Abstract
We report results from a study of human and canine mucosal cells, investigated by infrared micro-spectroscopy, and analyzed by methods of multivariate statistics. We demonstrate that the infrared spectra of individual cells are sensitive to the stage of maturation, and that a distinction between healthy and diseased cells will be possible. Since this report is written for an audience not familiar with infrared micro-spectroscopy, a short introduction into this field is presented along with a summary of principal component analysis.
Keywords: Infrared micro-spectroscopy, Oral mucosa (buccal) cell, Canine ectocervical cell, Principal component analysis (PCA), Spectral cytology
1. Introduction
In this paper, we wish to explore the possibilities of using objective, spectroscopic measurements and unsupervised methods of multivariate statistics for the analysis of exfoliated cells. The long-term aim of this research is to develop automatic methods to aid cytology in the identification of abnormal cells, based on infrared microspectroscopy (IR-MSP). IR-MSP monitors the composition of a cell in terms of the major biochemical components by probing the vibrational infrared spectrum. Since IR spectroscopy monitors the inherent signature of the cell’s components, no stain or agents are required to distinguish normal from diseased cells, or to increase the contrast between tissue types.
IR-microspectra are collected from areas as small as ca. 10×10 μm2, and comprising as little as 10−10 g of sample. IR-MSP has been utilized to study a number of normal and pathological tissues; for example, the detection of dysplasia and neo-plasia in cervical tissue [1–3], the distinction and grading of malignant tumors [4–6] and the identification of metastatic cancers in lymph nodes have been reported [7]. In these applications, the anatomical structure of normal and diseased tissues has been reproduced by spectral results interpreted by completely unsupervised methods of data analysis, such as hierarchical cluster analysis (HCA) or artificial neural nets (ANN).
The application of IR-MSP to cytology has seen less progress, although several research groups began to utilize IR spectroscopy to study exfoliated human cervical cells in the early 1990s. However, this sample system was not an easy one to start such a new and pioneering method, since samples of cervical cells are often contaminated by erythrocytes, lymphatic cells such as polymorphonuclear lymphocytes (PMNs), cervical mucous and bacteria [8–10]. Furthermore, even the exfoliated cervical cells themselves are mixtures of squamous and glandular (columnar) epithelial cells.
In order to alleviate this problem, we decided to investigate cells of much higher homogeneity, such as cultured cells [11], or carefully collected and purified exfoliated cells. However, even these samples demonstrate very high spectral variance, and it is only now that the causes of the spectral heterogeneity have been understood, and the spectra analyzed by statistical methods that emphasize common features and de-emphasize irrelevant variations in the spectra. In this paper, we present the first carefully controlled results of a study of epithelial cell types that may serve as model systems for the analysis of cervical cells: human oral mucosa cells, and canine cervical cells. The former of these models was selected since oral mucosa cells are readily available and are quite similar in morphology and composition to squamous ectocervical cells. Canine cervical cells (collected from the cervix of animals being spayed) are morphologically and developmentally very similar to human cervical cells. Collection of both of these cell types posed no risks to the donors, and could be accomplished under the auspices of a local IRB. Most importantly, the cell types reproduce the problems encountered with cervical samples very well, and thus, represent ideal model systems to work out methodology of data collection and analysis. Since these cells are quite similar, one also can test the sensitivity of spectroscopic methods to distinguish and identify these cell types.
2. Background
Since this article is written for a non-spectroscopic audience, a brief review of the principles of Fourier transform infrared spectroscopy (FT-IR) and infrared-micro-spectroscopy (IR-MSP) will be presented. In FT-IR and IR-MSP, low energy photons with wavelengths from about 3 to 50 μm (3300 to 200 cm−1) are used to excite the vibrational motion of covalently bonded moieties. In principle, each molecule has its own specific vibrational spectral pattern or spectral fingerprint [12]. The major cellular constituents, the proteins, exhibit the most prominent spectral features in cells and tissues. Although many of a cell’s proteins exhibit very similar vibrational spectra due to their overall structural and chemical similarities, it is, nevertheless, possible to differentiate proteins, and determine many details such as degree of hydration, secondary structure and other features from the infrared spectra [13]. In addition, spectral signatures of the other cellular components, such as DNA, RNA, phospholipids, carbohydrates, etc. are well established and distinguishable from those of intact cells and tissue [14].
IR-MSP is a technique that was developed in the 1970s and 1980s, mostly for the semiconductor industry and material science fields, and was applied to biomedical problems in the early 1990s [15]. The IR beam probing the sample at the microscope’s focal plane is focused to a size of about 10 μm (restricted by the diffraction limit). Consequently, infrared spectra of microscopic objects can be acquired with a spatial resolution of about 10 μm, and spectral data from a single human cell may be acquired. The main advantage of the IR-MSP diagnostic method over standard cytology is that it may be implemented as an automated procedure that is fast, reliable, operator-independent and cost-effective. IR-MSP offers the advantage over other spectral techniques in that no probe groups (such as stains, fluorescent markers attached to antibodies or to DNA-binding drugs) are required to detect specific changes in cells, since infrared spectroscopy monitors the inherent spectral signatures of the cellular components.
