Abstract
In an epizootiologic survey of 122 rodents captured in Vladivostok, Russia, antibodies positive for hantavirus were found in Apodemus peninsulae (4/70), A. agrarius (1/39), and Clethrionomys rufocanus (1/8). The hantavirus sequences identified in two seropositive A. peninsulae and two patients with hemorrhagic fever with renal syndrome (HFRS) from the Primorye region of Far East Russia were designated as Solovey and Primorye, respectively. The nucleotide sequences of the Solovey, Primorye, and Amur (obtained through GenBank) sequences were closely related (>92% identity). Solovey and Primorye sequences shared 84% nucleotide identity with the prototype Hantaan 76-118. Phylogenetic analysis also indicated a close relationship between Solovey, Primorye, Amur, and other viruses identified in Russia, China, and Korea. Our findings suggest that the Korean field mouse (A. peninsulae) is the reservoir for a hantavirus that causes HFRS over a vast area of east Asia, including Far East Russia.
Keywords: Hantavirus, Apodemus peninsulae, hemorrhagic fever with renal syndrome, HFRS, Far East Russia
Currently, at least 20 serotypes and genotypes of the Hantavirus genus (family: Bunyaviridae) have been identified worldwide. Rodents are the natural reservoir for hantaviruses, although one virus strain has been isolated from the house shrew (Suncus murinus), an insectivore (1). A unique characteristic of hantaviruses is the close association between the virus type and its natural reservoir (2).
Hantaviruses cause two forms of human disease, hemorrhagic fever with renal syndrome (HFRS), and hantavirus pulmonary syndrome (HPS); human infection occurs after the inhalation of aerosolized rodent excreta. HFRS is manifested as high fever, renal dysfunction, and hemorrhage; HPS is characterized by an acute progressive pulmonary edema and a fatality rate of about 40%. Among the hantaviruses that cause HFRS in Eurasia are Hantaan virus (HTNV), Seoul virus (SEOV), Puumala virus (PUUV), and Dobrava-Belgrade virus (DOBV) (3), which are carried by the striped field mouse (Apodemus agrarius), Norway rat (Rattus norvegicus) and black rat (R. rattus), bank vole (Clethrionomys glareolus), and yellow-necked field mouse (A. flavicollis), respectively. DOBV was also found in A. agrarius in Europe (4,5). Sin Nombre virus (SNV), New York virus (NYV), Black Creek Canal virus (BCCV), Bayou virus (BAYV), Andes virus (ANDV), and other related viruses cause HPS in the New World and are carried by the deer mouse (Peromyscus maniculatus), white-footed mouse (P. leucopus), cotton rat (Sigmodon hispidus), marsh rice rat (Oryzomys palustris), and Oligoryzomys longicaudatus, respectively (2,6). Although the known genotypes and serotypes have increased in number with advances in the knowledge of epidemiology and epizootiology of hantavirus infection (2), some still-unidentified hantaviruses carried by specific rodent hosts may exist. HFRS is generally known to be endemic to Far East Russia. However, the genetics of hantaviruses that are pathogenic for humans are not well defined. Reed voles (Microtus fortis) in Far East Russia were found to harbor two novel hantaviruses, Khabarovsk virus (KHAB) and Vladivostok virus (7,8). Another hantavirus, Topografov virus (TOPV), was isolated from brown lemmings (Lemmus sibiricus). The correlation between these three viruses and their pathogenicity for humans are not yet known (9).
A recent study reported two novel hantaviruses, designated as Amur (AMR) and Far East (FE), that were identified from HFRS patients in Far East Russia (10). The natural reservoir of AMR genotype seems to be A. peninsulae, according to a recent study on nucleotide sequence comparisons by Yashina et al. (11).
In 1999, we carried out an epizootiologic survey in a suburb of Vladivostok, Russia, to determine the characteristics of hantaviruses circulating in Far East Russia and to examine the possibility that A. peninsulae is a carrier of pathogenic hantaviruses. We detected antibodies to hantaviruses in A. peninsulae, and the viral genome characteristics were extremely similar to the newly identified genotype, AMR (10). Using phylogenetic analysis to characterize the sequences of viruses identified from HFRS patients and A. peninsulae, we were able to corroborate the assumption of Yashina et al. (11). We also found that A. peninsulae-related viruses are pathogenic for humans and are distributed over a large area of east Asia that includes Far East Russia.
Materials and Methods
We collected sera and organs from wild rodents captured during 1999. We also collected sera and autopsy materials from HFRS patients in two rural villages in the Primorye region of Russia, located 400 km and 600 km from Vladivostok.
Rodent sera were screened for antibodies to HTNV and PUUV or both by indirect immunofluorescent antibody assay (IFA). Vero E6 cells infected with the Hantaan 76-118 strain of HTNV or the Sotkamo strain of PUUV were used as antigen slides. Diluted sera (1:16 and 1:64) were spotted onto the antigen slides and incubated at 37°C for 1 h. After three washes with phosphate-buffered saline (PBS), protein G-conjugated fluorescein isothiocyanate (FITC) (Zymed Laboratories, Inc., San Francisco, CA) was spotted onto the slides. After incubation at 37°C for 1 h, the slides were washed and observed by fluorescence microscopy. Scattered, granular fluorescence in the cytoplasm of infected Vero E6 cells was considered a positive reaction. Antibodies in HFRS patient sera were detected by the same protocol, except for the substitution of FITC-conjugated antihuman immunoglobulin (Ig) G (ICN Pharmaceuticals, Inc., Aurora, OH).
Total RNA was extracted from lung tissues of seropositive A. peninsulae with Isogen (Nippon Gene Co., Ltd., Osaka, Japan), which is based on the acid guanidium-phenol-chloroform technique, according to manufacturer’s instructions. Similarly, total RNA was extracted from lung, liver, kidney, spleen, and brain tissues of HFRS patients. Reverse transcription (RT) was carried out at 42°C for 30 min by using Superscript II and random primer (Gibco-BRL, Rockville, MD). Full-length S segments were amplified with Platinum Taq (Gibco-BRL) and HTNV–full S primer for 30 polymerase chain reaction (PCR) cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 68°C for 2 min. Amplification of M segments was identical to that of S segments, except for the use of M genome-specific primers (Table 1). Part of the M segment (232 nucleotides) and the entire S segment (except for the 5´ and 3´ ends) were sequenced with primers specific for HTNV or SEOV or both. Amplification of the partial M segment was achieved only with nested PCR. The PCR-amplified products were separated by using a Rapid Gel Extraction kit (Gibco-BRL) according to the manufacturer’s instructions. Purified DNA fragments were cloned into the PCR 2.1 vector provided in the TA cloning kit (Invitrogen Corporation, Carlsbad, CA). The ligated products were transformed into Top 10 competent cells (Invitrogen Corporation) and purified with a Miniprep kit (QIAGEN GmbH, Hilden, Germany). DNA sequencing was performed with the ABI-PRISM Dye Terminator Sequencing kit (Applied Biosystems, Foster City, CA) and an ABI 373-A genetic analyzer.
