Abstract
The E1-like superfamily is central to ubiquitin (Ub) conjugation, biosynthesis of cysteine, thiamine and MoCo and several secondary metabolites. Yet, its functional diversity and evolutionary history is not well-understood. We develop a natural classification of this superfamily and use it to decipher the major adaptive trends occurring in the evolution of the E1-like superfamily. Within the Rossmann fold, E1-like proteins are closest to NAD(P)/FAD-dependent dehydrogenases and S-AdoMet-dependent methyltransferases. Hence, their phosphotransfer activity is an independent catalytic “invention” with respect to such activities seen in other Rossmannoid folds. Sequence and structure analysis reveals a striking diversity of residues and structures involved in adenylation, sulfotransfer and substrate-binding between different E1-like families, allowing us to predict previously uncharacterized functional adaptations. E1-like proteins are fused to several previously undetected domains, such as a predicted sulfur transfer domain containing a novel superfamily of the TATA-binding protein fold, different types of catalytic domains, a novel winged helix-turn-helix domain and potential adaptor domains related to Ub conjugation. Based on these fusions we develop a generalized model for the linking of E1 catalyzed adenylation/thiolation with further down-stream reactions. This is likely to involve a dynamic interplay between the E1 active sites and diverse fused C-terminal domains. We also predict participation of E1-like domains in previously uncharacterized bacterial secondary metabolism pathways, new cysteine biosynthesis systems, such as those associated with archaeal O-phosphoseryl tRNA, metal-sulfur cluster assembly (e.g. in nitrogen fixation) and Ub-conjugation. Evolutionary reconstructions suggest that the last universal common ancestor (LUCA) contained a single E1-like domain possessing both phosphotransfer and thiolating activities and participating in multiple sulfotransfer reactions. The E1-like superfamily subsequently expanded to include 26 families clustering into three major radiations. These are broadly involved in ubiquitin activation, cofactor and cysteine biosynthesis, and biosynthesis of secondary metabolites. In light of this we present evidence that in eukaryotes other E1-like enzymes, such as Urm1, were independently recruited for Ubl conjugation, probably functioning without conventional E2-like enzymes.
INTRODUCTION
Modification of eukaryotic proteins by ubiquitin (Ub) and ubiquitin-like proteins (Ubls) is a three step cascade catalyzed by the E1, E2 and E3 enzymes 1,2. The E1 enzyme initiates the process via adenylation of the carboxy-terminal glycine residue of the Ub/Ubl polypeptide. Remarkably, a similar reaction occurs in the biosynthesis of thiamine and molybdenum or tungsten cofactors (MoCo/WCo), where a homolog of the E1 enzyme, ThiF or MoeB, adenylates the C-terminus of a ubiquitin-like protein, ThiS or MoaD 3–8. Upon modification, the trajectories of the ubiquitin-like proteins are very different in the conjugation and cofactor biosynthesis pathways. In the ubiquitin modification system, Ub/Ubl is covalently conjugated to a lysine on the protein substrate via trans-thiolation reactions between E1, E2 and in some cases E3 enzymes (i.e. E3s with HECT domains). In contrast, in cofactor biosynthesis pathways, the adenylated C-terminus of the ThiS or MoaD protein is further modified, using a sulfur donor, to a thiocarboxylate, which then serves as a sulfur donor during the biosynthesis of cofactors. E1-like enzymes are also present in other sulfur incorporation steps involved in biosynthesis of certain siderophores (e.g. quinolobactin), peptide antibiotics, small molecule first messengers and cysteine in prokaryotes 9–15.
Recently, there have been several advances in the deciphering of the structure and mechanisms of the E1-like superfamily (hereinafter referring to E1, MoeB, ThiF and all other homologous proteins that are closer to them than to any other superfamily of enzymes) 4,5,7,8,16,17. Site-directed mutagenesis and X-ray crystallography of different members of the E1-like superfamily has supported a comparable role for various conserved residues, albeit pointing to subtle differences in the catalyzed reactions 17,18. Despite their diversity, active E1-like enzymes share overarching biochemical themes related to adenylation and sulfotransfer. This aspect roused our interest in exploring the natural history of the E1-superfamily of enzymes, both in the context of the Ub-conjugation systems and sulfur metabolism at large. In particular, we wanted to address the following issues: 1) Relationships of the E1-like superfamily to other superfamilies within the Rossmann fold and understanding the key modifications that resulted in the evolution of its extant biochemical activities. 2) Conserved sequence and structural features common to all members of the fold and assessing how variations to this core set of features affect functional properties such as substrate choice and reaction mechanism. 3) The complete set of contextual associations of the E1-like domain, such as domain architectures and conserved gene-neighborhoods. 4) Implications of these contextual associations for the interplay between the adenylation and thiolating activities of E1-like domains and interactions with proteins catalyzing preceding or subsequent reactions. 5) Determining the evolutionary radiations of the E1-like superfamily, establishing its major adaptive trends and inferring any novel biochemical roles they might have acquired.
