Abstract
The genetic diversity of 134 serogroup X Neisseria meningitis isolates from Africa, Europe, and North America was analyzed by multilocus sequence typing and pulsed-field gel electrophoresis. Although most European and American isolates were highly diverse, one clonal grouping was identified in sporadic disease and carrier strains isolated over the last 2 decades in the United Kingdom, the Netherlands, Germany, and the United States. In contrast to the diversity in the European and American isolates, most carrier and disease isolates recovered during the last 30 years in countries in the African meningitis belt belonged to a second clonal grouping. During the last decade, these bacteria have caused meningitis outbreaks in Niger and Ghana. These results support the development of a comprehensive conjugate vaccine that would include serogroup X polysaccharide.
Keywords: molecular epidemiology, genotyping, meningococcal meningitis, Africa
Bacterial meningitis due to Neisseria meningitidis (meningococcus) causes epidemics in Africa usually associated with serogroup A meningococci. Sporadic cases, outbreaks, and hyperendemic disease in Europe and the United States are usually caused by serogroups B and C (1). Occasionally, however, endemic disease and outbreaks are caused by bacteria belonging to other serogroups, including W135, Y, and X. Serogroup X N. meningitidis was described in the 1960s (2,3), and serogroup X meningitis has been observed in North America (4), Europe (5,6), Australia (7), and Africa (8,9). Serogroup X outbreaks have been reported in Niger (10,11) and Ghana (12). In some cases, serogroup X disease has been associated with a deficiency of particular complement components (13,14) or with AIDS (15).
Asymptomatic nasopharyngeal carriage of N. meningitidis is common, and in only a small percentage of colonized persons do the bacteria invade the bloodstream and cerebrospinal fluid to cause disease. Meningococcal populations are highly diverse, and lineages of meningococci with increased capacity to cause invasive disease are thought to arise periodically and spread, sometimes globally (16). Relatively few of these hyperinvasive lineages or clonal groupings are responsible for most meningococcal disease worldwide (17). These clonal groupings diversify during spread (18,19), primarily as a result of frequent horizontal genetic exchange (19–21). However, many variants are isolated only rarely or from a single country and are not transmitted further because of bottlenecks associated with geographic spread and competition (19,22). The population structure of N. meningitidis is effectively panmictic as a result of frequent horizontal genetic exchange (23), but that of some groupings, such as epidemic serogroup A meningococci, is largely clonal (24). The population structure of serogroup X meningococci has not yet been investigated in detail.
After an epidemic of serogroup A disease in 1997-1998 in northern Ghana (9), we conducted a longitudinal carriage study to investigate the dynamics of meningococcal carriage during an interepidemic period (12). We observed a sharp increase in nasopharyngeal carriage of serogroup X meningococci by healthy persons, accompanied by several cases of serogroup X meningitis. To investigate the phylogenetic relationships of these bacteria, we compared the isolates from Ghana with other serogroup X meningococci isolated during recent decades in Africa, Europe, and North America.
Materials and Methods
Bacterial Strains
We analyzed 134 N. meningitidis isolates of serogroup X by pulsed-field gel electrophoresis (PFGE) (130 isolates) or multilocus sequence typing (MLST) (41 isolates). Of these bacteria, 102 were isolated in Africa from 1970 to 2000: from meningitis patients (9 isolates) and healthy carriers (70 isolates) in Ghana, 1998-2000; from healthy carriers in Mali in 1970 (9 isolates) and 1990-91 (4 isolates); and from patients in Chad (1995, 1 isolate), Niger (1997-1998, 4 isolates), and Burkina Faso (1996-1998, 5 isolates). Six isolates were not tested serologically; the other 96 were NT:P1.5.
In addition, 32 serogroup X strains isolated from 1988 to 2000 in the United Kingdom (22 isolates), Germany (3 isolates), United States (4 isolates), France (1 isolate), Norway (1 isolate), and the Netherlands (1 isolate) were included in the analysis. The 26 strains subjected to further testing had diverse serotypes (2b, 4, 4/21, 14, 16, 21, 22) and serosubtypes (nonsubtypable [NST], P1.5, P1.5,10, P1.7, P1.12, P1.14, P1.15, P1.16) in various combinations.