Table 1. Primers used for reverse transcription-polymerase chain reaction and/or sequencing of S and M genome segments of hantaviruses.
| Gene | Primer name | Primer sequence (5´–3´) | Position |
|---|---|---|---|
| S segment | M13 Fw | ctggccgtcgttttac | |
| PEN 215 S Fw | gaattgaaagacaattggc | 215–233 | |
| KPS3a | tc(a/c)agcatgaaggc(a/t)gaagagat | 592–703 | |
| PEN 780 SFw | acagaggcaggcagctttag | 780–799 | |
| PEN 1042 S Fw | gcaggatatgcggaatacaa | 1042–1061 | |
| HTNV 1390 S Fw | attgcactattattatcagg | 1390–1409 | |
| HTNV Full S | ttctgcagtagtagtag(t)a(g)ctccctaa | ||
| PEN180 S Rv | ttccctgtctgttaatgctc | 180–199 | |
| PEN 585 S Rv | tgggcaaggacacatagaga | 585–604 | |
| PEN 946 Rv | atgatggtgactcgatgtct | 946–965 | |
| PEN 1160 S Rv | gttgtattcccattgactgt | 1160–1179 | |
| HTNV 1493 SRv | cacccacaacggattaactg | 1493–1512 | |
| M13 Rv | caggaaacagctatgac | ||
| M segment | HS1a | ac(a/c)tgtca(c/a)tttgg(a/t)gaccc | 2636–2655 |
| HS2a | tcaca(g/a)gcctttattga(g/t)gt | 3072–3091 | |
| HS3a | t(t/c)aggaa(ga)aaatg(tc)aactttgc | 2715–2736 | |
| HS4a | acacc(a/t)gaaccccaggc(a/c)cc | 3000–3019 | |
| M13 Fw | ctggccgtcgttttac | ||
| M13 Rv | caggaaacagctatgac |
aPrimers designed by Yashina et al.
We used the ClustalX program package (version 1.81; available from: URL: ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/) to generate the phylogenetic trees by using the neighbor-joining method with 1000 bootstrap replicates. Hantavirus sequences used in the comparisons were obtained from GenBank. The S and M genome sequences used in this study are listed in Table 2.
Table 2. Hantavirus sequences used in this studya.
| Virus type | Strain | Source | Country |
Accession nos. |
References | ||
|---|---|---|---|---|---|---|---|
| Region | Location | M | S | ||||
| HTNV | |||||||
| SL/AP61/1999 | Apodemus peninsulae | Far East Russia | Solovey | AB071185 | AB071183 | This report | |
| SL/AP63/1999 | A.peninsulae | Far East Russia | Solovey | AB071186 | AB071184 | This report | |
| PRI/H1/2000 | Human | Primorye | Cavalerovo | AB071187 | —b | This report | |
| PRI/H2/2000 | Human | Primorye | Cavalerovo | AB071188 | — | This report | |
| AMR/680 | A.peninsulae | Far East Russia | Khabarvosk | AF332571 | — | 11 | |
| AMR/1166 | A.peninsulae | Far East Russia | Khabarvosk | AF332569 | — | 11 | |
| AMR/1169 | A.peninsulae | Far East Russia | Khabarvosk | AF332570 | — | 11 | |
| AMR/4234 | Human | Far East Russia | Amursk | AF172422 | — | 10 | |
| AMR/4309 | Human | Far East Russia | Amursk | AF172423 | — | 10 | |
| AMR/4313 | Human | Far East Russia | Korphovsky | AF172424 | — | 10 | |
| H8205 | Human | China | — | AB030232 | — | — | |
| HTNV261 | — | China | Heilongjiang | — | AF252259 | — | |
| Z10 | Human | China | Zhejiang | AB027076 | AB027108 | 12 | |
| Chen4 | Human | China | Anhui | — | AB027101 | 12 | |
| Maaji1 | A. agrarius | Korea | — | — | AF321094 | Lee PW c | |
| Maaji-2 | Human | Korea | — | — | AF321095 | Lee PW c | |
| HTN 76-118 | A. agrarius | South Korea | — | M14627 | M14626 | 13,14 | |
| Q32 | — | China | Guizhou | — | AB027097 | 12 | |
| HV114 | A. agrarius | China | Hubei | L08753 | AB027110 | 12,15 | |
| A9 | A. agrarius | China | Jiangsu | AF035831 | — | 16 | |
| Hojo | Human | South Korea | — | D00376 | — | 17 | |
| FE/7866 | Human | Far East Russia | Razdolnoye | AF172439 | — | 10 | |
| NC167 | Niviventer confucianus | China | Anhui | AB027115 | AB027523 | 12 | |
| H3 | Human | China | Hubei | — | — | 18 | |
| H5 | Human | China | Heilongjiang | — | — | 18 | |
| A3 | A. agrarius. | China | Zhejiang | AB027055 | — | 12 | |
| B78 | Human | China | Shandong | AB027056 | AB027093 | 12 | |
| Q36 | A. agrarius | China | Guizhou | AB027057 | AB027094 | 12 | |
| Q7 | A. agrarius | China | Guizhou | AB02058 | AB027095 | 12 | |
| Q20 | A. agrarius | China | Guizhou | AB027059 | AB027096 | 12 | |
| Niongxia-A | A. agrarius | China | Niongxia | AB027060 | — | 12 | |
| Q10 | A. agrarius | China | Guizhou | AB027062 | AB027098 | 12 | |
| A16 | A. agrarius | China | Sanxi | AB027063 | AB027099 | 12 | |
| Q37 | A. agrarius | China | Guizhou | AB027064 | AB027100 | 12 | |
| Q33 | A. agrarius | China | Guizhou | AB027065 | AB027102 | 12 | |
| Bao9 | A. agrarius | China | Heilongjiang | AB027066 | AB027103 | 12 | |
| Jiang13 | A. agrarius | China | Heilongjiang | AB027067 | AB027104 | 12 | |
| Bao14 | A. agrarius | China | Heilongjiang | AB027068 | AB027105 | 12 | |
| Bao10 | A. agrarius | China | Heilongjiang | AB027069 | AB027106 | 12 | |
| Lee | Human | South Korea | — | D00377 | — | 17 | |
| 62HTNV | — | — | — | AB027070 | — | 12 | |
| 6B | — | — | — | AB027071 | — | 12 | |
| Vaccine | — | — | — | AB027072 | — | 12 | |
| H2 | — | North Korea | — | AB027073 | AB027107 | 12 | |
| HN26-L | A. agrarius | China | Hainan | AB027074 | — | 12 | |