RESULTS AND DISCUSSION
Identification and classification of E1-like families
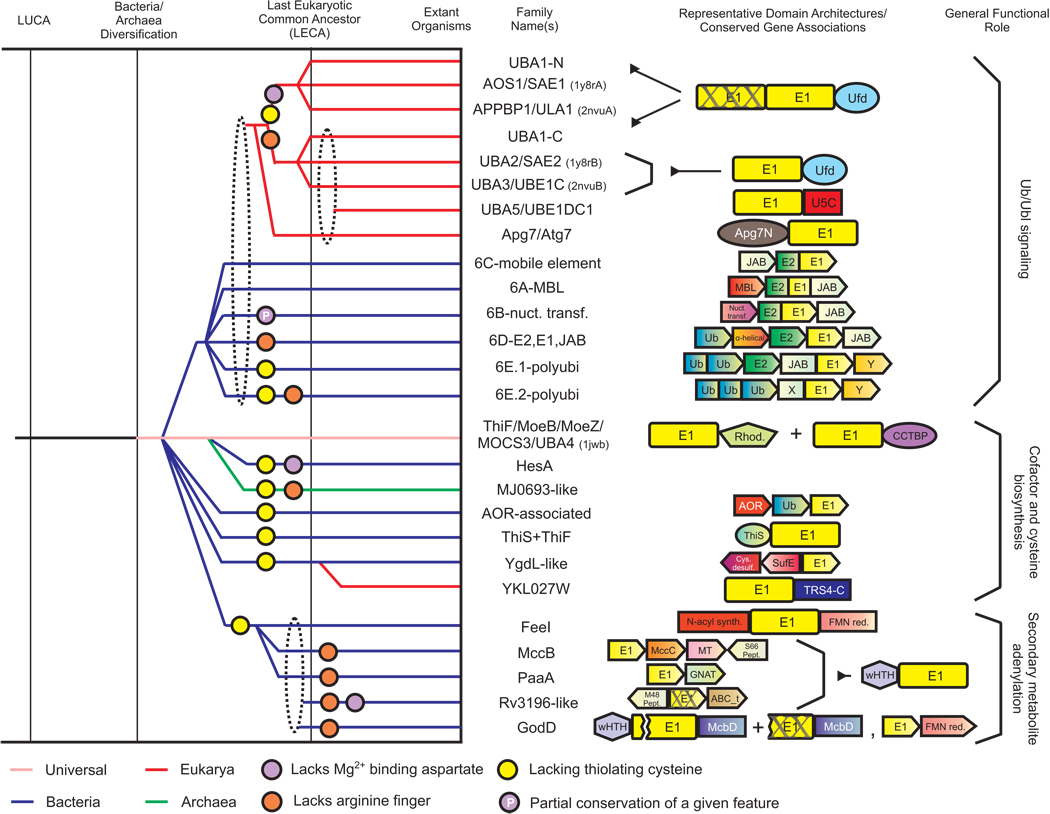
To address the above issues we performed a comprehensive analysis to systematically identify members of the E1-like superfamily. We first transitively searched the PDB database with available 3D structures of the E1 superfamily by using the FSSP program to detect structurally related modules. We then aligned the modules recovered in these searches with the MUSTANG program to obtain a structural alignment of the E1 superfamily with their related structures. This alignment enabled us to objectively identify the features distinguishing the E1 modules from other related Rossmannoid domains (see below). We used several representative sequences of E1 superfamily proteins as seeds to initiate sequence profile searches against the NCBI NR (non-redundant) database with the PSI-BLAST program (see Materials and Methods for details on searches). Sequences detected in these searches were used to prepare a multiple alignment and a Hidden Markov model (HMM) derived from it was used to further search genomic databases using the HMMer package. As a result we exhaustively recovered E1 proteins from the NR database and classified them by means of phylogenetic tree analysis, uniquely shared sequence motifs and structural features (see Materials and Methods for details). Consequently we delineated 26 distinct families of E1-like domains. We also identified their key structural features, established their phyletic patterns, identified conserved gene-neighborhoods (predicted operons) and domain architectures and collated their biochemical functions where available. The reconstructed evolutionary history and natural classification of the E1-like superfamily based on this information is summarized in Figure 1 and Table 1 (For further details refer supplementary material).
Figure 1. Evolutionary history and contextual information for the E1 superfamily.
Individual families are listed to the right of the diagram with solid horizontal lines tracing the inferred evolutionary depth of each family across several key evolutionary transition events represented by the labeled vertical lines. Horizontal lines are color-coded (see key) by observed phyletic distributions. Horizontal lines connecting to a dashed ellipse indicates the family descended from any one of the lineages bundled by the ellipse. Colored circles placed at points along the horizontal lines indicate loss of an ancestral sequence feature in lineage (see key at the bottom of the figure). Representative domain architectures and conserved gene neighborhoods of the families are shown to the right of the family name. Colored polygons represent individual protein domains, while boxed arrows represent individual genes in conserved gene neighborhoods. Breaks within a domain indicate a loss of one or more structural elements. Inactive domains are marked with an ‘X’. General functional roles of the different families are listed to the right. Abbreviations: Rhod., Rhodanese domain; CCTBP, Cysteine-containing TBP-like domain; AOR, Aldehyde ferredoxin oxidoreductase; desulf., desulfurase; FMN red., flavindependent oxidoreductase; Pept., peptidase; GNAT, GNAT-type acetyltransferase; ABC_t, ABC transporter; X, predicted novel peptidase domain; Y, predicted metal-binding domain.
Table 1.
Secondary structure features of major E1 domain structural categories.
| General Description of Function2 | Family Name(s)2 | Secondary structure features/conserved residues common to the E1 domain1 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nh | S1 | H1 | S2 | CC | DNRK* | H2 | S3 | H3 | S4 | D* | H4 | +* | ee | S5 | S6 | I1 | |||
| Inactive Eukaryotic Ub/Ubl-associated versions | UBA1-N | Nh | S1 | H1 | S2 | CC | DD---- | H2 | S3 | H3 | S4 | -- | H4 | -- | -- | S5 | S6 | eeeeee | e |
| AOS1/SAE1 (1y8rA) | Nh | S1 | H1 | S2 | CC | DD---- | H2 | S3 | H3 | S4 | -- | H4 | -- | -- | S5 | S6 | ecc | e | |
| APPBP1/ULA1 (2nvuA) | Nh | S1 | H1 | S2 | CC | DD---- | H2 | S3 | H3 | S4 | -- | H4 | -- | -- | S5 | S6 | ehhhhhhhhe | ||
| Active Eukaryotic Ub/Ubl-associated versions | UBA1-C | Nh# | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | ||
| UBA2/SAE2 (1y8rB) | ---- | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | |||
| UBA3/UBE1C (2nvuB) | Nh# | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | ee | S5 | S6 | |||
| UBA5/UBE1DC1 | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | |||
| APG7/ATG7 | Nh(Rf1) | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | cc | ||
| Prokaryotic Ub/Ubl-associated | 6C-mobile element | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | -- | -- | S5 | S6 | ||
| 6A-MBL-assoc. | Nh(Rf2) | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | -- | -- | S5 | S6 | |||
| 6B-nuct. transf.-assoc. | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | d | H4 | R | -- | S5 | S6 | cc | ||
| 6D-E2, E1, JAB-assoc. | ---- | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | cc | ||
| 6E.1-polyUb-assoc. | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | |||
| 6E.2-polyUb-assoc. | Nh# | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R/K | -- | S5 | S6 | |||