3. Methods
3.1. Cell collection and sample preparation
Oral mucosa (buccal) cells were harvested from volunteer graduate and undergraduate students in the department under a local IRB protocol. Before sample collection, the oral cavity was rinsed with mouthwash. Subsequently, the inside of the cheek was gently swiped with a sterile polyester swab. Cells were shaken off the swabs in sterile, buffered saline solution (BSS), repeatedly centrifuged, washed with BSS, and deposited onto the sample substrate. The aspect of sample substrates will be discussed in Section 3.2. We estimate that a single swiping of the oral cavity results in about 105 to 106 exfoliated cells. Visual microscopic inspection of the cell samples produced indicates good homogeneity of the slides: over 99% of all cells are large squamous cells with small, well-delineated nuclei and a large cytoplasmic area. These cells typically are between 60 and 100 μm in diameter.
Canine cervical cells were collected in collaboration with a local veterinarian clinic. During spaying of dogs, the ovaries, and the uterus including the cervix, are surgically removed. These organs were immediately placed in BSS, refrigerated, and transported to our laboratory. Cervical cells were exfoliated from the exposed cervices using miniature dental brushes. These experiments were performed with permission from the Animal Welfare Committee of Hunter College. The exfoliated canine cervical cells were further processed like the human oral mucosa cells.
Exfoliated cells were deposited onto spectral substrates by one of two different methods developed originally for cytology: the CytoSpin™ and the ThinPrep™ methods. The aim of either deposition techniques is to produce sparse monolayers of cells such that all cells are well separated from their nearest neighbors, providing an unobstructed view (or measurement apertures) of each cell. This is quite different from the “smears” produced from cervical brushes, where cells are smeared onto microscope slides, which may produce thick clumps of cells that are not suitable for automatic analysis.
All results presented here were collected from samples prepared by the CytoSpin™ method, which is referred to as “liquid-based” methods since the cells are deposited from a cell suspension, rather then from a brush or other exfoliation device. In the CytoSpec method, ca. 0. 5 mL of cell suspension, with a concentration of about 5×104 cells/mL, are placed into a special, conical funnel, which is clamped against the microscope slide. Between the funnel and the slide is a layer of wicking paper with a 5 mm diameter hole. The entire assembly is placed into a special rotor of a centrifuge, and spun between 800 and 1200 rpm for about 10 min. The centrifugal force presses the liquid onto the slides, where it is wicked away by the absorbent layer of paper, leaving the cells on the substrate. A typical exfoliated cell sample prepared by this method is shown in Fig. 1A. The sampling area, 5 mm in diameter, contains about 103 cells with more than a cells size distance between cells.
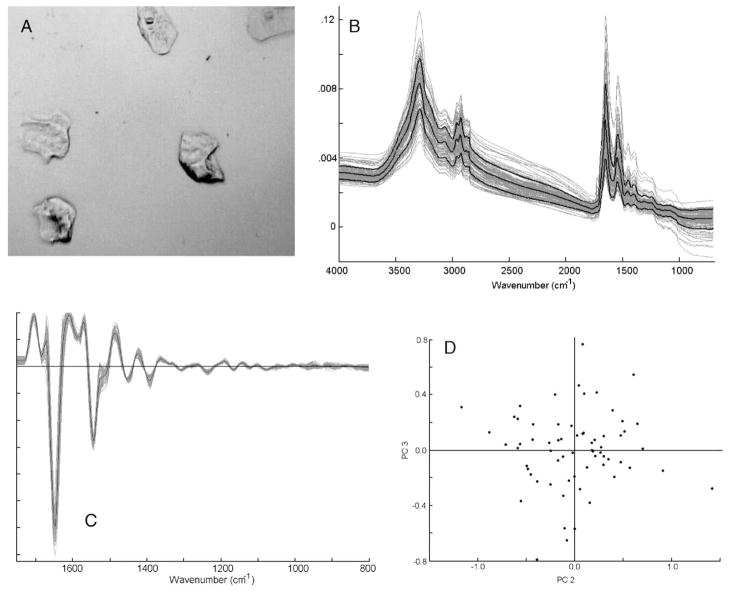
Fig. 1.
Panel A: photomicrograph of a sample of oral mucosa cells, prepared by the CytoSpin method (see text for detail). Panel B: mid-infrared (800–4000 cm−1) spectra of ca. 60 individual oral mucosa cells from one donor. The heavy black lines denote the mean spectrum, and the standard deviation spectra. Panel C: second derivative spectra of data set shown in Panel B, expanded between 800 and 1800 cm−1 Panel D: Scores plot (PC2 vs. PC3) of data set shown in Panel C. Each dot represents one spectrum.
