Abstract
The range and precision of limb movements are dependent on the specific patterns of muscles and tendons. To facilitate analyses of tendon and muscle phenotypes we compiled a description of these tissues in the forelimb of developing mouse embryos. Individual tendons, muscles and ligaments were annotated in a series of transverse sections through the forelimb of an embryo at day 18.5 of embryonic development (E18.5). Transverse sections present a distinctive and highly reproducible pattern of the muscles and tendons at different limb levels that can be used as a simple reference in analyses of mutant phenotypes. A comparable set of sections from an embryo at E14.5 was included to highlight structural features that change during the maturation of the musculoskeletal system. The ability to define the precise position of transverse sections along the proximal-distal axis of the limb may also be useful in studies of other features in developing limbs.
Keywords: Tendon, muscles, forelimb, mouse embryo
Introduction
Vertebrate limbs evolved to perform a wide range of movements combining fine mechanical skills and the capacity to attenuate the intensity and strength in the application of force. Movement is dependent on the skeleton that provides solid support and articulation, and force that is generated by muscle contraction and transmitted by tendons to the skeleton. The repertoire and range of movement is therefore determined by structural features of individual muscles, the trajectory of the tendons that transmit the force generated by the muscles and the details and precise position of the insertion of tendons into the respective skeletal elements.
Interest in the tissues of the musculoskeletal system is evident from the remarkable number of studies addressing cell fate decisions in the differentiation of muscles and skeletal tissues. These studies identified complex signaling and transcription networks that regulate these cellular events and the major transcription factors that orchestrate these processes such as Sox9 and Runx2 for the cartilage and bone respectively (reviewed in Karsenty and Wagner, 2002), and the family of Myogenic Regulatory Factors that feature prominently in muscle differentiation (reviewed in Bryson-Richardson and Currie, 2008). Recent studies initiated a similar line of research for tendons, identifying the transcription factor Scleraxis (Scx) as a distinctive marker for tendon cells (Schweitzer et al., 2001; Brent et al., 2003) with a major role in tendon differentiation (Murchison et al., 2007), and FGF signaling as a potent inducer of the tendon cell fate (reviewed in Tozer and Duprez, 2005).
Progress in studies of the patterning of the musculoskeletal system is significantly more lopsided. The mechanisms that regulate limb patterning were the subject of numerous studies and in most cases, limb pattern was determined by the effects on skeletal development, resulting in the description and analysis of the effects of a large number of mutations on skeletal patterning. The focus on skeletal patterning was largely due to easy procedures for generation of skeletal preparations and the relative simplicity of the skeletal pattern that enables a detailed analysis in preparations of whole limbs. In stark contrast, the pattern of the muscles and tendons is rarely included in mutant analyses because of the absence of simple procedures to visualize these tissues and the relative complexity of their patterns. The patterning of muscles and tendons is however crucial to the normal functioning of the musculoskeletal system and unraveling the nature of activities that direct muscle or tendon patterning would be of great importance in studies of congenital conditions that result in disruptions of musculoskeletal function.
In studies of tendon development during mouse embryogenesis we have found that transverse sections through the forelimb provide a simple and distinct pattern of muscle and tendon distribution along the proximal-distal axis of the limb. These patterns are useful in analysis of tendon and muscle phenotypes in developing embryos and for identification of specific perturbations in these tissues. We therefore devised a description of these tissues in a set of 24 sections that correspond to the major patterns in the distribution of muscles and tendons from the tip of the digits and up to the elbow. To facilitate use of this data set, the annotated forelimb sections were associated with their specific positions along the proximal-distal axis. Importantly, the resolution and annotation of the data included in this work were all geared for the development of a tool for phenotypic analysis and do not represent an effort for a comprehensive description of musculoskeletal anatomy.
Results and Discussion
Organization and annotation of muscle and tendon patterns
Scx is a distinctive marker of tendon cells through development (Schweitzer et al., 2001; Brent et al., 2003) and detection of its expression represented a crucial step in description of tendon development (reviewed in Tozer and Duprez, 2005). To facilitate the study of tendon phenotypes a tendon reporter, ScxGFP, was also generated, in which the expression of EGFP is driven by regulatory elements of the Scx gene (Pryce et al., 2007). The expression of GFP in the tendons of ScxGFP mice and embryos provided a robust depiction of the tendons in tissue sections.