| Luyao | Human | China | Shandong | — | AB027109 | 12 | |
| B659 | Human | China | Shandong | S72339 | — | 18 | |
| Hu | Human | China | Hubei | AB027077 | AB027111 | 12 | |
| Q83 | — | — | Guizhou | AB027078 | — | 12 | |
| B256 | — | — | — | AB027079 | AB027112 | 12 | |
| Thailand | Bandicota indica | Thailand | — | L08756 | — | — | |
| Topografov | Lemmus sibericus | Far East Russia | Siberia | AJ011647 | — | 9 | |
| SEOV | |||||||
| L99 | Rattus losea | China | Jiangxi | AF035833 | AF288299 | — | |
| SR11 | R. norvegicus | Japan | Sapporo | M34882 | M34881 | 19 | |
| Gou3 | R. rattus | China | Zhejiang | AB027521 | AB027522 | 12 | |
| NM39 | R. norvegicus | China | Neimeng | AB027080 | — | 12 | |
| HB55 | Human | China | Henan | AF035832 | — | 17 | |
| Wan | Human | China | Jiangsu | AB027081 | — | 12 | |
| J12 | Human | China | Jieling | AB027082 | — | 12 | |
| Henan94 | R. norvegicus | China | Henan | AB027083 | — | 12 | |
| Shanxi | — | — | — | AB027084 | — | 12 | |
| HN71-L | R. norvegicus | China | Hainan | AB027085 | — | 12 | |
| Guang199 | — | — | — | AB027086 | — | 12 | |
| Beijing-Rn | R. norvegicus | China | Beijing | AB027087 | — | 12 | |
| c3 | Human | China | Hebei | AB027088 | — | 12 | |
| Hebei4 | Cricetulus barabensis | China | Hebei | AB027090 | — | 12 | |
| SD227 | — | China | Shangdong | AB027091 | — | 12 | |
| SD10 | R. norvegicus | China | Shangdong | AB027092 | — | 12 | |
| Hbei1 | Human | China | Hubei | S72343 | — | 17 | |
| Seoul | R. norvegicus | South Korea | — | S47716 | — | 20 | |
| Tchoupitoulas | R. norvegicus | North America | — | U00473 | — | 21 | |
| B-1 | R. norvegicus | Japan | — | X53861 | — | 22 | |
| Girard Point | R. norvegicus | North America | — | U00464 | — | — | |
| DOBV | |||||||
| DOB/SLOV | A. flavicollis | Slovenia | — | L33685 | L41916 | 23 | |
| DOB/SAA | A. agrarius | Estonia | — | AJ009774 | AJ009773 | 4 | |
| SNV | SNV | Peromyscus maniculatus | North America | — | L25783 | L25784 | 24 |
| PUUV | PUU/Sot | Clethrionomys glareolus | Finland | — | X61034 | — | 25 |
| Kamiiso | C. rufocanus | Japan | Kamiiso | AB011631 | — | 8 | |
| KHAB | Khabarovsk | Microtis fortis | Far East Russia | Khabarvosk | AJ011648 | — | 9 |
aAbbreviations used: HTNV and HTN, Haantan virus; SL, Solovey; PRI, Primorye; AMR, Amur; SEOV, Seoul virus; DOB and DOBV, Dobrava-Belgrade virus; SLOV, Slovenia; SAA, Saarema; SNV, Sin Nombre virus; PUUV, Puumala virus; and KHAB, Khabarovsk virus. b —, not reported/not used in this study. cPers. comm.
Formalin-fixed lung, liver, kidney, and brain tissues from an HFRS patient who died of acute renal failure were observed under light microscopy and subjected to immunohistochemical analysis with monoclonal antibodies against Hantaan virus.
Results
We carried out the epizootiologic survey on 122 rodents captured in a suburb of Vladivostok; results of serologic screening of rodent sera by IFA are shown in Table 3. Identified rodent species included (70) A. peninsulae, (39) A. agrarius, (8) C. rufocanus, (3) M. fortis, and (2) Tamias sibiricus. Screening by IFA showed that one A. agrarius (2.5%), four A. peninsulae (5.7%), and one C. rufocanus (12.5%) had antibodies to HTNV or PUUV or both. HTNV-antibody titers ranged from 1:32 to 1:512. All the seropositive rodents, except for C. rufocanus, lacked antibody against PUUV (Table 4). Lung tissues from seropositive A. peninsulae were subjected to RT-PCR to amplify the virus genomes. Two of the four rodents with high IFA titers to HTNV (1:256 and 1:512) were positive by PCR for both the S and M segments of hantavirus.
Table 3. Serologic screening by immunofluorescent antibody assay for Haantan virus and Puumala virus antibodies in rodents, Vladivostok, Russiaa.
| Rodent species | No. of sera tested | Positives by IFA (%) |
|
|---|---|---|---|
| HTNV | PUUV | ||
| Apodemus peninsulae | 70 | 4(5.7) | 0 |
| A. agrarius | 39 | 1(2.5) | 0 |
| Clethrionomys rufocanus | 8 | 1(12.5 | 1(12.5) |
| Microtus fortis | 3 | 0 | 0 |
| Tamias sibiricus | 2 | 0 | 0 |
| Total | 122 | 6(4.9) | 1(0.8) |
aAbbreviations used: IFA, immunofluorescent antibody assay; HTNV, Haantan virus; PUUV, Puumala virus.
Table 4. Haantan virus and Puumala virus antibody titers determined by immunofluorescent antibody assay and polymerase chain reaction results.
| Species | Sample number | IFA antibody titer |
PCR | |
|---|---|---|---|---|
| HTNV | PUU | |||
| Apodemus peninsulae | 47 | 256 | <16 | -b |
| A. peninsulae | 61 | 512 | <16 | +c |
| A. peninsulae | 63 | 256 | <16 | + |
| A. peninsulae | 74 | 64 | <16 | - |
| A. agrarius | 10 | 32 | <16 | NA |
| Clethrionomys rufocanus | 32 | 256 | 256 | ND |
aAbbreviations used: HTNV, Haantan virus; PUUV, Puumala virus; IFA, immunofluorescent antibody assay; PCR, polymerase chain reaction; NA, not available; ND, not done. b-, negative. c+, positive.