3.2. Substrate and fixation issues
The choice of substrates for data acquisition is a crucial issue in infrared micro-spectroscopy. Glass or quartz (SiO2) cannot be used in IR-MSP for sample substrates, since the Si–O vibrations produce a broad, strong absorbance which render quartz opaque below 2500 cm−1. However, specially coated glass-slides can be used in reflection IR-MSP. These slides, known as “low-e” (low emissivity) slides consist of a thin layer of Ag atoms on glass, overcoated by a tin oxide layer. The coating is sufficiently thin to be transparent in the visible, but highly reflective in the infrared region. Thus, any cell on these slides can be inspected by visual microscopy, and IR spectral data can be collected, in reflectance, from these cells.
Fixation is a major issue for spectral cytology. Exfoliated cells, such as the oral mucosa cells to be discussed in detail below, can be used unfixed after removal from the Cytospin™ instrument. Once deposited, the cells were found to be stable for many days; this was established by examining spectra days and even weeks apart. Neither morphologic nor spectroscopic changes could be observed as a function of time. It is well known that dry fixation is a particularly mild form of fixation that preserves structure and composition of biological samples such as cells and tissues. The process of precipitating and drying proteins appears to have a similar effect to fixation in that it renders the protein matrix insoluble and relatively inert. Formalin-fixation, using a 5% buffered aqueous solution of formaldehyde, for less than 30 s produces spectra that are indistinguishable from unfixed cells. This was demonstrated by analyzing data sets of hundreds of fixed and unfixed spectra by multivariate methods (unpublished results).
3.3. Data acquisition
Spectra were collected using a Perkin-Elmer (Perkin Elmer Corp, Sheldon, Connecticut) Spectrum One/Spotlight 300 Fourier Transform IR micro-spectrometer in single point mode. Detection is provided by a 100×100 μm2 photoconductive HgCdTe (MCT) detector, operated at liquid nitrogen temperature, which covers the entire IR spectral range from 7000 to 650 cm−1.
The objective provides an image magnification of 6× at a numerical aperture of 0.58. Visual image collection via a CCD camera is completely integrated with the microscope stage motion and IR spectra data acquisition. Visible images of the deposited cells are collected under white light LED illumination, and “quilted” together to give a picture of the entire sample deposition area. Hundreds of individual cells are selected from this sample image on screen, and for each cell, the position, aperture size and aperture orientation are defined. The aperture size is selected to straddle the cell, typically between 40×40 to 80×80 μm2. Cell position and apertures are stored for each cell. Data acquisition of all stored positions proceeds automatically. For single cell spectra, 128 to 256 interferograms, collected at 4 cm−1 spectral resolution, were co-added, requiring 1 to 2 min per spectrum.
The microscope and the optical bench are continuously purged with purified, dry air. In addition, the sample area in the focal plane of the microscope has been enclosed in a sample chamber, which is purged as well.
3.4. Data processing
The analysis of spectra from individual cells was carried out by principal component analysis (PCA). PCA is a well-established multivariate method ideally suited to distinguishing small, re-occurring spectral variations in large data sets containing uncorrelated variations. Refer to Adams for a discussion on PCA [16]. We selected PCA over other methods, since it is a completely unsupervised method, to establish whether or not spectra of cells group into classes due to cell type, donor identity or disease, among other factors.
For PCA, the entire spectral data set, containing n spectra, is written into one matrix S. In this matrix, each column represents one spectrum S(ν) of m intensity data points. The spectral vectors normally are pre-processed by calculating the 2nd derivative and vector-normalizing all spectral data.
The intensity covariance matrix is constructed from the spectral matrix S according to
| (1) |
or
| (2) |
C is a (m·m) matrix, in which the off-diagonal terms Ckl express the covariance between intensity values at wavelengths νk and νl, summed over all spectra. Diagonalization of the intensity covariance matrix, according to
| (3) |
yields the eigenvector matrix P, from which “principal components” (PC) Z are calculated according to
| (4) |
In Eq. (3), the superscript T refers to the transpose of the matrix P. The eigenvalues Λ express the variance contained in each of the principal components. Thus, from the viewpoint of linear algebra, the principal components are the original spectra expressed in a coordinate system rotated to maximize the variance of the original spectra. Subsequently, we may express each of the original spectra S(ν) in terms of the new principal components
| (5) |
or
| (6) |
where the “scores”, α, are given by
| (7) |
As described so far, the procedure does not seem to simplify the problem at all. However, in the spectral data sets of individual cells one finds that a large fraction of the total spectral variance is contained in the first few eigenvectors. Typically, 5–8 eigenvectors contain more than 99% of the variance. Thus, the spectral expansion given by summation in Eq. (6) can be truncated after the p’th term, where p is the number of relevant loading vectors or principal components. Then, the score matrix α, which determines how much each principal component contributes to each spectrum, will have the dimension (p·n). This step reduces the size of the problem significantly, since all spectra are expressed in terms of a few (typically 5–8) basis functions and a “score vector” of about p entries.