Molecular Typing of Bacteria
PFGE was done by digesting chromosomal DNA prepared in agarose blocks with NheI and SpeI as described (22), and MLST by sequencing gene fragments of abcZ, adk, aroE, fumC, gdh, pdhC, and pgm, also as described (16; http://www/mlst.net). The detailed MLST results and sources of isolates have been deposited in a public database http://www.mlst.net). Additional MLST data for 31 isolates in 30 sequence types were obtained with permission from http://www.mlst.net.
Data Analysis
A neighbor-joining tree was constructed by using the numbers of MLST allele differences with Bionumerics 2.0 (25).
Results
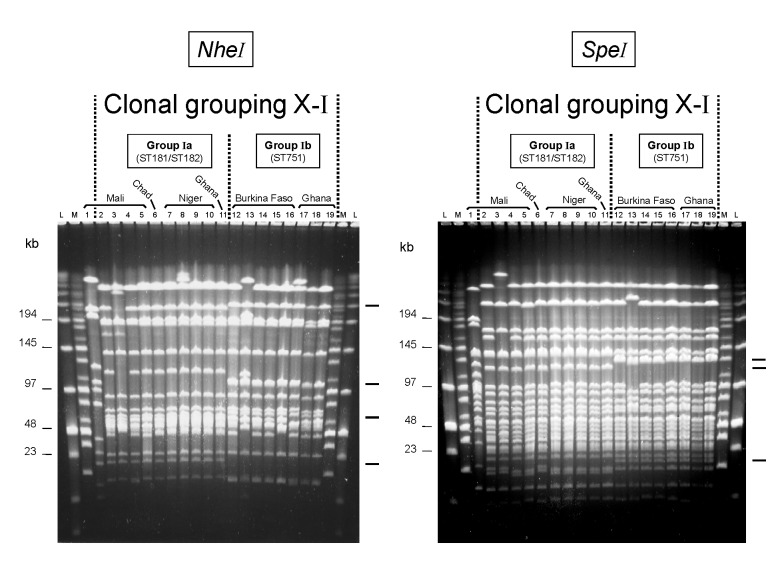
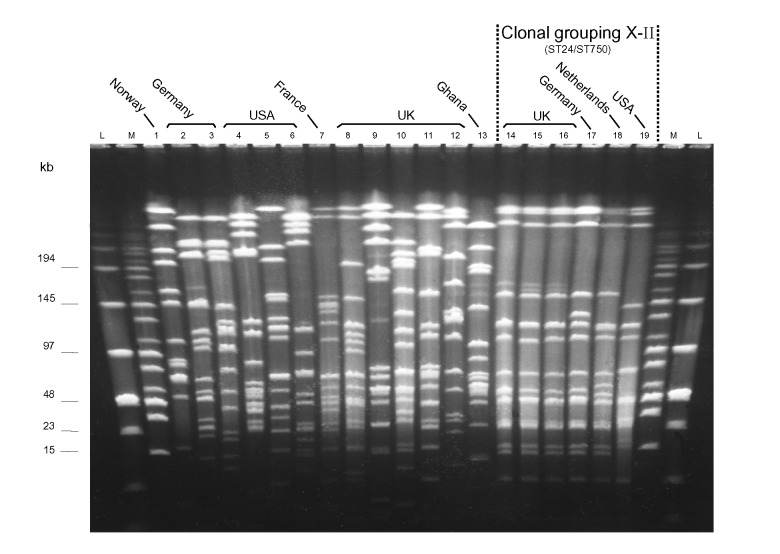
PFGE with two discriminatory rare-cutting enzymes NheI and SpeI) was used to identify groups of closely related strains in 134 isolates of serogroup X N. meningitidis from countries in Africa, Europe, and North America. All but 3 of 102 isolates from Africa had similar PFGE patterns (Figure 1, clonal grouping X-I). In contrast, 19 of 32 isolates from Europe and North America had distinct PFGE patterns (Figure 2) that differed from those of the African isolates. However, among the latter 32 strains, similar PFGE patterns were observed for 13 isolates from the United Kingdom, Germany, the Netherlands, and the United States (Figure 2, clonal grouping X-II).