We obtained the clinical histories of two fatal cases of HFRS in the Primorye region. The patients, who lived in villages 400 km and 600 km from Vladivostok, died 8–13 days after the onset of illness; gastrointestinal bleeding and acute renal failure were the causes of death. Serologic screening showed that both patients were positive for hantaviral antibodies. Antibody titers to HTNV and SEOV were apparently higher than to PUUV. We used lung, liver, kidney, spleen, and brain tissues of these HFRS patients for RT-PCR analysis; the lung and kidney tissues of patient no. 1 and the spleen tissue of patient no. 2 were positive for hantaviral M segment.
To examine the histopathologic changes in HFRS patients, we used light microscopy to examine sections of formalin-fixed lung, liver, kidney, spleen, and brain tissues from patient no. 2, who had died of acute renal failure (Figure 1). The kidney was the only tissue that showed the recognizable histopathologic changes. Salient changes included interstitial edema with mild infiltration of mononuclear cells (Figure 1, small arrow) and degeneration of renal tubules (Figure 1, large arrow) in the cortex (Figure 1, A). Although proteinaceous casts and exudates were observed in the lumina of renal tubules (Figure 1, arrowhead), there were no apparent glomerular changes. In addition, a prominent well-defined necrotic lesion (Figure 1, asterisk) was noted in the medulla (Figure 1, B). Viral antigens were not detected in these specimens by using monoclonal antibodies to HTNV.
Figure 1.
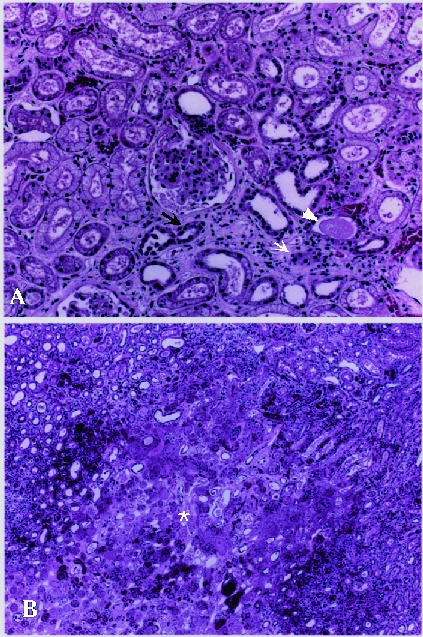
Histopathologic changes in kidney tissue from a patient with hemorrhagic fever with renal syndrome, Primorye region. Changes include interstitial edema with mild infiltration of mononuclear cells (small arrow) and degeneration of renal tubules (large arrow) in cortex. Proteinaceous casts and exudate (arrowhead) are seen in lumina of renal tubules (A). No apparent glomerular changes. Most prominent change in the medulla is well–defined necrotic lesion (asterisk) (B).
The entire S segments of the viruses from two seropositive A. peninsulae were amplified and sequenced. We designated these segments as Solovey/AP61/1999 and Solovey/AP63/1999 based on the name of the village closest to the survey point, the rodent species from which the sample was taken, and the year in which the epizootiologic survey was done. We compared the coding regions of these sequences with those of other hantaviruses (Table 5). The S segments of the two Solovey sequences had 99.0% and 98.8% identities in nucleotide and amino acid sequences, respectively. Solovey sequences and Hantaan viruses had 78.2%–84.5% nucleotide sequence identity and 86.7%–93.3% amino acid sequence identity, regardless of their source or geographical origin. Lower nucleotide sequence identities were seen than in Solovey sequences and other viruses: DOBV (73.6%), SEOV (73.9%), and SNV (63.9%).
Table 5. Comparison of nucleotide (open reading frame) and amino acid of S genome between those from Apodemus peninsulae and other hantavirusesa.
| Nucleotide and amino acid identities %b |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL/AP61 | SL/AP63 | HTNV261 | Z10 | Chen4 | Maaji-1 | HTNV 76-118 | Q32 | NC1167 | SR11 | GOU3 | Dob/Slo | |
| SL/AP61 | 99.0 | 84.5 | 83.5 | 83.4 | 82.9 | 82.7 | 82.3 | 78.3 | 73.7 | 73.7 | 72.9 | |
| SL/AP63 | 98.8 | 84.2 | 83.5 | 83.4 | 82.9 | 82.8 | 81.5 | 78.2 | 73.9 | 73.8 | 72.2 | |
| HTNV261 | 91.9 | 91.5 | 85.6 | 85.7 | 83.0 | 88.6 | 84.7 | 78.9 | 74.1 | 73.6 | 72.6 | |
| Z10 | 91.9 | 91.5 | 92.9 | 89.1 | 83.6 | 85.9 | 87.5 | 79.8 | 75.3 | 74.2 | 73.3 | |
| Chen4 | 93.0 | 92.5 | 93.2 | 96.2 | 82.8 | 85.8 | 90.3 | 78.7 | 73.2 | 74.2 | 73.4 | |
| Maaji-1 | 91.5 | 90.8 | 90.8 | 91.3 | 93.0 | 82.9 | 82.1 | 78.2 | 74.2 | 73.0 | 74.2 | |
| HTNV76-118 | 92.2 | 91.5 | 94.9 | 92.9 | 93.7 | 91.0 | 84.4 | 78.2 | 74.6 | 73.8 | 74.0 | |
| Q32 | 92.7 | 92.3 | 93.7 | 94.4 | 96.0 | 91.8 | 93.2 | 79.1 | 73.1 | 74.3 | 73.8 | |
| NC167 | 87.2 | 86.7 | 85.3 | 85.8 | 85.3 | 84.8 | 86.9 | 85.1 | 75.3 | 73.6 | 72.7 | |
| SR11 | 75.0 | 74.5 | 74.1 | 73.9 | 74.6 | 74.3 | 74.8 | 74.1 | 77.2 | 87.8 | 73.7 | |
| GOU3 | 75.7 | 75.5 | 75.0 | 74.8 | 76.2 | 74.3 | 74.8 | 76.7 | 76.7 | 91.5 | 73.1 | |
| Dob/Slo | 76.4 | 76.4 | 76.8 | 75.7 | 77.6 | 76.6 | 75.5 | 77.2 | 76.0 | 73.1 | 73.1 | |
aValues in bold show the close identities between the two Solovey sequences. Abbreviations used: SL, Solovey; HTNV, Haantan virus; Dob, Dobrova; Slo, Slovenia. bValues above the diagonal and to the right show nucleotide identities; those below the diagonal and to the left show amino acid identities.