Similar spectra exhibit similar scores α, which may be used to discriminate, or group, spectra. This is accomplished by plotting the values αi and αj (that is, the contribution of PCi and PCj to each spectrum) against each other. In these “scores plots”, shown in later figures, each data point represents one spectrum. If grouping is observed, there are quantifiable and significant variations in the spectra, which can be used to construct discriminant algorithms for distinguishing cell types, state of differentiation and maturation of cells, and disease.
4. Results and discussion
4.1. Infrared spectral results of oral mucosa cells
We first present a review of spectral results of exfoliated oral mucosa cells from one donor, before discussing spectral differences observed between donors. The former aspects were reported in detail in the literature [17].
4.1.1. Heterogeneity of spectra from one donor
Fig. 1, Panel B, shows individual spectra of about 60 oral mucosa (buccal) cells, along with their mean spectrum, and the standard deviation from the mean. We observed large variations in the overall amplitude of the spectra, broad undulating features centered around 2300 cm−1, and distinct spectral variations in the low frequency region.
These features were discussed in detail [18], and will only be summarized here. The broad, undulating features observed relatively weakly in the spectra of entire cells were previously attributed to Mie scattering [19] of the cellular nuclei. The overall intensity variations are most likely due to variations in the thickness of the cell, as well as the nucleus/cytoplasm (N/C) ratio. In the cytoplasm of a dried epithelial cell, typically ca. 2–3 μm thick, the amide I vibration typically exhibits absorbance values of ca. 0.05 OD units. The nucleus maintains a more spherical shape, with an average thickness of about 3–50 μm, and a much higher protein concentration. The nuclear spectra are dominated by protein features, and often exhibit absorbance values of 0.3–0.5 OD units in the amide I band. A simple calculation shows that the cross section of the nucleus, exhibiting an absorbance of 0.5 OD units, averaged over the cross section of a cell’s cytoplasm with an absorbance of 0.05 OD units, contributes about 20% of the total protein signal, although the area of the nucleus is only about 2.5% of the cell’s area. Thus, a small decrease in the nuclear size (for example, from 10 to 7 μm) will cause an overall change in protein signal by about 10%. Thus, small variations in nuclear size will influence the total absorption intensity to a large extent. Thus, we believe that the large variations in spectral amplitudes of cells result from variations in the size of the nucleus as compared to the size of the entire cell. In cytology, this effect is referred to as the nuclear/cytoplasmic ratio (N/C), which is a commonly applied measure of a cell’s health and activity.
In spite of the spectral variations shown in Fig. 1B, cellular spectra can be analyzed by multivariate statistical methods that emphasize common features and de-emphasize irrelevant variations in the spectra. To this end, it is advantageous to pre-process the data. This pre-processing includes computation of the second derivative of spectral intensity I with respect to wavenumber, ∂2I/∂nu;2, normalization and expansion. The derivative spectra exhibit nearly completely flat baselines, and collapsed band width. Spectral expansion emphasizes the most diagnostic “mid-IR” region between 800 and 1800 cm−1, and eliminates a large spectral range (1800–2800 cm−1), which contains no vibrational spectroscopic information. Subsequently, spectra are vector normalized, i.e., the squared integrated spectral intensity is set to unity. This last step removes the variation in spectral intensity discussed above. Expanded, derivatized and normalized spectra of the original data set (Fig. 1B) are shown in Fig. 1, Panel C. A scores plot of this data set, obtained via PCA, is shown in Fig. 1, Panel D. In this plot, each dot represents one spectrum, and correlates the variance between all spectral data. The symmetric shape of this plot demonstrates that the data set exhibits no significant trends.
The causes for variations of spectral patterns in the low frequency (<1400 cm−1) region, observed for very few cells, may have a variety of causes. In principle, this spectral range is very sensitive to cellular proliferation; however, since oral mucosa cells are expected to be divisionally inactive, we believe that other causes are responsible for the observed variations. Pyknosis and/or necrosis of the cells, which are being studied in detail in the authors’ laboratory, may contribute to the spectral differences. However, after the data pre-processing described above, the variations of these spectra is minimized as well.