| Cofactor/cysteine biosynthesis | ThiF/MoeB/MoeZ/MOCS3 (1jwb) | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | ||
| HesA | Nh | S1 | H1 | S2 | CC | RDRR | H2 | S3 | H3 | S4 | -- | H4 | R | -- | S5 | S6 | |||
| This+ThiF | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | K | -- | S5 | S6 | |||
| AOR-assoc. | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R | -- | S5 | S6 | |||
| YdgL-like | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | K | -- | S5 | S6 | |||
| YKL027W | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | K | -- | S5 | S6 | |||
| MJ0693-like | ---- | S1 | H1 | S2 | CC | DD--K | H2 | S3 | H3 | S4 | d | H4 | -- | -- | S5 | S6 | |||
| Secondary metabolite adenylation | FeeI | Nh | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | R/K | -- | S5 | S6 | ||
| GodD | ---- | s1 | h1 | s2 | ---- | --------- | ---- | ---- | H3 | S4 | d | H4 | -- | -- | S5 | S6 | |||
| Rv3196-like | ---- | S1 | H1 | S2 | cc | --------- | h2 | s3 | H3 | S4 | d | H4 | R | -- | S5 | S6 | |||
| MccB | Nh# | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | -- | -- | S5 | S6 | |||
| PaaA | Nh# | S1 | H1 | S2 | CC | DNRK | H2 | S3 | H3 | S4 | D | H4 | -- | -- | S5 | S6 | |||
| General Description of Function1 | Family Name(s)1 | Secondary structure features/conserved residues common to the E1 domain2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CxxC1 | Ime | C* | I2 | H5 | ExxK | S7 | S8 | CxxC2 | ||
| Inactive Eukaryotic Ub/Ubl-assoc. versions | UBA1-N | --------- | ----- | -- | hhh | H5 | ExxK | S7 | S8 | -------- |
| AOS1/SAE1 (1y8rA) | --------- | ----- | -- | hhh | H5 | Exxk | S7 | S8 | -------- | |
| APPBP1/ULA1 (2nvuA) | --------- | ----- | -- | hhh | H5 | ExxK | S7 | S8 | -------- | |
| Active Eukaryotic Ub/Ubl-assoc. versions | UBA1-C | --------- | ----- | C | hhhhhhhhh | H5 | ExxK | S7 | S8 | -------- |
| UBA2/SAE2 (1y8rB) | CxxC1 | ----- | C | hhhhh | H5 | exxk | S7 | S8 | CxxC2 | |
| UBA3/UBE1C (2nvuB) | CxxC1 | ----- | C | hhh | H5 | ExxK | S7 | S8 | CxxC2 | |
| UBA5/UBE1DC1 | CxxC1 | ----- | C | -- | H5 | NxxK | S7 | S8 | CxxxxC2 | |
| Apg7/Atg7 | CxxC1 | ----- | C | -- | H5 | Exx-- | S7 | S8 | CxxC2 | |
| Prokaryotic Ub/Ubl-assoc. versions | 6C-mobile element | CxxC1 | hhh | C | ee | H5 | ExxK | S7 | S8 | CxxC2 |
| 6A-MBL-assoc. | --------- | ----- | C | -- | H5 | -------- | S7 | S8 | -------- | |
| 6B-nuct. transf.-assoc. | cxxc1 | ----- | C | -- | H5 | -------- | S7 | S8 | cxxc2 | |
| 6D-E2, E1, JAB-assoc. | --------- | ----- | C | -- | H5 | -------- | S7 | S8 | -------- | |
| 6E.1-polyUb-assoc. | cxxc1 | ----- | -- | -- | H5 | Exxk | S7 | S8 | cxxc2 | |
| 6E.2-polyUb-assoc. | -------- | ----- | -- | -- | H5 | Nxx-- | S7 | S8 | -------- | |
| Cofactor/cysteine biosynthesis | ThiF/MoeB/MoeZ/MOCS3 (1jwb) | CxxC1 | ----- | C | -- | H5 | ExxK | S7 | S8 | CxxC2 |
| HesA | CxxC1 | ----- | -- | -- | H5 | ExxK | S7 | S8 | CxxC2 | |
| This+ThiF1y | --------- | ----- | -- | -- | H5 | -------- | ---- | ---- | -------- | |
| AOR-assoc. | --------- | ----- | -- | -- | H5 | exxk | S7 | S8 | -------- | |
| YdgL-like | --------- | ----- | -- | -- | H5 | -------- | S7 | S8 | -------- | |
| YKL027W | --------- | ----- | -- | -- | H5 | -------- | S7 | S8 | -------- | |
| MJ0693-like | --------- | ----- | -- | -- | H5 | -------- | S7 | S8 | -------- | |
| Secondary metabolite adenylation | FeeI | --------- | ----- | -- | -- | H5 | -------- | S7 | S8 | cxxc2 |
| GodD | CxxC1 | ----- | -- | -- | H5 | -------- | S7 | S8 | CxxC2 | |
| Rv3196-like | CxxC1 | ----- | -- | -- | H5 | -------- | S7 | S8 | CxxC2 | |
| MccB | CxxC1 | ----- | -- | -- | H5 | D/Exx-- | S7 | S8 | CxxC2 | |
| PaaA | CxxC1 | ----- | -- | -- | H5 | ExxK | S7 | S8 | CxxC2 | |
Abbreviations: assoc., associated; nuct. transf., Nucleotidyl transferase; MBL, Metallo-β-Lactamase domain; Ub, Ubiquitin.
Abbreviations: S, conserved strand; H, conserved helix; Nh, N-terminal conserved 2 helices which house arginine finger; #, lack of arginine finger in Nh; Rf1, lacks arginine in canonical position but possesses two other highly conserved arginine residues: 1) different position in middle helix of the N-terminal three-helix bundle and 2) in the extended insert between S7 and S8; Rf2, lacks arginine in canonical position but possesses a conserved arginine in the variable region connecting the two subdomains of a metallobetalactamase domain in its conserved gene neighborhood 36; CC, extended coil region housing adenylation active site; +, conserved positively-charged Ub/Ubl-recognition residue; I1, insert region occurring before crossover loop; CxxC1, first Mg2+ chelating motif; CxxC2, second Mg2+ chelating motif; ExxK, conserved motif found in H5; Ime, insert found in 6c lineage; I2, insert region occurring after crossover loop; e, insert in extended conformation (strand-like); h, insert in helical conformation; cc, long coil insert
marks residues essential for catalysis; --, indicates a feature is absent in the lineage. Capital letters in a column denote conserved residues; lower case letters under these columns indicates only partial conservation of the residue in the lineage.