The identification of the muscles and tendons of the forelimb is presented below in three components: (1) Two tables that include the nomenclature for the muscles, tendons and ligaments in the limb (Table 1&2). (2) A depiction of the major tendons in whole limbs including a key for the position of each transverse section along the proximal-distal axis of the forelimb (Fig. 1). (3) Transverse sections through the forelimb with annotations that identify the muscles, tendons and ligaments (Figs. 2&3).
Table 1.
| Muscle/Tendon # | Muscle/Tendon Name |
|---|---|
| 1 | Interosseous |
| 2 | Lumbrical |
| 3 | Flexor Digitorium Sublimis (FDS) |
| 4 | Flexor Digitorium Profundus (FDP) |
| 5 | Extensor Digitorium Communis (EDC) |
| 6 | Extensor Indicis Proprius |
| 7 | Extensor Pollicis |
| 8 | Extensor Carpi Ulnaris |
| 9 | Extensor Carpi Radialis Longus |
| 10 | Extensor Carpi Radialis Brevis |
| 11 | Extensor Digiti Quarti |
| 12 | Extensor Digiti Quinti |
| 13 | Flexor Carpi Radialis |
| 14 | Flexor Carpi Ulnaris |
| 15 | Palmaris Longus |
| 16 | Vinculum |
| 17 | Pronator Quadratus |
| 18 | Pronator Teres |
| 19 | Supinator |
| 20 | Thenar |
| 21 | HypoThenar |
Table 2.
| Ligament # | Ligament Name |
|---|---|
| 1 | Collateral Ligament Distal Interphalangeal joint (DIP) |
| 2 | Collateral Ligament Proximal Interphalangeal joint (PIP) |
| 3 | Collateral Ligament Metacarpophalangeal joint (MP) |
| 4 | Palmar Metacarpal Ligaments |
| 5 | Palmar Carpometacarpal Ligaments |
Fig. 1.
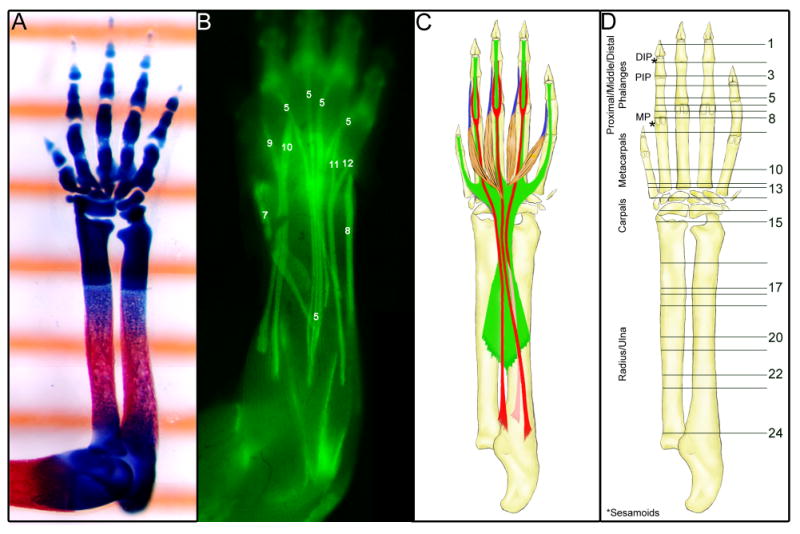
The tendons of the forelimb at E18.5.
(A) A ventral view of a skeletal prep of a forelimb from an embryo at E18.5 captured over a ruler showing 1mm gradation marks.
(B) A dorsal view of a skinned forelimb of an E18.5 ScxGFP embryo. The extensor tendons are identified with a number that identifies them in the tendon table (Table 1).
(C) Schematic drawing of the major flexor tendons in the forelimb at E18.5. Green – Flexor Digitorium Profundus tendon; Red – Flexor digitorium Sublimis tendon; Blue – Lumbrical muscles and tendons.
(D) A schematic drawing of the ventral side of the forelimb that serves to illustrate the position of sections in Figs. 2&3. DIP – Distal interphalangeal joint; PIP – proximal interphalangeal joint; MP – Metacarpophalangeal joint.