4.1.2. Variations of spectral results from five donors
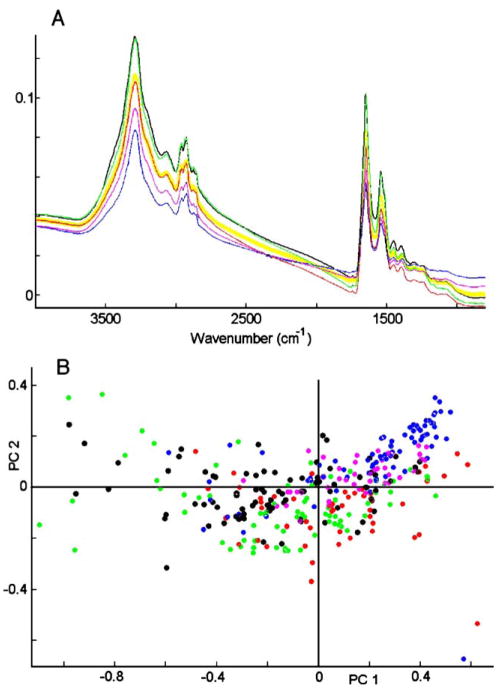
We now turn to the discussion of the analysis of oral mucosa cells from different donors. In total, we collected samples from about a dozen volunteers within the Chemistry Department at Hunter College. The results shown in Fig. 2 are typical for the spectral variance observed between donors. In Panel A, we depict the average cell spectra from 5 donors; each average is derived from between 55 and 70 spectra. In addition, the average of all 320 spectra is shown by the yellow trace. These results demonstrate that the overall spectral intensities of cells from different donors, as well as their average band shapes, vary significantly. The variance is also borne out in the scores plot, shown in Fig. 2, Panel B. This plot, in which each dot represents one complete spectrum, is non-symmetrical, indicating major spectral differences along PC 1, which is mostly due to overall intensity variations. Similar groupings of cellular spectra were observed when other scores were plotted.
Fig. 2.
Panel A: spectra of 320 oral mucosa cells, averaged separately for each of 5 donors, (shown as black, blue, green, red and purple traces), and average of all 5 donors (yellow). Panel B: scores plot (PC1 vs. PC2) of data set shown in Panel A. Each dot represents one spectrum.
At first, such donor-to-donor variations appear troublesome, since they indicate that spectra of cells group primarily by donor, and might mask effects of disease or other factors one may want to study spectroscopically. However, previous efforts to use infrared spectral methods to identify and classify prokaryotic cells reported the methodology of reducing the variation between repeat measurements, or slightly different origins of samples [20]. As described before, such spectral variations can be minimized by suitable data pre-processing that removes overall intensity and baseline variations.
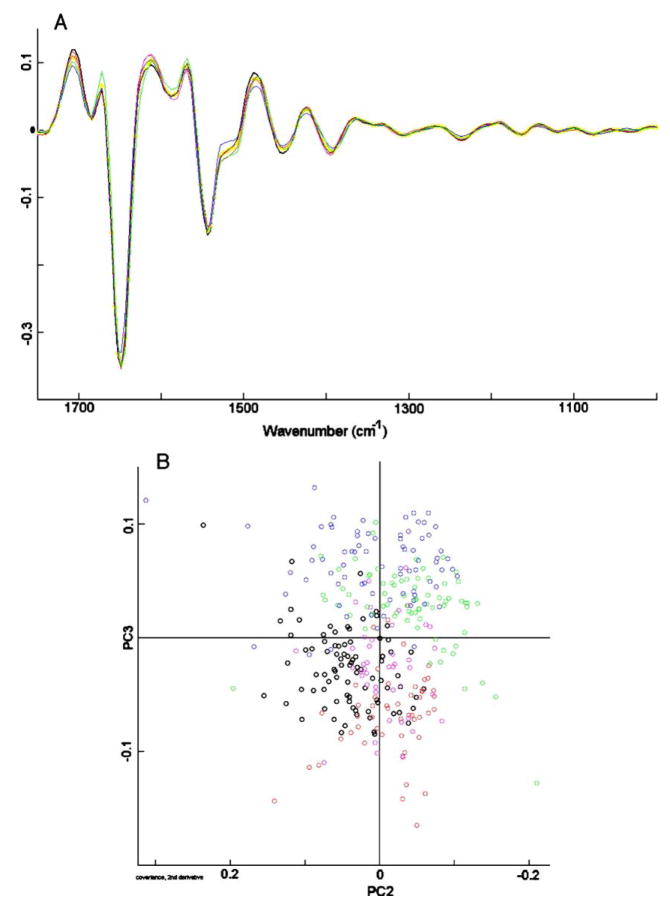
This is shown in Fig. 3, Panel A, which shows the data of Fig. 2 after expansion, vector normalization and second derivative computation. Vector normalization minimizes intensity variations between the spectral data set, whereas the derivatization removes baseline drifts. The resulting spectral traces are quite similar for different donors, and yield a scores plot, shown in Fig. 3, Panel B, in which the variances along PC2 and PC3 are much smaller than those shown in Fig. 2B, and are symmetric about the origin. However, there still exist some systematic differences, mainly for the cells marked with blue and black circles. The origin of this spectral difference is not clear at this point, but the spectra of cells from other donors tend to fall within the range set by the five donors shown in Fig. 3B. Furthermore, the donor-to-donor differences are smaller than those observed when another cell type is added to the PCA analysis, for example the canine cells to be discussed below (Section 4.3).