Figure 1.
Two groups of pulsed-field gel electrophoresis patterns among NheI- and SpeI-digested chromosomal DNA from selected serogroup X Neisseria meningitidis strains isolated in Africa. Lane: strain: 1: D93 (ST188); 2: 1970; 3: 3187; 4: 3529; 5: D5; 6: LNP13407; 7: LNP14964; 8: LNP15040; 9: 97013; 10: 97014; 11: Z9413; 12: LNP14297; 13: LNP15061; 14: BF2/97; 15: BF5/97; 16: BF1/98; 17: Z7091; 18: Z8336; 19: Z9291. Molecular weight markers were loaded in the flanking lanes as indicated (L: low-range marker; M: midrange marker); their molecular weights are indicated at the left. Characteristic band differences are indicated on the right.
Figure 2.
Pulsed-field gel electrophoresis patterns of NheI-digested chromosomal DNA of selected serogroup X Neisseria meningitidis isolates from Europe and the United States, plus a prototype isolate from Ghana (lane 13). Lane: strain: 1: E26; 2: X4571; 3: X4890; 4: M2526; 5: M4222; 6: M3772; 7: LNP17351; 8: J88-603; 9: K89-1395; 10: L92-1413; 11: M98-253172; 12: M00-240465; 13: Z9291; 14: M98-252848; 15: M98-252718; 16: M99-240899; 17: X5967; 18: 860060; 19: M4370.
Forty-one isolates, each representing a distinct PFGE pattern, were analyzed by MLST. For bacteria from which multiple isolates with a similar PFGE pattern had been detected, we tested at least one representative from each year and country of isolation. Together with other data in the MLST WEB site (http://www.mlst.net), 39 distinct sequence types (STs) have been found in 50 serogroup X meningococci. The general structure of a neighbor-joining tree of allelic differences resembles a bush, with little phylogenetic structure (Figure 3). However, isolates with similar PFGE patterns were assigned to closely related STs. All 29 clonal grouping X-I isolates analyzed by MLST were in STs ST181, ST182, or ST751 (Figure 3), which differ by one to three of the seven gene fragments (Table). Similarly, all five clonal grouping X-II isolates were in STs 24 and 750, which differ by one of the seven gene fragments (Table). The three unusual African isolates (strain designations D87, D91, and D93) were in ST188, which is very distinct from STs of clonal grouping X-I (Figure 3). These results show that numerous serogroup X isolates from Africa and nearly half the serogroup X isolates from Europe and North America belong to two clonal groupings, while other serogroup X isolates from Europe or North America are quite diverse.
Figure 3.
Neighbor-joining phenogram of allelic identities in 39 multilocus sequence typing (MLST) sequence types from serogroup X Neisseria meningitidis. Numbers at nodes are the percentages of 1,000 bootstrap replicates in which these nodes appeared. Only nodes with percentages >50% were included. The two boxes indicate clonal groupings that were detected by MLST or pulsed-field gel electrophoresis.
Table. Multilocus sequence typing results of two serogroup X Neisseria meningitidis clonal groupings.