Basic structural features and higher order relationships of the E1-like superfamily
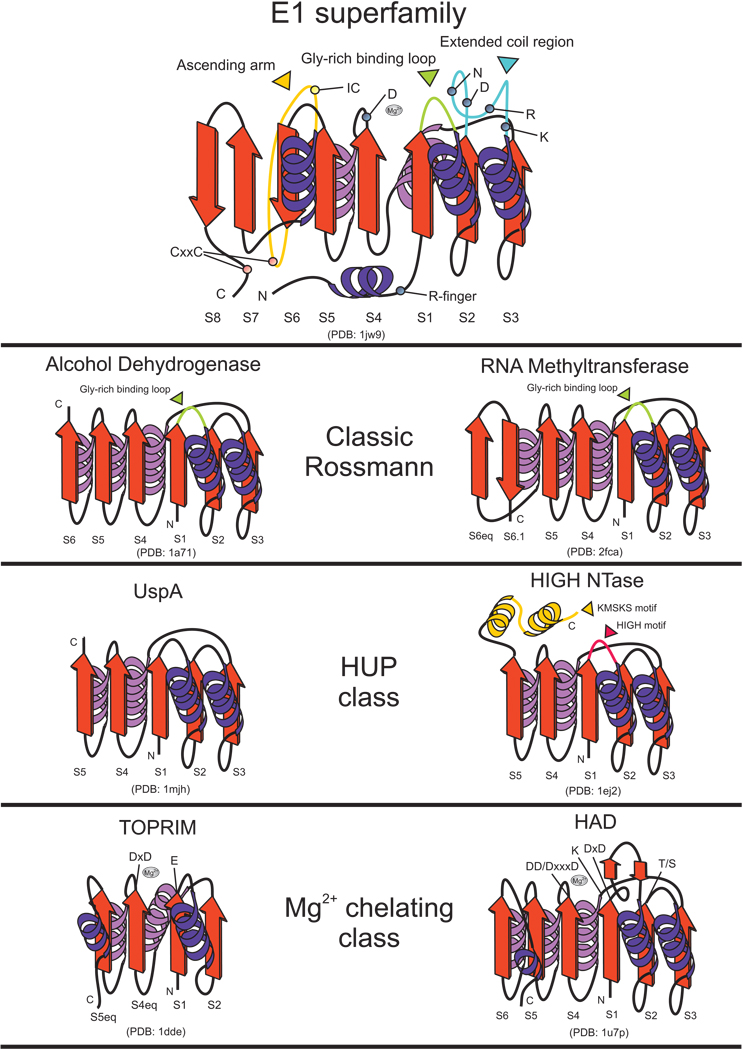
The E1-like domain adopts a 3-layered α/β sandwich fold with a central, eight-stranded β-sheet, with strand order 87654123 (hereafter referred to as S1–S8) (Fig. 2) and helices packing against either face of the sheet. The core of the domain is a Rossmann fold comprised of the β-α units defined by strands S1 to S5 and corresponding helices labeled H1–H4. E1-like domains possess at least three other well-studied structural elements and several subtler sequence features distinguishing them from other superfamilies containing a Rossmann fold. The first of these structural elements is a dyad of helices N-terminal to the Rossmann fold. This unit often contains a conserved arginine that projects into the active site of the opposite monomer of the homo- or hetero- dimer and appears to stabilize the hyper-charged pentavalent phosphorus during the phosphotransfer reaction 7. Thus, it is equivalent to the arginine finger seen in the P-loop NTPases; hence, we refer to this feature as the “arginine finger” hereinafter (Table 1) 4,7,19,20. The second distinctive feature is an extended loop between S2 and H2 containing several polar residues (usually D, N, R and K) strongly conserved across most E1 families (Table 1). Along with the arginine finger from the dimerizing partner, these residues are necessary for catalyzing the adenylation reaction 7,17. The third feature unique to E1-like domains is the extension to the core Rossmann fold, which includes strands S6–S8 with S6 and S8 being anti-parallel to the other strands (Figure 2). This unique extension contains a characteristic linker between strands S6 and S7 which has been termed the “crossover loop” 4 (Figure 2). This structure has one or more helical elements and harbors a strongly conserved cysteine which is required for thiolation reactions catalyzed by these enzymes (henceforth thiolating cysteine). Catalytically active E1-like domains also possess a conserved aspartate in S4 which is involved in coordinating Mg2+ and probably orienting Mg2+-ATP, analogous to the aspartate from the Walker B motif of the P-loop NTPase fold 21. Further, a highly conserved arginine after H4 makes polar contacts with the polypeptide backbone of the Ub/Ubl substrate, perhaps directing the Ub/Ubl tail to the adenylating active site.
Figure 2. Topology diagrams and comparison with other Rossmann-like proteins.
Representative cores of Rossmann-like domains belonging to different classes of the fold are depicted as cartoons. Inserts and other lineage-specific features are depicted and labeled with various other colors. Gray spheres represent the magnesium ions in various active sites. Residues experimentally shown to contribute to catalytic activity in the given representatives are labeled. Strand numbers are given at the bottom of each strand; “eq” refers to strands that are spatially equivalent to strands in the E1 superfamily.
A poorly understood feature of several families of E1-like domains is a pair of CxxC motifs that coordinate a zinc ion. One of the CxxC motifs is present in the “crossover loop” and the other in the poorly-structured coil region following S8 (Table 1, Supplementary material). Crystal structures 4,5,8 suggest that the chelated Zn2+ holds the portion of the “crossover loop” downstream of S6 away from the core sheet, thereby forming an arch to allow the C-terminal tail of the substrate (Ub/Ubl) to access the adenylating active site. However, the CxxC motifs are independently and sporadically lost in many families (Table 1). In some cases its loss correlates with absence of catalytic activity (e.g. inactive eukaryotic E1 families; Fig. 1) or absence of a peptide substrate (e.g. FeeI, see below). But in other cases there is no evidence for such a correlation, suggesting that alternative interactions might have taken the role of the structural zinc to stabilize the “crossover loop” region. Even less understood is a strongly conserved ExxK motif in H5 (Supplementary Material). We observed that in available crystal structures glutamate and lysine residues of the ExxK motif form a salt-bridge and contact the tip of the S7–S8 hairpin from the opposite subunit in the E1-like domain dimers. Hence, these residues might play a role in stabilizing and orienting the interface of subunits in the dimer.