Fig. 3.
Panel A: second derivative spectra of data set shown in Fig. 2, Panel A, expanded between 800 and 1800 cm−1 Panel B: scores plot (PC2 vs. PC3) of data set shown in Panel A. Each circle represents one spectrum.
These data demonstrate that it is absolutely essential to carry out any comparative studies such that all unwanted and irrelevant variations are minimized, or employ sufficiently large data sets to average out unwanted variations. We illustrate this aspect of sampling variations in the following example. To investigate the effect of cell fixation, we carried out an experiment in which cells were collected from one volunteer, and data of the unfixed cells were collected immediately. Several days later, cells from the same volunteer were collected, formalin-fixed, and spectra were acquired immediately. PCA subsequently separated the cells clearly between fixed and un-fixed cells. However, when two cell samples were collected at the same time from one volunteer, and one sample was fixed, whereas the other was left un-fixed, the spectral differences disappeared, and PCA scores showed a randomly distributed plot. The originally observed “differences” between the fixed and un-fixed cells may have been due to different cleansing procedures of the oral cavity, slightly different exfoliation procedures, as well as some yet unexplored time variation of the cells. These aspects need to be taken into account in all proposed methods to utilize IR spectroscopic methods for diagnostic applications.
Although this high sensitivity of IR spectroscopy may seem a bit disconcerting, we found that the systematic variations between normal and diseased cells, or between different epithelial cell types, are much larger than the deviations described above between donors and/or acquisition date. This will be demonstrated by the results shown in Section 4.3.
4.2. Infrared spectral results of canine cervical cells
Here, we report results from another model system of epithelial cells. Canine cervical cells were selected as a model since quite homogeneous cell samples can be obtained by gently scraping the cervix from spayed dogs with a miniature dental brush, and vortexing the cells from the brushes in BSS. Visual inspection of these cells revealed a fairly homogeneous population of ecto-cervical cells, measuring about 25 to 35 μm in size, on average. Sample preparation and data collection proceeded as in the case of oral mucosa cells.
The variation in the spectra of cells collected from different animals is similar to that observed between human donors. The spectra of human oral mucosa cells, and of canine cervical cells, were found to be nearly indistinguishable by eye. PCA reveals good homogeneity of the spectra, and no systematic variations between them. In view of the previous discussion, these results can be expected. However, it is now important to demonstrate that the data pre-treatment, and the principal component analysis described above, preserved the differences between the two different kinds of epithelial cells.
4.3. Comparison between human and canine mucosal cells
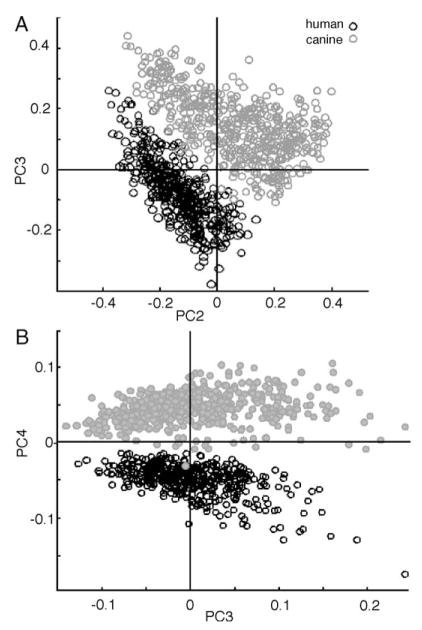
Morphologically, human oral mucosa cells and canine cervical cells are very similar. Furthermore, in terms of their biochemical composition, which ultimately determines the observed IR spectra, these cells are expected to be very similar. Thus, we thought it an important test of the methodology whether or not the spectra of these cells could be distinguished. We included all available data subsets to minimize factors such as different collection times from influencing the analysis. In total, 7 sets human oral mucosa cells (comprising 427 spectra), and 5 sets from animals (560 spectra) were combined into one data set, and analyzed via PCA. The scores plots (PC2 vs. PC3) of uniformly pre-processed data are shown in Fig. 4A, and the PC3 vs. PC4 plot in Fig. 4B. The separation between canine cervical and human buccal cells, shown in Fig. 4, is excellent, both in the PC2 vs. PC3 and the PC3 vs. PC4 plots. However, at this point, the spectral features responsible for the differentiation are not understood. The shape of the two clusters is very different from those shown in Fig. 3. This is fully expected because PCA uses the entire variance of the data set, expressed as the covariance matrix, and determines the principal components from the eigenvectors of this matrix. When new data are added to a data set, the overall variance changes, which in turn will change the eigenvectors and the scores plots from the subsequent PCA analysis. Thus, the changes in the cluster shape between Fig. 3B and the human cell cluster in Fig. 4A and B are fully expected.