To explore the genetic diversity of hantaviruses identified in A. peninsulae in more detail, we sequenced the partial M segment of the G2 region (232 nt). We also sequenced the partial M segments of genetic lineages identified in the two HFRS patients from the Primorye region, designated as Primorye/H1/2000 and Primorye/H2/2000. The M segment of Solovey and Primorye sequences were compared with those of other hantaviruses (Table 6). Nucleotide sequence identities among these sequences were between 92.2% and 98.2%; amino acid sequence identities were almost identical (98.7%–100%). We also compared the M segment sequences of Solovey and Primorye with those of AMR genetic lineage, recently identified in HFRS patients and A. peninsulae in Far East Russia (10,11). The nucleotide and amino acid identities between Solovey, Primorye , and AMR lineages were 91.3%–98.3% and 93.5%–98.7%, respectively. The M segment sequences of Solovey, Primorye , and AMR lineages were compared with that of H8205, isolated from an HFRS patient in China. In this case, the nucleotide sequence identities were 93.5%–96.1%, and the amino acid sequence identities were 94.8%–100%. Lower nucleotide identities were seen with HTNV (78.8%–86.2%), SEOV (79.3%–81.4%), and DOBV (75.8%–77.1%). This high level of sequence identity among Solovey, Primorye, AMR, and H8205 sequences suggests that some patients acquired the infection from the Korean field mouse (A. peninsulae) in Far East Russia and China. Our results also suggest that this genetic lineage is widely distributed throughout east Asia.
Table 6. Comparison of nucleotide (bases 2737–2969)a and amino acid of M genome between those from Primorye patients, Apodemus peninsulae, and other hantaviruses.
| Nucleotide and amino acid identities % b |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SL/ AP61 | SL/ AP63 | AMR/ 1169 | PRI/ H1 | PRI/ H2 | H8205 | AMR/ 4313 | HV 114 | A9 | HTNV 76-118 | Hojo | FE | NC167 | DOB/ Slo | SR- 11 | PUU | |
| SL/AP61c | 99.5 | 97.8 | 96.1 | 98.2 | 94.8 | 94.3 | 86.2 | 85.7 | 84.4 | 82.7 | 82.7 | 79.3 | 79.3 | 79.7 | 60.3 | |
| SL/AP63 | 100 | 97.8 | 92.2 | 94.3 | 94.8 | 94.3 | 85.7 | 85.3 | 84.0 | 82.3 | 83.1 | 78.8 | 80.1 | 81.4 | 60.7 | |
| AMR/1169 | 94.8 | 94.8 | 96.5 | 98.7 | 95.6 | 95.6 | 86.6 | 86.2 | 84.9 | 83.1 | 81.4 | 79.7 | 80.1 | 79.3 | 60.3 | |
| PRI/H1 | 100 | 100 | 94.8 | 96.9 | 93.5 | 92.2 | 84.0 | 83.6 | 83.1 | 82.3 | 80.6 | 79.3 | 78.8 | 79.3 | 60.3 | |
| PRI/H2 | 98.7 | 98.7 | 93.5 | 98.7 | 94.8 | 94.3 | 85.7 | 85.3 | 84.0 | 82.3 | 81.4 | 78.8 | 79.3 | 78.8 | 59.4 | |
| H8205 | 100 | 100 | 94.8 | 100 | 98.7 | 91.3 | 83.6 | 83.1 | 85.3 | 84.9 | 80.6 | 77.1 | 79.3 | 77.1 | 60.7 | |
| AMR/4313 | 98.7 | 98.7 | 93.4 | 98.7 | 97.4 | 98.7 | 85.7 | 85.3 | 83.6 | 81.8 | 82.7 | 78.0 | 78.0 | 78.8 | 59.9 | |
| HV114 | 93.5 | 93.5 | 88.3 | 93.5 | 92.2 | 93.5 | 92.2 | 99.5 | 86.6 | 84.4 | 87.9 | 78.4 | 75.8 | 83.1 | 51.9 | |
| A9 | 93.5 | 93.5 | 88.3 | 93.5 | 92.2 | 93.5 | 92.2 | 98.9 | 86.2 | 84.0 | 87.5 | 78.0 | 75.4 | 81.8 | 50.6 | |
| HTNV76118 | 94.8 | 94.8 | 89.6 | 94.8 | 93.5 | 94.8 | 93.5 | 97.4 | 96.1 | 94.6 | 88.7 | 79.7 | 78.4 | 76.7 | 59.9 | |
| Hojo | 94.8 | 94.8 | 89.6 | 94.8 | 93.5 | 94.8 | 93.5 | 97.4 | 96.1 | 100 | 87.9 | 78.0 | 78.8 | 76.7 | 51.5 | |
| FE | 92.2 | 92.2 | 87.0 | 92.2 | 90.9 | 92.2 | 90.9 | 87.9 | 87.5 | 97.4 | 98.7 | 75.8 | 73.7 | 78.4 | 59.9 | |
| NC167 | 86.8 | 86.8 | 80.5 | 86.8 | 85.5 | 86.8 | 85.5 | 89.5 | 88.2 | 90.8 | 90.8 | 88.2 | 75.4 | 77.5 | 49.3 | |
| DOB/Slo | 88.3 | 88.3 | 83.1 | 88.3 | 87.0 | 88.3 | 87.0 | 88.3 | 87.0 | 87.0 | 87.0 | 84.4 | 81.6 | 75.0 | 59.9 | |
| SR11 | 83.1 | 83.1 | 79.2 | 83.1 | 81.8 | 83.1 | 81.8 | 83.1 | 81.8 | 81.8 | 81.8 | 83.1 | 80.3 | 80.5 | 56.0 | |
| PUUV | 53.2 | 53.2 | 53.2 | 53.2 | 51.9 | 53.2 | 51.9 | 62.9 | 62.5 | 51.9 | 61.6 | 53.2 | 61.6 | 49.4 | 61.2 | |
aBased on Haantan 76-118. bValues above the diagonal and the right show nucleotide identities; those below the diagonal and to the left show amino acid identities. cValues in bold show the close identities between those sequences. Abbreviations used: SL, Solovey; AMR, Amur; PRI, Primorye; HTNV, Haantan virus; FE, Far East virus; DOB, Dobrova; Slo, Slovenia; PUUV, Puumala virus.