| ST | Allele numbers | Country (no. of isolates) | Year | ||||||
|---|---|---|---|---|---|---|---|---|---|
| AbcZ | Adk | aroE | FumC | gdh | pdhC | Pgm | |||
| 24 | 2 | 5 | 2 | 7 | 15 | 20 | 5 | Netherlands (1), United States (1) | 1986, 1993 |
| 750 | 2 | 5 | 2 | 9 | 15 | 20 | 5 | United Kingdom (2), Germany (1) | 1998-1999 |
| 181 | 10 | 3 | 15 | 7 | 5 | 41 | 31 | Mali (6), Chad (1), Niger (2), Ghana (1) | 1970-2000 |
| 182 | 10 | 3 | 15 | 26 | 5 | 41 | 31 | Mali (4) | 1970 |
| 751 | 10 | 3 | 15 | 7 | 8 | 41 | 6 | Burkina Faso (3), Ghana (12) | 1996-2000 |
Serologic Results
African isolates of clonal grouping X-I were NT:P1.5. The 11 North American and European isolates of clonal grouping X-II for which serologic data were available were 21:P1.16. Diverse serotype and serosubtype patterns were found for the other isolates from North America and Europe
The PFGE patterns distinguished two finer groups (Ia and Ib) in clonal grouping X-I, which differ consistently in four NheI and three SpeI fragments (Figure 1). All 14 group Ia strains tested were either ST181 or ST182, which differ at one of the seven gene fragments (Table). All 15 group Ib strains tested were ST751, which differs from ST181 and ST182 at two to three loci (Table). Group Ia included 10 isolates from Mali (1970-1990), 4 isolates from Niger (1997-1998), and the sole isolate from Chad (1995), as well as one of 79 isolates from Ghana (2000). All five isolates from Burkina Faso (1996-1998) and 78 of 79 isolates from Ghana (1998-2000) were in group Ib.
Discussion
The general population structure of N. meningitidis is panmictic as a result of the frequent import of alleles from unrelated Neisseriae (20,23). Furthermore, several MLST studies have demonstrated that meningococci from healthy carriers are highly diverse (16,26). Phylogenetic trees of different housekeeping genes from N. meningitidis are no more congruent with each other than with random trees (27). Our results for sequence typing of housekeeping genes of serogroup X meningococci also fit this pattern. Phylogenetic analysis of allele differences resulted in a bushlike tree that does not seem to contain any deep phylogenetic information. However, two clonal groupings were found in this otherwise panmictic group of bacteria. The same isolates were assigned to both clonal groupings by two independent methods, MLST and PFGE, indicating that these assignments reflect real genetic relationships and do not depend on the methods used.
Similar concordant genetic relationships were discerned in epidemic serogroup A N. meningitidis by multilocus enzyme electrophoresis (MLEE), random amplification of polymorphic DNA (RAPD), and MLST; population genetic analyses confirmed that the population structure of these bacteria is clonal (28). Concordant groupings were also discerned by MLEE and MLST among the so-called hypervirulent serogroup B and C isolates of the ET-5 complex, ET-37 complex, lineage III, and cluster A4 (16). Although their apparent clonality may reflect an epidemic population structure (23), this possibility has been excluded for epidemic serogroup A meningococci (28). Therefore, multiple clonal groupings exist in N. meningitidis, even though the population structure of most of the species is panmictic.
The population structure of subgroup III serogroup A meningococci seems to represent continual, sequential replacement of fit genotypes by related variants during periods of several years to decades (19). In subgroup III, nine genoclouds, each consisting of a frequent genotype plus its rarer, less fit variants, have been identified during 3 decades of pandemic spread. Our PFGE data for clonal grouping I of serogroup X suggest that clonal grouping X-I also has a genocloud structure. Two sets of PFGE variants (group Ia and Ib), which might each represent a genocloud, were detected in different countries (Mali, Chad, and Niger; and Burkina Faso and Ghana, respectively). Additional analyses of polymorphic genes are necessary to clarify the uniformity of these groups and to test the similarity of their population structure compared with that of subgroup III.
Both serogroup X clonal groupings described here were isolated over decades, on multiple occasions, and from diverse locations. Clonal grouping X-I (1970-2000) was isolated from different countries in West Africa, and clonal grouping X-II (1986-1999) was isolated from Europe and North America. For clonal grouping X-I in Ghana, the disease rate in healthy carriers was estimated to be 3/10,000 (12). Clonal grouping X-1 is thus of considerably lower virulence than serogroup B ET-5 complex bacteria (disease/carrier rate of 2,100/10,000 [29]) or serogroup A subgroup III bacteria during a postepidemic period in a vaccinated population (100/10,000 [9]).
The relationship between bacterial fitness and clonality has not yet been investigated extensively in natural isolates. Variation in virulence between bacterial genotypes leads to more uniformity in disease isolates than in carriage organism in Streptococcus pneumoniae (30) and Staphylococcus aureus (31). However, our data suggest that the clonal structure of certain meningococcal genotypes need not reflect virulence but rather is associated with genotypes that are particularly fit at colonizing the nasopharynx and spreading from person to person.