Fig. 2.
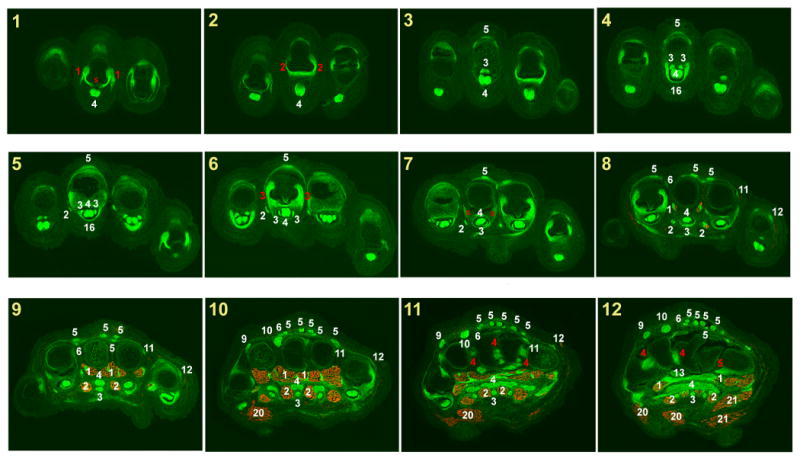
Muscles and tendons of the forelimb at E18.5.
Successive cross sections of a forelimb from an E18.5 ScxGFP embryo stained for MHC. In all panels dorsal is up and anterior is to the left.
Panel numbers indicate the position of each section in the illustration in Fig. 1D.
White numerals – Tendon or muscle number in Table 1.
Red numerals – Ligament number in Table 2.
Red S – sesamoid bone.
Fig. 3.
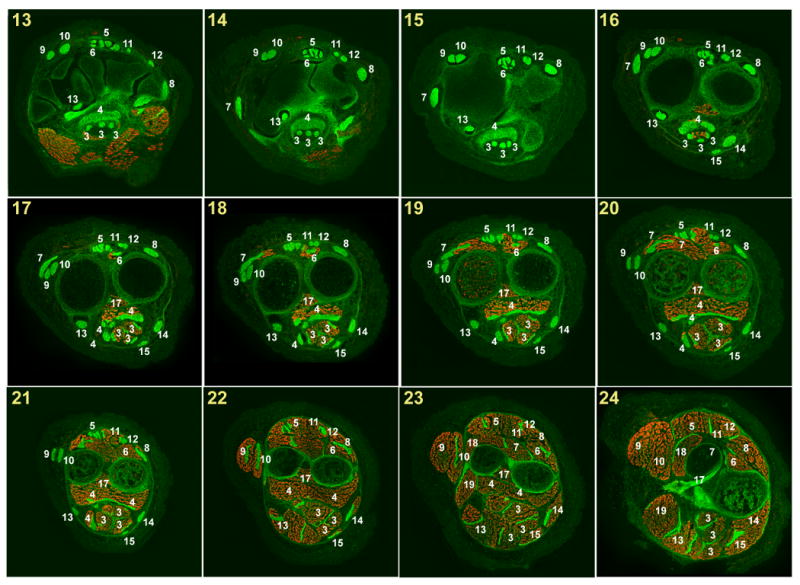
Muscles and tendons of the forelimb at E18.5 (2).
Successive cross sections of a forelimb from an E18.5 ScxGFP embryo stained for MHC. In all panels dorsal is up and anterior is to the left.
Panel numbers indicate the position of each section in the illustration in Fig. 1D.
White numerals – Tendon or muscle number in Table 1.
Red numerals – Ligament number in Table 2.
Red S – sesamoid bone.
The forelimb of the mouse embryo at E18.5 extends approximately 6mm from elbow to the tips of the digits and the relative size of the different parts of the limb can be appreciated in a skeletal preparation that was captured overlying a ruler in which the gradations are spaced 1mm apart (Fig. 1A). Depictions of the major tendons in a whole limb in Fig. 1 were added to provide a context for the distribution of tendons and muscles as seen in the specific section planes in Figs. 2&3. The limbs in the different panels were drawn or photographed to scale so that the section planes in Fig. 1D correspond with the specific tendon pattern in Fig. 1B&C. The extensor tendons were represented in a picture of a skinned forelimb from a ScxGFP embryo at E18.5 (Fig. 1B). The flexor tendons are more overlapping and stacked vertically and therefore could not be similarly captured from a ScxGFP embryo. The major flexor tendons were therefore represented in a schematic drawing (Fig. 1C).