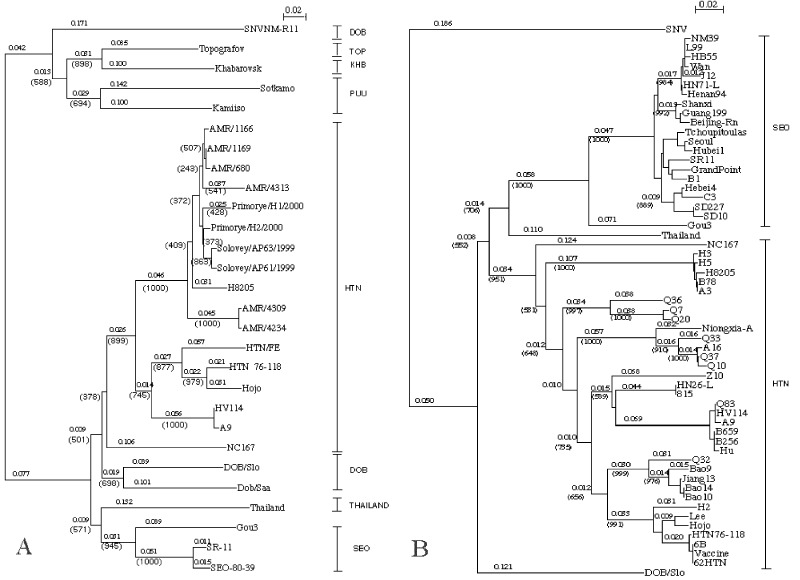
The M segments of Solovey, Primorye, and AMR sequences formed a common phylogenetic lineage with high bootstrap support values, regardless of viral origin (Figure 2, A). Furthermore, H8205 shared a common lineage with Solovey and Primorye sequences. Another phylogenetic analysis, based on a different region of the M segment, showed that Chinese virus isolates (H8205, H3, H5, and B78) formed a distinct lineage within the Hantaan clade (Figure 2, B). The phylogenetic tree constructed for the S sequences (Figure 3) showed that Solovey sequences formed a single cluster, together with Maaji1 (a Korean isolate) and B78, in a common lineage with high bootstrap support values within the Hantaan clade.
Figure 2.
Phylogenetic trees of hantavirus (A) partial M (nt 2736–2968) and (B) partial M (nt 2001–2301) segments. Trees were constructed by using ClustalX (ver. 1.81) program. Numbers above the branches are distances and those in parentheses are bootstrap support values for 1000 replicates.
Figure 3.
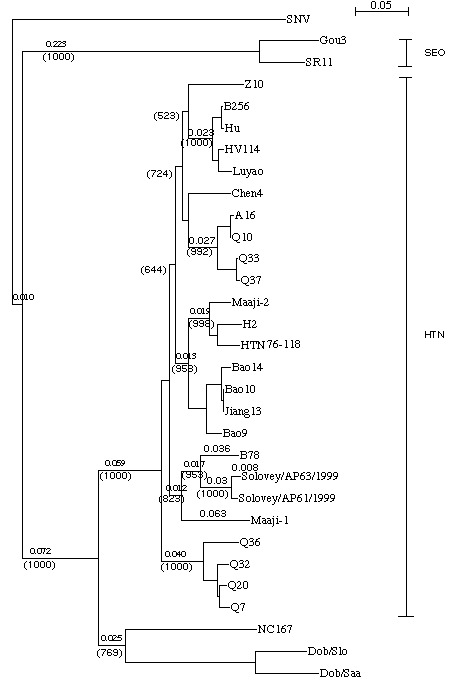
Phylogenetic tree of hantavirus partial S (nt 1216-1666) segments. Tree was constructed by using ClustalX (ver. 1.81) program. Numbers above the branches are distances and in parentheses are bootstrap support values for 1000 replicates.
To identify signature amino acids for each virus type, we compared the deduced partial amino acid sequences of their G2 regions using ClustalX multiple-sequence alignment (Figure 4). The presence of leucine or isoleucine at amino acid position (aa) 903 was unique to HTNV except for AMR lineage. The signature amino acids for SEOV were leucine at aa 918 and valine, isoleucine, and serine at aa 955-957. The signature amino acids for AMR lineage were methionine at aa 932 and aspartic acid at aa 967.
Figure 4.

Multiple alignment of partial deduced amino acid sequences of G2 region of hantaviruses. Amino acid sequences analyzed by using ClustalX (ver. 1.8) program. Amino acid positions indicated above sequences based on Haantan 76–118. First line shows the deduced amino acid of Dobrova/Saarema. Dots represent amino acids that are identical to those at corresponding positions in Dobrova/Saarema sequence. Amino acids that differ from those in the sequence are indicated at relevant positions. Hyphens are used in areas where amino acid sequence is not available. Signature amino acids are shaded.
Discussion
Each hantavirus serotype or genotype is generally associated with a specific rodent host, and various rodent species act as reservoir animals and sources of human infection. Since contact between rodents and humans occurs frequently during agricultural and forestry activities, most infections have been reported in rural areas. However, an urban epidemic of HFRS caused by SEOV has also been reported (26). A large number of rodent species may serve as reservoir animals for pathogenic hantaviruses. For example, few researchers suspected that P. maniculatus could transmit highly virulent hantavirus to humans until SNV was identified (27,28). Later studies showed that the other viral agents of HPS such as NYV, BCCV, BAYV, and ANDV, were carried by P. leucopus, S. hispidus (29), O. palustris (30), and O. longicaudatus (31), respectively. We emphasize the importance of discovering the characteristics of hantaviruses found in endemic areas and identifying the primary hosts.
Although Far East Russia has long been considered an HFRS-endemic area, few reports describe the hantaviral sequences in this region, and information on reservoir animals carrying pathogenic hantaviruses is limited. Our studies therefore focused on determining the genetic characteristics of hantaviruses circulating in this geographic area. We identified A. peninsulae as the natural reservoir rodent for a hantavirus pathogenic for humans in Far East Russia. We also identified hantavirus sequences designated as Solovey and Primorye in A. peninsulae and HFRS patients, respectively; genetic analysis showed that these sequences were very closely related to each other. This information and the pathological findings from the HFRS case in which Primorye sequence was identified strongly suggest that the virus of Solovey sequence is the causative agent of HFRS. The nucleotide sequence and phylogenetic analysis also showed that Solovey and Primorye sequences were most closely related to AMR and H8205 sequences from patients in Russia and China, but were clearly distinguishable from the prototype of Hantaan virus. Genetic and phylogenetic analysis indicated that Solovey and Primorye sequences were closely related to AMR, Maaji1, H8205, and B78 sequences, viruses derived from distant areas. While Solovey sequences were identified in a suburb of Vladivostok and PRI sequences in two villages 400 km and 600 km from Vladivostok, the H8205 and B78 viruses were derived in China, and Maaji1 was isolated in Korea. A. peninsulae is distributed in the same region where A. agrarius is prevalent in Korea (PW Lee, pers. comm.). Recently, AMR sequences were found in both HFRS patients and A. peninsulae (11). We suggest that some of the viruses circulating in the area of this study cause severe HFRS and are carried by the same host species, A. peninsulae. Comparison of the deduced hantaviral amino acid sequences showed that aspartic acid and methionine represented signature amino acids for AMR genetic lineage, regardless of the region in which the virus was identified or its origin (Figure 4). These signature amino acids may be used to distinguish AMR genetic lineage from other hantaviruses. We conclude from our results that A. peninsulae carries a hantavirus that is pathogenic for humans. Since A. peninsulae is widely distributed in Far East Russia, China, Korea, and Japan, this hantavirus and associated cases of HFRS may also be widely distributed.