The wide conservation of the above structural features across the entire E1-like superfamily suggests that they represent the ancestral condition for this domain. Our sequence and structure comparisons indicated that among the Rossmannoid superfamilies, E1-like domains are closest to NAD(P)/FAD-dependent dehydrogenases and S-AdoMet-dependent methyltransferases (see SCOP: http://scop.mrc-lmb.cam.ac.uk/scop/). These proteins show a congruent structural core spanning the β-α units determined by strands 1 to 5 and display a glycine rich loop between strands S1 and H1 which is involved in nucleotide binding (ATP in the E1-like superfamily). They also share a frequently conserved aspartate residue at the end of strand 2, and a characteristic aspartate or asparagine residue at the end of S4 (Figure 2). Thus, these three superfamilies are distinct from another monophyletic assemblage of Mg2+ chelating Rossmann-fold domains that unites several superfamilies with phosphoesterase activity, namely haloacid dehalogenases (HAD), receiver domains, DHH phosphoesterases, TOPRIM domains and PIN/5′-3′ nucleases. This latter assemblage is characterized by two acidic residues in their active site 19 (Figure 2). The E1-like superfamily, NAD(P)/FAD-dependent dehydrogenases and S-AdoMet-dependent methyltransferases are also distinct from the HUP assemblage, which unites the Rossmannoid catalytic domains of Class-I aminoacyl tRNA synthetases and related nucleotidyl transferases, PP-ATPases, the USPA superfamily, and photolyases 22,23 (Figure 2). The above-described three features specific to the E1-like domains appear to have been central to their acquisition of phosphotransfer/adenylation and thiolation activity (see below, Figure 2, and Table 1). Thus, phosphotransfer activity of the E1-like domain represents an “invention” that occurred independently in an ancestral version of the Rossmann fold resembling the nucleotide-binding version in NAD(P)/FAD-dependent dehydrogenases and S-AdoMet-dependent methyltransferases.
Diversification of the active site and dimer interface within the E1 superfamily
A case-by-case consideration showed that several E1-like families have developed unique family-specific features, including modifications of catalytic or substrate-binding sites (Table 1). In contrast to the major prokaryotic versions (ThiF and MoeB) which function as homodimers, eukaryotic E1-like domains often function as heterodimers 24. This appears to have emerged concomitant with a certain “division of labor” between the two subunits of the dimer. Members of the eukaryotic E1 families UBA1-N, AOS1/SAE1 and APPBP1, are by themselves catalytically inactive but supply the arginine finger to the active site. Conversely, the UBA1-C, SAE2/UBA2 and UBA3 families lack an arginine finger, but constitute rest of the active site of the dimer. The resulting asymmetry in the location of the active site with respect to the dimer interface appears to be critical for positioning the E2 polypeptide (via binding to UFD; see below) during trans-thiolation.
The eukaryotic Apg7/Atg7 family, and the prokaryotic families 6A, 6B, 6D and 6E, HesA, MJ0693-like, and a group of related bacterial families Rv3196, GodD, MccB and PaaA show different variations, each affecting a subset of the residues and structural features influencing phosphotransfer (Table 1 and Fig. 1). Of these a cluster of related families (MJ0693-like, Rv3196, GodD, MccB and PaaA), the prokaryotic group of 6A, 6D and 6E families and the eukaryotic Apg7/Atg7 family lack the N-terminal arginine finger. In the latter case adenylation and trans-thiolation of Apg12 (an Ubl) is experimentally supported 25, suggesting that these catalyze typical E1 reactions despite lacking the conventional arginine finger. Consistent with this, we detected structurally plausible candidates for alternative arginine fingers elsewhere in the same polypeptide in the Apg7/Atg7 family or from other potentially interacting proteins (in the bacterial 6A), which might substitute for the canonical arginine finger (See Table 1 for details).
Emergence of distinct arginine fingers has been previously observed in P-loop NTPases, where arginine fingers have independently evolved on multiple occasions, and are either provided from within the protein or from distinct polypeptides interacting with the NTPases 26,27. The bacterial HesA family lacks the Mg2+-coordinating aspartate, and along with the archaeal MJ0693 family displays substitutions in some of the conserved residues in the loop between S2 and H2. However, the HesA family maintains the arginine finger suggesting that it might still possess catalytic activity, perhaps with lower efficiency or function as a heterodimer.
A spectrum of further structural alterations is seen in the Rv3196 and GodD families (Table 1). In several members of both these families the entire loop between S2 and H2, along with H2 and S3 has been independently lost. Further, in the GodD family, the glycine-rich loop between S1 and H1 and in some cases the entire N-terminal region including S3 has been lost. Despite these alterations, most members of these families retain the Mg2+-coordinating aspartate and the remaining C-terminal portion of the E1-like domain (Table 1, supplementary material). This feature along with certain unique aspects of their domain architecture suggests that, despite their dramatic remodeling, this cluster of related families are likely to perform a catalytic function in conjunction with other fused domains or through dimerization with conventional E1-like domains (see below).
We also detected great diversity in regions that are known or predicted to participate in peptide substrate interaction and dimerization. This includes the region in the vicinity of the thiolating cysteine, between S6 and H5 (the “crossover loop” region), containing the pocket that interacts with Ub/Ubl and E2 in Ub/Ubl adenylating families 28,29. Diverse inserts with different predicted secondary structures are observed in this region in the MccB, Apg7/Atg7, prokaryotic 6B, 6C and 6D and eukaryotic E1 families (Table 1). These inserts are organized around and atop the active sites of E1-like domains, reminiscent of the cap domains inserted into the core Rossmannoid fold of the HAD superfamily. In the HAD superfamily they have been shown to influence substrate recognition, access to the active site and catalytic efficiency 19. By analogy, it is possible that these inserts in the E1-like superfamily correlate with distinct substrate specificities of the respective families. Another region that shows great sequence diversity is the β-hairpin formed by strands S7 and S8 (see supplementary material). Given the role of this hairpin in dimerization (see above), this diversity might correlate with differences in the dimer interface of different families. Crystal structures of eukaryotic E1 proteins show that this region also contacts the exposed face of the Ub/Ubl proteins 30. Hence, this region has also possibly diversified to recognize cognate Ubl substrates 30.