The annotation of the muscles, tendons and ligaments was performed on a series of 12 µm cryosections from a forelimb of an E18.5 ScxGFP embryo. ScxGFP signal marked the tendons and ligaments and the muscles were highlighted by staining for Myosin Heavy Chain. A set of 24 sections that represent the major patterns of tendons and muscles along this axis were chosen for presentation in Figs. 2&3.The panels were numbered, and panel numbers correspond to the section planes as they appear in Fig. 1D.
The tendon and muscle patterns are for the most identical in all digits and metacarpals, but because of the differences in digit length, similar structures appear in different digits at different section planes. To avoid clutter, annotations were added for the structures as they appear in the middle digit, which is the longest digit and therefore the one in which structural features appear first in a series of sections that progresses from distal to proximal. Tendon identities in the other digits can be recognized by locating the corresponding structure in the middle digit.
The major tendons of the forelimb
Analysis of a tendon phenotype is frequently dependent on the ability to conceptualize the trajectory of the tendons and changes that may have occurred in this trajectory or in a specific position along the tendon. We therefore include a limited description of the trajectories of some major tendons.
The major extensor tendon, the Extensor Digitorium Communis (EDC) inserts at the distal Interphalangeal (DIP) joint and the individual EDC tendons extend along the dorsal side of each digit and metacarpal bone and coalesce to traverse the wrist in a dedicated synovial compartment that provides protection from friction and compressive forces. To enhance the connection between the tendons and their muscles the tendons do not terminate at the junction with the EDC muscles and extend along the individual EDC muscles almost up to the wrist (Fig. 1B). Other extensor tendons insert in more proximal positions of only one or two digits, but all extensor tendons coalesce close to the wrist and cross the wrist in specialized synovial compartment (see also accompanying manuscript by Staverosky et al.).
The most robust flexor tendon, the Flexor Digitorium Profundus (FDP), inserts in the ventral side of the DIP to coincide with the distal sesamoid bone in all digits (Fig. 1C green). The individual FDP tendons extend along the digits and metacarpal bones and fuse when they reach the level of the carpal bones to traverse the wrist as a single expanded tendon. More proximally, the tendons separate and integrate with individual heads off the FDP muscle. The Flexor Digitorium Sublimis (FDS) inserts more proximally at the base of the Proximal Interphalangeal (PIP) joint (Fig 1C red). The FDS tendons then split and the two FDS tendons in each digit wrap around the FDP tendon and flatten so that they rejoin at the level of the Metacarpophalangeal joint (MP) in of a thin cup-like structure (Fig 2-8). Proximal to the sesamoid bones of the metacarpal the FDS tendons become rounded and extend ventral (and superficial) to the FDP tendons across the wrist. The FDS muscles are the muscles that extend most distally in the forelimb (Fig 3-16) but the tendons again continue along the length of the muscles almost up to the muscle origin in the elbow. Finally, the Lumbrical muscles are muscles that reside intrinsic to the hand. Contrary to most muscles the origin of these muscles is not in a skeletal element, but at the FDP tendon at the level of the carpals. The Lumbrical muscles extend below and between the metacarpal bones and send a short tendon that begins at the level of the MP joint and insert at the lateral side of the PIP joint (Fig 1C blue).
The tendons and muscles of the forelimb at E14.5
Induction of tendon progenitors begins in the limb bud at E9.5, but overtly distinct tendons are first detected only at E13.5, and a mature tendon system is first seen at E14.5 (Schweitzer et al., 2001; Murchison et al., 2007). The tendons and muscles at E14.5 include most features of the mature musculoskeletal system. Various features, however, are incomplete or different at E14.5 and the final pattern is achieved only at E16.5. It is therefore important to take the developmental stage of analyzed tissues into consideration and recognize the features that differ in WT embryos at these earlier stages so that such features will not be mistaken for a phenotypic consequence.