In the kidney tissue of one HFRS patient (no. 2) from Primorye region, we detected pathologic changes typical of severe HFRS caused by hantavirus infection (32–35). We also detected and sequenced the partial M segment in the spleen of the same patient. However, we could not detect the viral antigen in the kidney samples, possibly because of low levels of the virus in the kidneys of this patient. Nested PCR allowed the amplification of viral M segments from the spleen, but not from kidney, of this patient.
Through epizootiologic, clinical, pathologic, and sequencing studies, we identified a hantavirus carried by A. peninsulae as one of the causative agents of HFRS. We think that this information may be helpful in preventing human infections in East Asia. Controversy persists over whether A. peninsulae carries a distinct virus type or a subtype of HTNV. A similar question arises with Dobrava/Slovenia and Dobrava/Saaremaa, which are carried by A. flavicollis and A. agrarius, respectively. The S segment identities between Dobrava/Slovenia and Dobrava/Saaremaa (both obtained from GenBank for comparison purposes) were 87.8% (nucleotide) and 92.7% (amino acid). Similarly, the nucleotide and amino acid sequence identities of the S segments of Solovey sequences and HTN 76-118 were 82.7% and 92.2%, respectively. We suggest that Solovey sequences belong to a sublineage within the HTNV clade.
Acknowledgments
We thank Kimiyuki Tsuchiya of Miyazaki Medical College and Masahiro Iwasa and Hitoshi Suzuki of Hokkaido University for providing rodent information.
This study was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Culture, and Sport, Japan (projects 1357529 and 13660311) and by Health Science Grants for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor, and Welfare, Japan.
Biography
Ms. Lokugamage is a doctoral candidate studying the epidemiology of hantaviruses at the Laboratory of Public Health, Graduate School of Veterinary Medicine of Hokkaido University, Sapporo, Japan. She has also served as a faculty lecturer at the School of Veterinary Medicine, University of Peradeniya, Sri Lanka.
Footnotes
Suggested citation for this article: Lokugamage K, Kariwa H, Hayasaka D, Cui BZ, Iwasaki T, Lokugamage N, et al. Genetic characterization of hantaviruses transmitted by the Korean field mouse (Apodemus peninsulae), Far East Russia. Emerg Infect Dis [serial online] 2002 Aug [date cited];8. Available from: URL: http://www.cdc.gov/ncidod/EID/vol8no8/01-0494.htm
References
- 1.Tang YW, Xu ZY, Zhu ZY, Tsai TF. Isolation of haemorrhagic fever with renal syndrome virus from Suncus murinus, an insectivore. Lancet. 1985;1:513–4. 10.1016/S0140-6736(85)92108-7 [DOI] [PubMed] [Google Scholar]
- 2.Plyusnin A, Vapalahti O, Vaheri A. Hantaviruses: genome structure, expression and evolution. J Gen Virol. 1996;77:2677–87. 10.1099/0022-1317-77-11-2677 [DOI] [PubMed] [Google Scholar]
- 3.Clement J, Heyman P, McKenna P, Colson P, Avsic-Zupanc T. The hantaviruses in Europe: from the bedside to the bench. Emerg Infect Dis. 1997;3:205–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nemirov K, Vapalahti O, Lundkvist A, Vasilenko V, Golovljova I, Plyusnina A, et al. Isolation and characterization of Dobrava hantavirus in the striped field mouse (Apodemus agrarius) in Estonia. J Gen Virol. 1999;80:371–9. [DOI] [PubMed] [Google Scholar]
- 5.Scharninhausen JJ, Meyer H, Pfeffer M, Davis DS, Honeycutt RL. Genetic evidence of Dobrava virus in Apodemus agrarius in Hungary. Emerg Infect Dis. 1999;5:468–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters CJ, Gary LS, Levy H. Spectrum of hantavirus infection: Hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome. Annu Rev Med. 1999;50:531–45. 10.1146/annurev.med.50.1.531 [DOI] [PubMed] [Google Scholar]
- 7.Horling J, Chizhikov V, Lundkvist A. Khabarovsk virus: a phylogenetically and serologically distinct hantavirus isolated from Microtus fortis trapped in Far East Russia. J Gen Virol. 1996;77:687–94. 10.1099/0022-1317-77-4-687 [DOI] [PubMed] [Google Scholar]
- 8.Kariwa H, Yoshimatsu K, Sawabe J, Yokota E, Arikawa J, Takashima I, et al. Genetic diversities of hantaviruses among rodents in Hokkaido, Japan and Far East Russia. Virus Res. 1999;59:219–28. 10.1016/S0168-1702(98)00141-5 [DOI] [PubMed] [Google Scholar]
- 9.Vapalahti O, Lundkvist A, Fedorov V, Conroy CJ, Hirvonen S, Plyusnina A, et al. Isolation and characterization of hantavirus from Lemmus sibiricus: evidence for host switch during hantavirus evolution. J Virol. 1999;73:5586–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yashina LN, Pastrshev NP, Ivanov LI, Slonova RA, Mishin VP, Kompanez GG, et al. Genetic diversities of hantaviruses associated with hemorrhagic fever with renal syndrome in the far east of Russia. Virus Res. 2000;70:31–44. 10.1016/S0168-1702(00)00203-3 [DOI] [PubMed] [Google Scholar]
- 11.Yashina L, Mishin V, Zdanovskaya N, Schmaljohn C, Ivanov L. A newly discovered variant of a Hantavirus in Apodemus peninsulae, Far Eastern Russia. Emerg Infect Dis. 2001;7:912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Yoshimatsu K, Ebihara H, Ogino M, Araki K, Kariwa H, et al. Genetic diversity of hantaviruses isolated in China and characterization of novel hantaviruses isolated from Niviventer confucianus and Rattus rattus. Virology. 2000;278:332–45. 10.1006/viro.2000.0630 [DOI] [PubMed] [Google Scholar]
- 13.Schmaljohn CS, Jening GB, Hay J, Dalrymple JM. Coding strategy of the S genome segment of Hantaan virus. Virology. 1986;155:633–43. 10.1016/0042-6822(86)90223-0 [DOI] [PubMed] [Google Scholar]