Loss of the thiolating cysteine within the E1 superfamily
Though the thiolating cysteine is strongly conserved in most catalytically active E1-like families, which transfer Ub/Ubl or thiolate ThiS/MoaD-like substrates, it appears to have been lost in multiple families (Table 1, Fig. 1 and supplementary material). Known or predicted Ub/Ubl-interacting E1-like families lacking the thiolating cysteine (Table 1) are the bacterial 6E family, a small prokaryotic E1-like family with an N-terminal fusion to a ThiS-like Ubl domain and the E1-like family that is functionally associated with tungsten-cofactor-utilizing aldehyde-ferredoxin oxidoreductases. Additionally, several archaeal E1-like proteins lack the thiolating cysteine. These are encoded in conserved gene neighborhoods along with genes for molybdopterin, thiamine and cysteine biosynthesis enzymes (Supplementary material). Site-directed mutagenesis studies on Escherichia coli MoeB showed that MoaD can be thiolated despite disruption of the thiolating cysteine, if other sulfur abstracting functional partners such as IscS or cysteine sulfinate desulfinase are available 6,31. Thus, in some of the above instances such extrinsic partners might provide an alternative to the thiolating cysteine. Two related uncharacterized families, namely the bacterial YgdL-like family and the eukaryotic YKL027W-like family possess an intact adenylating active site but have a divergent C-terminus lacking the structural Zn-chelating motifs and thiolating active site. Hence, they are also predicted to only catalyze the adenylation step. However, they might cooperate with other proteins in subsequent sulfotransfer reactions, possibly in conjunction with Ubls (see below).
The remaining families lacking the thiolating cysteine show no evidence for interaction with Ub/Ubl proteins and appear to be either purely adenylating enzymes or catalytically inactive (Fig. 1 and Table 1). Chief among these are the FeeI, MccB, GodD, Rv3196, and PaaA assemblage of bacterial families known or predicted to participate in biosynthesis of secondary metabolites, polypeptide antibiotics and small signaling peptides. The eponymous prototype of the FeeI family apparently adenylates a fatty acid in the formation of N-acyl tyrosine, a potential signal released by soil bacteria 32,33. Here, instead of the thiolating cysteine, the thiol group of a phosphopantetheinyl moiety attached to the acyl carrier protein FeeL forms a thiocarboxylate with the adenylated fatty acid. The MccB family is involved in biosynthesis of microcin C7-like peptides and appears to be the enzyme which adenylates the carboxy terminus of the microcin 14,15. GodD is involved in biosynthesis of goadsporin, an actinobacterial signaling peptide 13, and other members of this family might be involved in synthesis of other thiazole-, oxyazole- and lanthionine-containing peptides. A subset of proteins of the GodD family is predicted to be catalytically active and participate in adenylating steps in the synthesis of these modified peptides. Likewise, members of the uncharacterized PaaA family (which is closer to MccB) and a subset of the Rv3196 family are predicted to catalyze similar adenylation reactions in biosynthesis of peptide secondary metabolites.
Contextual information predicts novel biochemical functions in the E1-like superfamily
Four forms of contextual information are valuable in uncovering functional linkages and predicting biochemical interactions of uncharacterized proteins: 1) conserved gene neighborhoods or predicted operons; 2) domain architectures; 3) phyletic distribution profiles; 4) information regarding interacting partners gleaned from large-scale protein interaction maps. Gene neighborhoods and phyletic profiles are particularly useful in prokaryotes in determining the biochemical pathways to which the E1-like superfamily has been recruited 34,35 (Figure 1, 3 and Table 2).
Figure 3. Phylogenetic tree of the ThiF/MoaD/MOCS3 family along with along with domain architectures and gene neighborhoods.
The tree was constructed using the least-squares method followed by local rearrangement to obtain a maximum-likelihood tree. Closely-related branches have been combined into groups, whose sizes are scaled relative to the number of lineages within each group. Nodes in the tree with >70% bootstrap support are denoted by small gray circles. Groups are colored according to associations with the rhodanese and CCTBP domains (see key at bottom left). The taxonomy of the lineages within a group is also indicated (see key at bottom right). Neighborhoods and architectures are as in Fig. 1; genes encoding multi-domain proteins are shown as boxed arrows demarcated by horizontal lines. Additional abbreviations not found in Figure 1: Ubl, ubiquitin-like; CT_p, C-terminal processing serine peptidase; CS, cysteine synthase, MS, methionine synthase.
Table 2.
Summary of novel contextual associations
| E1 Family Name | Proteins encoded in conserved gene-neighborhoods | Comments |
|---|---|---|
| ThiF/MoeB/MoeZ/MOCS3 | Predicted bacterial cysteine/methionine biosynthesis gene clusters: JAB domain peptidase, cysteine synthase, O-acetylhomoserine sulfhydrylase/methionine lyase, SirA-like sulfur carrier/redox protein, PDZ domain-containing C-terminal serine peptidase. | These enzymes are involved in both cysteine and methionine biosynthesis. The SirA protein is likely to be a sulfur carrier in this process and contains a conserved cysteine. |
| ThiF/MoeB/MoeZ/MOCS3 | Predicted archaeal cysteine biosynthesis gene clusters: O-acetylserine/O-phosphoserine sulfhydrylase, Cys-RS and SEP-RS. | Implicated in tRNA associated cysteine synthesis |
| YdgL-like | SufE and a pyridoxal phosphate (PLP)-dependent enzyme. | SufE defines a family of sulfur-carrier proteins and the pyridoxal phosphate (PLP)-dependent enzyme is related to cysteine sulfinate desulfinase. SufE proteins and PLP-dependent enzymes are known to cooperate in transferring sulfur from cysteine to metal-sulfur clusters. |
| HesA | NifN, NifB/X, NifW, β-grasp fold (2Fe-2S) ferrodoxin. | The Nif proteins are implicated in formation of metal-sulfur clusters of the nitrogen fixation complexes. |
| FeeI | Always associated with N-acyl amino acid synthase. In some bacteria additionally with: FeeL, FeeK, FeeJ, FeeM. | FeeL: an acyl carrier protein, FeeK: phosphopantetheinylates ACP, FeeJ: ACP acyl transferase, FeeM: acyl adenylate synthase Constitute the complete biosynthetic pathway for N-acyl tyrosine. |
| MccB | microcin C7 permease ( MccC), LD-carboxypeptidase, AdoMet-dependent methyltransferase. | Likely to constitute the system required for peptidolytic processing, methylation and export of microcin produced by this gene cluster. |
| PaaA | ABC transporter, M50-type metallopeptidase, GCN5-like acetyltransferase, prolyl hydroxylase. | Likely to constitute the system required for peptidolytic processing, acetylation, hydroxylation and export of a secondary metabolite. |