To highlight the differences between the tendon and muscle systems at these two stages the tendons and muscles in a forelimb from an E14.5 embryo were visualized as described above and the sections presented in Fig. 4 were associated with the corresponding section level at E18.5. Many of the tendons appear similar in both stages and Fig. 4 therefore includes only 12 sections that represent mostly section levels that highlight major differences between these stages. While most tendons are already present at E15.5, the ligaments, sesamoid bones and digit vinculum, have not yet developed and begin to appear at E15.5 in a proximal to distal progression.
Fig. 4.
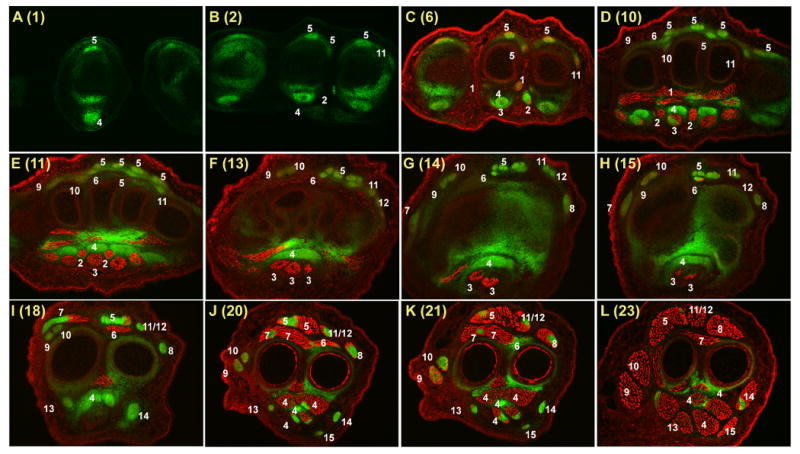
Muscles and tendons of the forelimb at E14.5.
Successive cross sections of a forelimb from an E14.5 ScxGFP embryo stained for MHC. In all panels dorsal is up and anterior is to the left.
The number in parentheses in each panel indicates the matching E18.5 section in Figs. 2&3 and the corresponding position of the section in the illustration in Fig. 1D.
White numerals – Tendon or muscle number in Table 1.
A major discrepancy between the two stages is however the absence of an FDS tendon at E14.5. FDS tendons are not seen in sections through the digits and only the cup-like structure at the level of the MP joint is seen (Fig. 4C). Surprisingly, the FDS tendons are also missing at the levels of the metacarpals where muscles occupy the position of the presumptive FDS tendons. These muscles are likely precursors of the mature FDS muscles and the unique features in the development of this tendon will be described in a separate study (Riordan and Schweitzer in preparation).
Practical use of the muscle and tendon Atlas
The compilation of the details of the muscles and tendons in the forelimb is presented in this study to facilitate analysis of the pattern of these tissues in a variety of mouse models. The tendons were conveniently visualized in this study using the ScxGFP reporter, but it is obviously important to be able to perform a similar analysis in tissues that do not contain the ScxGFP reporter. We are not aware at this time of an antibody to Scx or another tendon protein that yields a clean depiction of the tendons. In our laboratory, we therefore perform such studies by performing an in situ hybridization with a tendon specific probe. The most useful probes to delineate the tendons in our experience are Tenomodulin (Brandau et al., 2001), a transmembrane protein that provides a clean and robust signal restricted only to the tendons (Fig. 5A), and Col1a1, likely the most robust in situ hybridization probe for tendons (Murchison et al., 2007). ISH for Col1a1 labels other tissues as well thus broadening the scope of the phenotypic consequences that can be identified using this one staining (Fig. 5B). In situ hybridization can be followed, similar to the data presented in Figs 2&3, by antibody staining to MHC to label the muscles as well (Fig. 5A,B).
Fig.5.
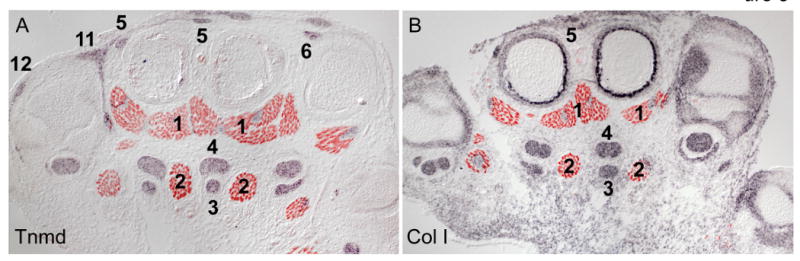
Other Methods for Tendon and muscle staining.