- 14.Schmaljohn CS, Schmaljohn AL, Dalrymple JM. Hantaan virus mRNA: coding strategy, nucleotide sequence, and gene order. Virology. 1987;157:31–9. 10.1016/0042-6822(87)90310-2 [DOI] [PubMed] [Google Scholar]
- 15.Xiao SY, Liang M, Schmaljohn CS. Molecular and antigenic characterization of HV114, a hantavirus isolated from a patient with haemorrhagic fever with renal syndrome in China. J Gen Virol. 1993;74:1657–9. 10.1099/0022-1317-74-8-1657 [DOI] [PubMed] [Google Scholar]
- 16.Shi XH, Liang MF, Hang CS, Gan S, McCaughey C, Eliott RM. Nucleotide sequence and phylogenetic analysis of the medium (M) genomic RNA segments of three hantaviruses isolated in China. Virus Res. 1998;56:69–76. 10.1016/S0168-1702(98)00065-3 [DOI] [PubMed] [Google Scholar]
- 17.Schmaljohn CS, Arikawa J, Hasty SE, Rasmussen L, Lee HW, Lee PW, et al. Conservation of antigenic properties and sequences encoding the envelope proteins of prototype Hantaan virus and two virus isolates from Korean haemorrhagic fever patients. J Gen Virol. 1988;69:1949–55. 10.1099/0022-1317-69-8-1949 [DOI] [PubMed] [Google Scholar]
- 18.Liang M, Li D, Xiao SY, Hang C, Rossi CA, Schmaljohn CS. Antigenic and molecular characterization of hantavirus isolates from China. Virus Res. 1994;31:219–33. 10.1016/0168-1702(94)90005-1 [DOI] [PubMed] [Google Scholar]
- 19.Arikawa J, La Penotiere HF, Iacono-Connors L, Wang MG, Schmaljohn CS. Coding properties of the S and the M genome segments of Sapporo rat virus: comparison to other causative agents of hemorrhagic fever with renal syndrome. Virology. 1990;176:114–25. 10.1016/0042-6822(90)90236-K [DOI] [PubMed] [Google Scholar]
- 20.Antic D, Lim BU, Kang CY. Molecular characterization of the M genomic segment of the Seoul 80-39 virus; nucleotide and amino acid sequence comparison with other hantaviruses reveal the evolutionary pathway. Virus Res. 1991;19:47–58. 10.1016/0168-1702(91)90093-B [DOI] [PubMed] [Google Scholar]
- 21.Xiao SY, Leduc JW, Chu YK, Schmaljohn CS. Phylogenetic analysis of virus isolates in the genus Hantavirus, family Bunyaviridae. Virology. 1994;198:205–17. 10.1006/viro.1994.1023 [DOI] [PubMed] [Google Scholar]
- 22.Isegawa Y, Fujiwara Y, Ohshima A, Fukunaga R, Murakami H, Yamanishi K, et al. Nucleotide sequence of the M genome segment of hemorrhagic fever with renal syndrome virus strain B-1. Nucleic Acids Res. 1990;18:4936. 10.1093/nar/18.16.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avsic-Zupanc T, Toney A, Anderson K, Chu YK, Schmaljohn C. Genetic and antigenic properties of Dobrava virus; a unique member of the Hantavirus genus, family Bunyaviridae. J Gen Virol. 1995;76:2801–8. 10.1099/0022-1317-76-11-2801 [DOI] [PubMed] [Google Scholar]
- 24.Chizhikov VE, Spiropoulou CF, Morzunov SP, Monroe MC, Peters CJ, Nichol ST. Complete genetic characterization and analysis and isolation of Sin Nombre virus. J Virol. 1995;69:8132–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vapalahti O, Kallio-Kokko H, Salonen EM, Brummer-Korvenkontio M, Vaheri A. Cloning and sequencing of Puumala virus Sotkamo strain S and M RNA segment: evidence for strain variation in Hantavirus and expression of the nucleocapsid protein. J Gen Virol. 1992;73:829–38. 10.1099/0022-1317-73-4-829 [DOI] [PubMed] [Google Scholar]
- 26.Lee HW, Lee PW, Tamura M, Tamura T, Okuno Y. Etiological relation between Korean hemorrhagic fever and epidemic hemorrhagic fever in Japan. Biken J. 1979;22:41–5. [PubMed] [Google Scholar]
- 27.Nichol ST, Spiropoulou CF, Morzunov S, Rollin PE, Ksiazek TG, Feldmann H, et al. Genetic identification of a hantavirus associated with an outbreak of acute respiratory illness. Science. 1993;262:914–7. 10.1126/science.8235615 [DOI] [PubMed] [Google Scholar]
- 28.Childs JE, Ksiazek TG, Spiropoulou CF, Krebs JW, Morzunov S, Maupin GO, et al. Serologic and genetic identification of Peromyscus maniculatus as the primary rodent reservoir for a new hantavirus in the southwestern United States. J Infect Dis. 1994;169:1271–80. [DOI] [PubMed] [Google Scholar]
- 29.Rollin PE, Ksiazek TG, Elliot LH, Ravkov EV, Martin ML, Morzunov S, et al. Isolation of black creek canal virus, a new hantavirus from Sigmodon hispidus in Florida. J Med Virol. 1995;46:35–9. 10.1002/jmv.1890460108 [DOI] [PubMed] [Google Scholar]
- 30.Ksiazek TG, Nichol ST, Mills JN, Groves MG, Wozniak A, McAdams S, et al. Isolation, genetic diversity, and geographic distribution of Bayou virus (Bunyaviridae; hantavirus). Am J Trop Med Hyg. 1997;57:445–8. [DOI] [PubMed] [Google Scholar]
- 31.Schmaljohn C, Hjelle B. Hantaviruses:a global disease problem. Emerg Infect Dis. 1997;3:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mustonen J, Helin H, Pietila K, Brummer-Korvenkontio M, Hedman K, Vaheri A, et al. Renal biopsy findings and clinicopathologic correlations in nephropathia epidemica. Clin Nephrol. 1994;41:121–6. [PubMed] [Google Scholar]
- 33.Grcevska L, Polenakovic M, Oncervski A, Zografski D, Gligic A. Different pathohistological presentations of acute renal involvement in Hantaan virus infection: report of two cases. Clin Nephrol. 1990;34:197–201. [PubMed] [Google Scholar]
- 34.Bren AF, Pavlovcic SK, Koselj M, Kovac J, Kandus A, Kveder R. Acute renal failure due to hemorrhagic fever with renal syndrome. Ren Fail. 1996;18:635–8. 10.3109/08860229609047688 [DOI] [PubMed] [Google Scholar]
- 35.Polenakovic M, Grcevska L, Gerasimovska-Tanevska V, Oncevski A, Dzikova S, Cakalaroski K, et al. Hantaan virus infection with acute renal failure. Artif Organs. 1995;19:808–13. 10.1111/j.1525-1594.1995.tb02432.x [DOI] [PubMed] [Google Scholar]