Although clonal grouping X-I bacteria are less virulent than serogroup A and B meningococci, they are still pathogenic. Most strains described here were isolated from asymptomatic carriers or patients with rare endemic cases. However, group Ia caused a meningitis outbreak with >60 cases in 1997 in Niger (11). Group Ib caused a smaller outbreak in 2000 in Ghana (12). These results suggest that X-I meningococci may even be capable of causing epidemics. Meningococci are naturally transformable, and horizontal DNA transfer is frequent in these bacteria (20–22). Meningococcal carriage is usually low in interepidemic periods in Africa (1,12,32,33), offering less opportunity for horizontal genetic exchange, which could account for the low genetic variability in serogroup X meningococci in Africa.
For more than a decade, many countries in the African meningitis belt have vaccinated extensively with A/C polysaccharide vaccines (34). Recently, mass vaccination with conjugated serogroup C vaccines has been implemented in the United Kingdom, and strong initial protection has been reported (35). However, if effective, these vaccines may well select for the spread of bacteria for which they are not protective (36), including unusual causes of disease such as serogroups Y, W135, and X. Capsule switching due to DNA transformation has been documented (37,38), and effective vaccination against serogroups A and C may select for capsule switch variants of fit genotypes expressing a capsular polysaccharide not included in the vaccination program. The recent outbreaks after the 2000 Hajj pilgrimage, caused by W135 ET-37 complex meningococci (39,40), may reflect exactly such selection. These findings support the development of comprehensive conjugate vaccines that include capsular polysaccharides from formerly rare causes of disease such as serogroup X.
Acknowledgments
We gratefully acknowledge the helpful comments and support of Alex Nazzar and Daniel Falush and the receipt of isolates from Dominique Caugant, Tanja Popovic, Ed Kaczmarski, and Mohamed-Kheir Taha. This publication made use of the Neisseria MultiLocus Sequence Typing website (http://neisseria.mlst.net) developed by Man-Suen Chan, funded by the Wellcome Trust, and located at the University of Oxford. We thank Keith Jolly for allowing the use of deposited data. Technical assistance by Santama Abdulai, Titus Teï, Susanne Faber, Marion Moebes, and Barica Kusecek is greatly appreciated.
This work was funded by the Stanley Thomas Johnson Foundation. Part of the work (P.K.) was supported by the research grant NI/6882-3 of the Internal Grant Agency of the Ministry of Health of the Czech Republic.
Biography
Dr. Gagneux is a postdoctoral research fellow at the Swiss Tropical Institute. His research interests focus on the epidemiologic and genetic characterization of Neisseria meningitidis and Mycobacterium tuberculosis.
Footnotes
Suggested citation: Gagneux S, Wirth T, Hodgson A, Ehrhard I, Morelli G, Kriz P, et al. Clonal Groupings in Serogroup X Neisseria meningitides. Emerg Infect Dis. [serial on the Internet]. 2002 May [date cited]. Available from http://www.cdc.gov/ncidod/EID/vol8no5/01-0227.htm
References
- 1.Achtman M. Global epidemiology of meningococcal disease. In: Cartwright KA, editor. Meningococcal disease. Chichester, UK: John Wiley, 1995:159-75. [Google Scholar]
- 2.Bories S, Slaterus KW, Faucon R, Audiffren P, Vandekerkove M. Peut-on individualiser deux nouveaux groupes sérologiques de Neisseria meningitidis? Med Trop. 1966;26:603–16. [Google Scholar]
- 3.Evans JR, Artenstein MS, Hunter DH. Prevalence of meningococcal serogroups and description of three new groups. Am J Epidemiol. 1968;87:643–6. [DOI] [PubMed] [Google Scholar]
- 4.Ryan NJ, Hogan GR. Severe meningococcal disease caused by serogroups X and Z. Am J Dis Child. 1980;134:1173. [DOI] [PubMed] [Google Scholar]
- 5.Pastor JM, Fe A, Gomis M, Gil D. [Meningococcal meningitis caused by Neisseria meningitidis of the X serogroup]. Med Clin (Barc). 1985;85:208–9. [PubMed] [Google Scholar]