| Rv319 | ABC transporter, Lon and Ste24-like peptidases. | Likely to constitute the system required for peptidolytic processing and export of a secondary metabolite. |
| GodD | ABC transporter, CAAX-like metallopeptidase, AdoMet-dependent methyltransferase, GCN5-like acetyltransferase, lantibiotic dehydratase, N-terminally truncated GodD E1-like version. | Likely to constitute the system required for peptidolytic processing, methylation, acetylation, thiazole and oxazole modification, and export of a peptide secondary metabolite. |
| E1 Family Name | Novel domain architectures and functional associations | Comments |
| ThiF/MoeB/MoeZ/MOCS3 | Majority are fused to rhodanese domains. A subset of members instead contains a C-terminal fusion to a CCTBP domain. | The CCTBP domain is predicted to be functionally equivalent to rhodanese domain and contains a conserved cysteine. |
| UBA4 | Genetic interactions suggest involvement of a complex comprised of Ncs2p, Ncs6p, Elp6 and Elp2 (also involved in tRNA thiouridylation) in conjugation of Urm1 to certain target proteins. | Ncs2p andNcs6p are PP-loop ATPases; Elp2 a WD40-type β-propeller protein; Elp6 is an unusual RecA superfamily P-loop NTPase with a modified Walker A motif |
| UBA1 family | Ciliate representatives of this family are fused to BRUCE-type E2 domains. | Function in E3-independent UB transfer in conjunction with fused E2 domains |
| UBA5/UBE1DC1 | Fused to UBA5 C-terminal (U5C) domain. | The U5C has three strands flanked by α-helices. It is predicted to function analogous to the UFD domain (Fig. 1). |
| Apg7/Atg7 | Fusion to Apg7 N-terminal domain. | This domain is an α+β domain that might recruit E2 partners. The Apg7 family might deviate from the usual pattern in utilizing an N-terminal domain in interactions with its functional partners. |
| YKL027W | Interaction with PINT domain subunit of proteosomal lid in high-throughput affinity-capture mass-spectrometry; C-terminal fusion to TRS4-C domain | The TRS4-C domain may recruit functional partner/s downstream of the adenylation reaction. |
| FeeI | Domains fusions usually with N-acyl amino acid synthase and occasionally also with flavindependent oxidoreductase domain. | Cooperates with N-acyl amino acid synthase in N-acyltyrosine biosynthesis. The oxidoreductase might further oxidatively modify this metabolite. |
| MccB | N-terminal fusion to wHTH; Domain fusion with zincin-like oligopeptidase in low GC Grampositive bacteria. | Peptidolytic processing of peptide metabolite by Zincin-like peptidase might be linked to its modification by adenylation. |
| PaaA | N-terminal fusion to wHTH. | |
| Rv3196 | N-terminal fusion to wHTH. | |
| GodD | N-terminal fusion to wHTH; C-terminal fusion with McbD domain ( also known as YcaO domain). | The McbD domain proteins are present in a wide range of bacteria and archaea. |
Functional implications of phyletic patterns and conserved gene neighborhoods
In prokaryotes the primary E1-like enzyme i.e. ThiF/MoeB ortholog is usually embedded in a gene neighborhood encoding thiamin biosynthesis genes, and less frequently in one containing molybdopterin biosynthesis genes 36. Recent characterization of cysteine biosynthesis in actinobacteria showed that the E1-like enzyme MoeZ adenylates a ThiS/MoaD-like Ubl which is then thiolated and used as a sulfur donor for the reaction catalyzed by cysteine synthase. This results in formation of a cysteine at the C-terminus of the Ubl which is then released by a JAB domain peptidase 9. In addition to the previously observed conserved linkage of the genes coding the Ubl, JAB peptidase and cysteine synthase in actinobacteria36 we uncovered novel gene neighborhood linkages between the ThiF/MoeB ortholog and several genes related to cysteine synthesis in several distant bacterial lineages (Figure 3). The linked genes encode several enzymes related to cysteine and methionine biosynthesis (Figure 1, 3 and Table 2; Supplementary Material). Interestingly, we noted that planctomycetes and several proteobacteria share a conserved gene neighborhood, which displays a PDZ-domain containing C-terminal-processing serine peptidase instead of the JAB peptidase. Thus, two unrelated types of peptidases might be utilized to release the newly synthesized C-terminal cysteine in different bacterial lineages. We also identified conserved gene neighborhoods in archaea linking ThiF/MoeB-like genes with those coding O-acetylserine/O-phosphoserine sulfhydrylase. However, the archaeal proteins consistently lacked a linked gene coding for a JAB peptidase, instead showing linkages with either the cysteinyl tRNA synthetase (e.g. Aeropyrum pernix) or O-phosphoseryl tRNA synthetase (e.g. Methanospirillum hungatei). This suggests that in both euryarchaea and crenarchaea E1-dependent biosynthesis of cysteine appears to be directly linked to tRNA aminoacylation. Linkage with the O-phosphoseryl tRNA synthetase suggests that in some methanogenic archaea (Methanocaldococcus and Methanospirillum) the E1-like enzyme-dependent mechanism might be active in in situ cysteine synthesis that occurs after charging of Sep-tRNA by the O-phosphoseryl tRNA synthetase 37.
Phyletic patterns suggest that in majority of prokaryotes a single E1-like protein is utilized in molybdopterin, thiamin and cysteine (if present) biosynthesis pathways. Thus, it appears that all these key adenylation and sulfotransfer reactions can be catalyzed by the same protein. However, on multiple occasions lineage-specific duplications have spawned dedicated paralogs functioning in particular pathways (Figure 1, 3). These include E1-like enzymes fused to or associated via predicted operons with JAB domain peptidases, E2 homologs and diverse Ubls, which are involved in siderophore biosynthesis, metal-sulfur cluster biogenesis or constitute predicted prokaryotic Ub-conjugation-like systems 12,36. We observed that the YdgL family shows gene neighborhood linkages with proteins predicted to participate in sulfur transfer during biosynthesis of metal-sulfur clusters38–40 (Figure 1 and Table 2; Supplementary Material). Hence, we predict that in metal-sulfur cluster synthesis YgdL proteins are likely to provide an initial adenylation step, which is followed by thiolation and thio-transfer mediated by SufE and a cysteine sulfinate desulfinase-like enzyme (Table 2). The HesA family is unique to nitrogen-fixing heterocyst-forming cyanobacteria and vesicle-forming actinobacteria. HesA genes are embedded in neighborhoods encoding proteins involved in formation of metal-sulfur clusters in nitrogen fixation complexes (Figure 1 and Table 1 and Supplementary Material). Thus, the HesA family too is likely to adenylate substrates prior to sulfotransfer in the biosynthesis of these complexes 41,42.
