler M., Powers S., et al. DLC1 is a chromosome 8p tumor suppressor whose loss promotes hepatocellular carcinoma. Genes & Dev. 2008;22:1439–1444. doi: 10.1101/gad.1672608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B.Z., Miller M.J., Keck C.L., Zimonjic D.B., Thorgeirsson S.S., Popescu N.C. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. [PubMed] [Google Scholar]
- Yuan B.Z., Jefferson A.M., Baldwin K.T., Thorgeirsson S.S., Popescu N.C., Reynolds S.H. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Thorgeirsson S.S., Popescu N.C. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]