Fig. 4.
Panel A: scores Plot (PC2 vs. PC3) of nearly 1000 human oral mucosa and canine cervical cells Panel B: scores plot (PC3 vs. PC4) of data set shown in Panel A. Each symbol represents one spectrum.
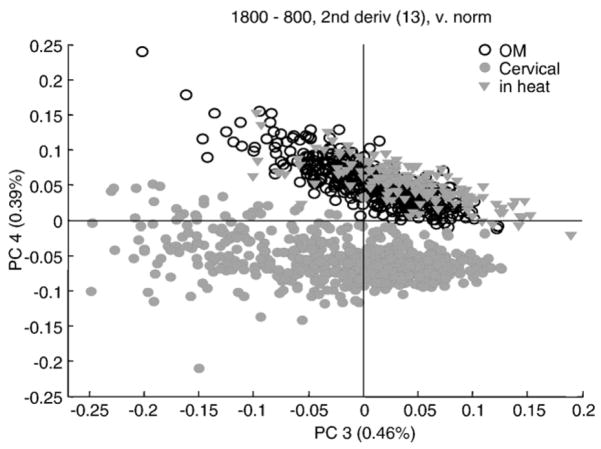
When the spectra of cells from a dog in estrus were incorporated into the PCA analysis, Fig. 5, they were found to fall exactly within the range of the human buccal cells, and were well separated from the spectra of cells of non-estrus dogs. The explanation of this spectral differentiation may be found in the level of maturity of the cells examined.
Fig. 5.
Scores plot (PC3 vs. PC4) of over 1000 human oral mucosa and canine cervical cells, including cervical cells from an estrus dog. Each circle represents one spectrum.
The human buccal cells, exfoliated from the oral mucosa, are mature squamous epithelial cells. These cells are typically between 50 and 60 μm on edge. For non-estrus dogs, the cervical cells are not completely mature, and are generally classified as intermediate squamous cells [21]. These cells are significantly smaller, about 25 to 35 μm on edge. During estrus, hormonal changes cause maturation of the canine cervical cells from intermediate to superficial cells. In this process, the cells enlarge significantly, to about the same size as mature human cervical or human buccal cells.
The spectral differentiation between immature (intermediate) canine cervical cells on one hand, and mature human buccal cells and mature canine cervical cells on the other, may then be due to the following causes. First of all, the similarity between the genetic make-up of mammalian and human cells (of similar cell type) make it improbable that the differences are due to species-specific changes. Thus, other causes need to be considered for the observed differentiation, in particular in view of the differentiation of estrus and non-estrus states of the dogs. This could, of course, be a direct effect of hormonal changes. In human cervical cells, for example, it is known that the glycogen content of the epithelial cells varies greatly with the menstrual cycle, and with the cell’s maturity. Thus, for human cells, the differentiation between intermediate and superficial layers of cervical epithelium is straightforward [9].
Canine cervical cells, however, do not exhibit noticeable glycogen peaks, and the differentiation of cells from estrus vs. non-estrus dogs is, most likely, not due to variations in glycogen. However, the size of the cell varies, and with it, the nucleus-to-cytoplasm (N/C) ratio. Since the nucleus contributes as much as 25% to the overall spectrum of the cell, variations in the N/C ratio will greatly affect the observed spectra. Thus, we believe that the patterns observed for human buccal, immature canine and mature canine cervical cells is a direct consequence of the N/C ratio of these cells.
Similarly, we have applied IR microspectral methods, coupled to principal component analysis, to distinguish healthy T-lymphocytes from cancerous T-cells, (anaplastic large T-cell lymphoma, cell line HA1). Here, the separation between the two cell types is better than the epithelial cell types, indicating that the methodology reported here can be applied for screening for cancerous cells. These results will be reported at a later date.
5. Conclusions
We find that infrared spectra collected from individual cells contain information on the cell type, level of maturity and state of disease that can be exploited for diagnostic purposes. Since the acquisition of data is carried out by a spectrometer and the interpretation by a computer algorithm, this method promises to be entirely objective. Prior studies have failed to recognize the variance in spectral data, and therefore, may not have employed suitable methods for the interpretation of the results.
Acknowledgments
Support of this research through a grant from the National Institutes of Health (CA 090346) is gratefully acknowledged.