In situ Hybridization with a Tnmd (A) and Col1A1I (B) probe was followed by antibody staining for MHC on transverse sections through the forelimb of an embryo at E17.5 at the level of the metacarpals corresponding to panel 10 in Figure 2. Numerals represent the muscle and tendon numbers in Table 1.
Finally, while the primary use of the data presented here will likely be in analysis of tendon and muscle phenotypes, the continuous changes of the muscle and tendon pattern along the proximal to distal axis provide a unique “stamp” to the different positions along this axis that therefore can also be used to identify the precise Proximal-Distal position in specific forelimb sections for other purposes.
Methods
Nomenclature
The nomenclature for the muscles, tendons and ligaments in mouse limbs in Tables 1&2 was largely derived from the two major resources that described rat anatomy (Greene, 1935; Popesko, 1992).
In situ Hybridization
Mouse embryos were fixed in 4%PFA (4 – 24hrs at 4°C), embedded in OCT (Tissue-Tek) and frozen sections (12μm) were processed for in situ hybridization as previously described (Murtaugh et al. 1999). A detailed protocol will be sent on request. Antisense RNA probe for Tenomodulin was described before (Brandau et al., 2001), and the Col1a1 probe was generated from pmColI-Bam, a plasmid that includes bases 3154 - 4446 from the murine Col1a1 cDNA as an EcoRI insert in Bluescript.
Immunohistochemistry
Immunohistochemistry was performed following the procedure in the M.O.M kit (Vector) either directly on cryosections or following the ISH procedure and monocolonal antibodies My32 mAb (1:400, Sigma) were used to detect the Myosin heavy chain, followed by detection with the Cy3 flurophore (Jackson). Images were captured using a Nikon Eclipse E800 compound microscope and a MicroPublisher cooled CCD camera from QImaging and the fluorescent and bright field images were merged in Photoshop.
Whole mount procedures
For whole mount visualization of the ScxGFP signal, tissues were skinned and observed directly or after 2-6 hrs fixation in 4%PFA. Pictures were captured on an MZFLIII dissecting microscope (Leica) with a DXM1200 camera (Nikon). Skeletal Prep was performed as previously described (Otto et al. 1997)
Acknowledgments
This work was supported by funding from the Shriners Hospitals for Children and by the NIH, grant R01 AR055640 from NIAMS.
References
- Brandau O, Meindl A, Fassler R, Aszodi A. A novel gene, tendin, is strongly expressed in tendons and ligaments and shows high homology with chondromodulin-I. Dev Dyn. 2001;221:72–80. doi: 10.1002/dvdy.1126. [DOI] [PubMed] [Google Scholar]
- Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell. 2003;113:235–248. doi: 10.1016/s0092-8674(03)00268-x. [DOI] [PubMed] [Google Scholar]
- Bryson-Richardson RJ, Currie PD. The genetics of vertebrate myogenesis. Nat Rev Genet. 2008;9:632–646. doi: 10.1038/nrg2369. [DOI] [PubMed] [Google Scholar]
- Greene EC. The Anatomy of the Rat. Braintree, MA: Braintree Scientific Inc.; 1935. [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Murchison ND, Conner DA, Keene DR, Olson EN, Tabin CJ, Schweitzer R. Regulation of tendon differentiation by Scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development. 2007;128:3855–66. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13:225–37. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bonedevelopment. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Popesko P, R V, Horak J. Rat & Mouse. Vol. 2. Wolfe Publishing; 1992. Colour Atlas of Anatomy of Small Laboratory Animals. [Google Scholar]
- Pryce BA, Brent AE, Murchison ND, Tabin CJ, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev Dyn. 2007;236:1677–1682. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128:3855–3866. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- Tozer S, Duprez D. Tendon and ligament: development, repair and disease. Birth Defects Res C Embryo Today. 2005;75:226–236. doi: 10.1002/bdrc.20049. [DOI] [PubMed] [Google Scholar]