- 6.Grahlow WD, Ocklitz HW, Mochmann H. Meningococcal infections in the German Democratic Republic 1971-1984. Infection. 1986;14:286–8. 10.1007/BF01643964 [DOI] [PubMed] [Google Scholar]
- 7.Hansman D. Meningococcal disease in South Australia: incidence and serogroup distribution 1971-1980. J Hyg (Lond). 1983;90:49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riou JY, Djibo S, Sangare L, Lombart JP, Fagot P, Chippaux JP, et al. A predictable comeback: the second pandemic of infections caused by Neisseria meningitidis serogroup A subgroup III in Africa, 1995. Bull World Health Organ. 1996;74:181–7. [PMC free article] [PubMed] [Google Scholar]
- 9.Gagneux S, Hodgson A, Ehrhard I, Morelli G, Genton B, Smith T, et al. Microheterogeneity of serogroup A (subgroup III) Neisseria meningitidis during an outbreak in northern Ghana. Trop Med Int Health. 2000;5:280–7. 10.1046/j.1365-3156.2000.00550.x [DOI] [PubMed] [Google Scholar]
- 10.Etienne J, Sperber G, Adamou A, Picq JJ. [Epidemiological notes: meningococcal meningitis of serogroup X in Niamey (Niger)]. Med Trop. 1990;50:227–9. [PubMed] [Google Scholar]
- 11.Campagne G, Schuchat A, Djibo S, Ousseini A, Cisse L, Chippaux JP. Epidemiology of bacterial meningitis in Niamey, Niger, 1981-96. Bull World Health Organ. 1999;77:499–508. [PMC free article] [PubMed] [Google Scholar]
- 12.Gagneux S, Hodgson A, Smith T, Wirth T, Ehrhard I, Morelli G, et al. Prospective study of a serogroup X Neisseria meningitidis outbreak in Northern Ghana. J Infect Dis. 2001. In press. [DOI] [PubMed] [Google Scholar]
- 13.Swart AG, Fijen CA, te Bulte MT, Daha MR, Dankert J, Kuijper EJ. [Complement deficiencies and meningococcal disease in The Netherlands]. Ned Tijdschr Geneeskd. 1993;137:1147–52. [PubMed] [Google Scholar]
- 14.Fijen CA, Kuijper EJ, Te BM, van de Heuvel MM, Holdrinet AC, Sim RB, et al. Heterozygous and homozygous factor H deficiency states in a Dutch family. Clin Exp Immunol. 1996;105:511–6. 10.1046/j.1365-2249.1996.d01-777.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morla N, Guibourdenche M, Riou JY. Neisseria spp. and AIDS. J Clin Microbiol. 1992;30:2290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. 10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caugant DA. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS. 1998;106:505–25. [PubMed] [Google Scholar]
- 18.Caugant DA, Froholm LO, Bovre K, Holten E, Frasch CE, Mocca LF, et al. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci U S A. 1986;83:4927–31. 10.1073/pnas.83.13.4927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu P, van der Ende A, Falush D, Brieske N, Morelli G, Linz B, et al. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc Natl Acad Sci U S A. 2001;98:5234–9. 10.1073/pnas.061386098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linz B, Schenker M, Zhu P, Achtman M. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol Microbiol. 2000;36:1049–58. 10.1046/j.1365-2958.2000.01932.x [DOI] [PubMed] [Google Scholar]
- 21.Kriz P, Giorgini D, Musilek M, Larribe M, Taha MK. Microevolution through DNA exchange among strains of Neisseria meningitidis isolated during an outbreak in the Czech Republic. Res Microbiol. 1999;150:273–80. 10.1016/S0923-2508(99)80052-7 [DOI] [PubMed] [Google Scholar]
- 22.Morelli G, Malorny B, Muller K, Seiler A, Wang JF, del Valle J, et al. Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol Microbiol. 1997;25:1047–64. 10.1046/j.1365-2958.1997.5211882.x [DOI] [PubMed] [Google Scholar]
- 23.Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90:4384–8. 10.1073/pnas.90.10.4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achtman M, van der Ende A, Zhu P, Koroleva IS, Kusecek B, Morelli G, et al. Molecular epidemiology of serogroup A meningitis in Moscow, 1969 to 1997. Emerg Infect Dis. 2001;7:420–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bionumerics 2.0. Sint-Martens-Latem, Belgium: Applied Maths; 2000.