References
- 1.Wood BR, Chiriboga L, Yee H, Quinn MA, McNaughton D, Diem M. FTIR mapping of the cervical transformation zone, squamous and glandular epithelium. Gynecol Oncol. 2004;93(4):59–68. doi: 10.1016/j.ygyno.2003.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steller W, Einenke J, Horn LC, Braumann UD, Binder H, Salzer R, Krafft C. Delimitation of a squamous cell cervical carcinoma using infrared microspectroscopic imaging. Anal Bioanal Chem. doi: 10.1007/s00216-005-0124-4. in press. [DOI] [PubMed] [Google Scholar]
- 3.Chiriboga L, Xie P, Yee H, Zarou D, Zakim D, Diem M. Infrared spectroscopy of human tissue. IV. Detection of dysplastic and neoplastic changes of human cervical tissue via infrared microscopy. Cell Mol Biol. 1998;44(1):219–229. [PubMed] [Google Scholar]
- 4.Lasch P, Haensch W, Naumann D, Diem M. Imaging of colorectal adenocarcinoma using FTIR micro-spectroscopy and cluster analysis. Biochim Biophys Acta. 2004;1688(2):176–186. doi: 10.1016/j.bbadis.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 5.Lasch P, Haensch W, Lewis EN, Kidder LH, Naumann D. Characterization of colorectal adenocarcinoma sections by spatially resolved FT-IR microspectroscopy. Appl Spectrosc. 2002;56:1–9. [Google Scholar]
- 6.Krafft C, Sobottka SB, Schackert G, Salzer R. Analysis of human brain tissue, brain tumors and tumor cells by infrared spectroscopic mapping. The Analyst. 2004;129(10):921–925. doi: 10.1039/b408934k. [DOI] [PubMed] [Google Scholar]
- 7.Romeo MJ, Diem M. Infrared spectral imaging of lymph nodes: strategies for analysis and artifact reduction. Vibr Spectrosc. 2005;38:115–119. doi: 10.1016/j.vibspec.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood BR, Quinn MA, Tait B, Romeo MJ, McNaughton D. A FTIR spectroscopic study to identify potential confounding variables and cell types in screening for cervical malignancies. Biospectroscopy. 1998;4:75–91. doi: 10.1002/(SICI)1520-6343(1998)4:2%3C75::AID-BSPY1%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 9.Chiriboga L, Xie P, Yee H, Vigorita V, Zarou D, Diem M. Infrared spectroscopy of human tissue: I. Differentiation and maturation of epithelial cells in the human cervix. Biospectroscopy. 1998;3:47–54. doi: 10.1002/(SICI)1520-6343(1998)4:1%3C47::AID-BSPY5%3E3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 10.Chiriboga L, Xie P, Vigorita V, Zarou D, Zakim D, Diem M. Infrared spectroscopy of human tissue: II. A comparative study of spectra of biopsies of cervical squamous epithelium and exfoliated cervical cells. Biospectroscopy. 1998;3:55–59. doi: 10.1002/(SICI)1520-6343(1998)4:1%3C55::AID-BSPY6%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Diem M, Romeo MJ, Matthaus C, Miljkovic M, Miller L, Lasch P. Comparison of Fourier transform infrared (FTIR) spectra of individual cells acquired using synchrotron and conventional sources. Infrared Phys Technol. 2004;45:331–338. [Google Scholar]
- 12.Diem M. Introduction to Modern Vibrational Spectroscopy. J. Wiley & Sons Interscience; New York: 1993. [Google Scholar]
- 13.Levin IW. Chemical, Biological and Industrial Applications of Infrared Spectroscopy. Wiley; New York: 1985. [Google Scholar]
- 14.Diem M, Boydston-White S, Chiriboga L. Infrared spectroscopy of cells and tissues: shining light onto an unsettled subject. Appl Spectrosc. 1999;53(4):148A–161A. [Google Scholar]
- 15.Humecki HJ. Practical Guide to Infrared Microscpectroscopy. Marcel Dekker; New York: 1995. [Google Scholar]
- 16.Adams MJ. Chemometrics in Analytical Spectroscopy. 2. Royal Society of Chemistry; Cambridge: 2004. [Google Scholar]
- 17.Romeo MJ, Mohlenhoff B, Diem M. Infrared microspectroscopy of human cells: Causes for the spectral variance of oral mucosa (buccal) cells. Vibrational Spectrosc. doi: 10.1016/j.vibspec.2006.04.009. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mohlenhoff B, Romeo MJ, Wood BR, Diem M. Mie-type scattering and non-Beer–Lambert absorption behaviour of human cells in infrared microspectroscopy. Biophys J. 2005;88(5):3635–3640. doi: 10.1529/biophysj.104.057950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mie G. Beitrage zur Optik Truber Medien, Speziell Kolloidaler Metal-losungen. Ann Phys ( Leipz ) 1908;25:377–452. [Google Scholar]
- 20.Helm D, Labischinski H, Schallen G, Naumann D. Classification and identification of bacteria by FT-IR spectroscopy. J Gen Microbiol. 1991;137:69–79. doi: 10.1099/00221287-137-1-69. [DOI] [PubMed] [Google Scholar]
- 21.Holst PA. Canine Reproduction: A Breeder’s Guide. Alpine Publications; Loveland: 1985. [Google Scholar]