- 26.Jolley KA, Kalmusova J, Feil EJ, Gupta S, Musilek M, Kriz P, et al. Carried meningococci in the Czech Republic: a diverse recombining population. J Clin Microbiol. 2000;38:4492–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes EC, Urwin R, Maiden MC. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 1999;16:741–9. [DOI] [PubMed] [Google Scholar]
- 28.Bart A, Barnabé C, Achtman M, Dankert J, van der Ende A, Tibayrenc M. Strong linkage disequilibrium between different genetic markers challenges the epidemic clonality model in Neisseria meningitidis serogroup A isolates. Infect Genet Evol. 2001;1:117–22. 10.1016/S1567-1348(01)00011-9 [DOI] [PubMed] [Google Scholar]
- 29.Cartwright KA, Stuart JM, Jones DM, Noah ND. The Stonehouse survey: nasopharyngeal carriage of meningococci and Neisseria lactamica. Epidemiol Infect. 1987;99:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith T, Lehmann D, Montgomery J, Gratten M, Riley ID, Alpers MP. Acquisition and invasiveness of different serotypes of Streptococcus pneumoniae in young children. Epidemiol Infect. 1993;111:27–39. 10.1017/S0950268800056648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day NPJ, Moore CE, Enright MC, Berendt AR, Maynard Smith J, Murphy MD, et al. A link between virulence and ecological abundance in natural populations of Staphylococcus aureus. Science. 2001;292:114–6. 10.1126/science.1056495 [DOI] [PubMed] [Google Scholar]
- 32.Blakebrough IS, Greenwood BM, Whittle HC, Bradley AK, Gilles HM. The epidemiology of infections due to Neisseria meningitidis and Neisseria lactamica in a northern Nigerian community. J Infect Dis. 1982;146:626–37. [DOI] [PubMed] [Google Scholar]
- 33.Hassan-King MK, Wall RA, Greenwood BM. Meningococcal carriage, meningococcal disease and vaccination. J Infect. 1988;16:55–9. 10.1016/S0163-4453(88)96117-8 [DOI] [PubMed] [Google Scholar]
- 34.Tikhomirov E, Santamaria M, Esteves K. Meningococcal disease: public health burden and control. World Health Stat Q. 1997;50:170–7. [PubMed] [Google Scholar]
- 35.Ramsay ME, Andrews N, Kaczmarski EB, Miller E. Efficacy of meningococcal serogroup C conjugate vaccine in teenagers and toddlers in England. Lancet. 2001;357:195–6. 10.1016/S0140-6736(00)03594-7 [DOI] [PubMed] [Google Scholar]
- 36.Maiden MC, Spratt BG. Meningococcal conjugate vaccines: new opportunities and new challenges. Lancet. 1999;354:615–6. 10.1016/S0140-6736(99)00252-4 [DOI] [PubMed] [Google Scholar]
- 37.Swartley JS, Marfin AA, Edupuganti S, Liu LJ, Cieslak P, Perkins B, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 1997;94:271–6. 10.1073/pnas.94.1.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogel U, Claus H, Frosch M. Rapid serogroup switching in Neisseria meningitidis. N Engl J Med. 2000;342:219–20. 10.1056/NEJM200001203420319 [DOI] [PubMed] [Google Scholar]
- 39.Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, et al. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet. 2000;356:2159. 10.1016/S0140-6736(00)03502-9 [DOI] [PubMed] [Google Scholar]
- 40.Popovic T, Sacchi CT, Reeves MW, Whitney AM, Mayer LW, Noble CA, et al. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg Infect Dis. 2000;6:428–9. [DOI] [PMC free article] [PubMed] [Google Scholar]