The FeeI, MccB, PaaA, Rv319, and GodD families are usually found in bacteria with complex organization or development such as actinomycetes, cyanobacteria and endospore-forming firmicutes. These families never show linkages to genes encoding Ubls or JAB peptidases. Instead, they show gene-neighborhood associations, which are consistent with their role in adenylation steps in biosynthesis of diverse secondary metabolites. FeeI is often in the neighborhood of a gene coding the N-acyl amino acid synthase with which it cooperates in the synthesis of N-acyl tyrosine (Figure 1 and Table 2; Supplementary Material)32,33. In the MccB, PaaA, Rv319 and GodD families we discovered several distinct gene-neighborhood associations encoding multiple enzymes reflective of the wide array of additional modifications with which adenylation might combine in secondary metabolite biosynthesis (Table 2). Additionally, a subset of the GodD neighborhoods contain a pair of adjacent E1-like genes, one of which encodes a full length version, while the other codes the N-terminally truncated version lacking S1 to S3 (Figure 1; supplementary material). These are predicted to physically interact to generate a dimer with a single active site.
Evidence from domain architectures: prediction of novel interactions related to sulfotransfer and Ub/Ubl conjugation
Experimental studies have suggested an important role for the interplay between different E1-like domains and the respective unrelated C-terminal domains in various reactions catalyzed by them. For example in MOCS3, the rhodanese domain fused to the C-terminus of the E1-like domain initiates sulfotransfer by forming a persulfide linked to its conserved cysteine. The Ubl (i.e. MoaD) adenylated by the E1-like domain is then attacked by this persulfide to form an acyl-disulfide linkage. This linkage is then reduced by the thiolating cysteine on of the E1-like domain of MOCS3 to release a thiolated MoaD 18,43,44. Likewise, many eukaryotic E1 families contain a C-terminal permuted ubiquitin-like domain, the UFD domain that recruits the E2 enzyme, delivering it to the trans-thiolating active site of the E1 enzyme 16. These observations suggested that interaction between C-terminal domains and the active sites on the E1-like domain might be a general theme required for linking successive reactions that follow the initial adenylation.
Across the three superkingdoms of life MoeB/ThiF-like proteins involved in cofactor and cysteine synthesis and their paralogs involved in siderophore biosynthesis are often fused to a rhodanese domain (Fig.1 and 3). However, E1-like proteins in low GC Gram-positive bacteria and sporadically in other bacterial and archaeal lineages lack a C-terminal rhodanese domain or even one encoded by a standalone neighboring gene. Interestingly, we found that many of these proteins had another C-terminal domain, which also occurs as a standalone protein in euryachaea (e.g. Pyrococcus furiosus PF0466). Using transitive sequence profile searches with the PSI-BLAST program and profile-profile comparisons with the HHpred program we established a statistically significant relationship (p-value=10−5 in profile-profile comparisons) between this domain and the TATA-box binding protein (TBP) domain 45. A multiple alignment of the domain revealed an absolutely conserved cysteine residue in the N-terminal strand (Fig. 4) and accordingly we term it the CCTBP (cysteine containing TBP-like) domain. Comparisons with the TBP structure suggest that the cysteine is present on the same face of the helix-grip fold of TBP that mediates contact with DNA 46, and is hence likely to be a surface residue available for persulfide bond formation. Thus, it is probable that in E1-like proteins that possess the CCTBP domain, it is functionally equivalent to the rhodanese domain. Consistent with this prediction, the CCTBP domain is found in contexts analogous to the rhodanese domain which is suggestive of a role in sulfur metabolism and metal-sulfur cluster assembly. For example, the CCTBP domain is fused to PP-loop ATPase domain in some archaea (e.g. MTH990 from Methanothermobacter). This is reminiscent of the fusion of the rhodanese domain and the PP-loop ATPase domain in the ThiI protein, where the two domains cooperate in successive adenylation and sulfur transfer steps during 4-thiouridine biosynthesis 47. The CCTBP domain is also fused to 4Fe-4S ferredoxins and DNA-binding helix-turn-helix domains in several archaea, and is found in the gene neighborhood of cysteine desulfurases and proteins involved in redox reactions (Fig. 4, Supplementary Material). These observations suggest that the CCTBP domain might also function in assembly of metal-sulfur clusters in ferredoxins as well as a redox sensor of single-component transcription factors.
Figure 4. Multiple sequence alignment of CCTBP-like and TATA-box binding protein domains.
Proteins are labeled to the left of each sequence by their gene names, species abbreviations, and gi numbers demarcated by underscores. Amino acid residues are colored according to side chain properties and degree of conservation within the alignment, set at 75% consensus. The secondary structure is indicated above the alignment. The conserved cysteine of the CCTBP domain is in yellow and shaded in red. The consensus abbreviations and coloring scheme are as follows: h, hydrophobic residues shaded yellow; s, small residues colored green; p, polar residues colored purple; +, positively charged residues colored blue and shaded gray; and b, big residues colored blue. The conserved glycine residue is colored light green. Species abbreviations are as follows: Abac: Acidobacteria bacterium; Aful: Archaeoglobus fulgidus; Amet: Alkaliphilus metalliredigenes; Anid: Aspergillus nidulans; Apen: Acetabularia peniculus; Aper: Aeropyrum pernix; AtCV1: Acanthocystis turfacea Chlorella virus 1; Atha : Arabidopsis thaliana; Bfir: Bacillus firmus; Blic: Bacillus licheniformis; Bmar: Blastopirellula marina; Bsp.: Bacillus sp.; Bsub: Bacillus subtilis; Bthu: Bacillus thuringiensis; CDes: Candidatus Desulfococcus; CKue: Candidatus Kuenenia; Cele: Caenorhabditis elegans; Cmaq: Caldivirga maquilingensis; Csym: Cenarchaeum symbiosum; Esib: Exiguobacterium sibiricum; Gkau: Geobacillus kaustophilus; Glam: Giardia lamblia; Hasp: Halobacterium sp.; Haur: Herpetosiphon aurantiacus; Hmob: Heliobacillus mobilis; Hsap: Homo sapiens; Linn: Listeria innocua; Lmon: Listeria monocytogenes; Lpla: Lactobacillus plantarum; Lreu: Lactobacillus reuteri; Mace: Methanosarcina acetivorans; Maeo: Methanococcus aeolicus; Mbar: Methanosarcina barkeri; Mbur: Methanococcoides burtonii; Mjan: Methanocaldococcus jannaschii; Mlab: Methanocorpusculum labreanum; Mmar: Methanoculleus marisnigri; Mmaz: Methanosarcina mazei; Mmus: Mus musculus; Moth: Moorella thermoacetica; Mthe: Methanosaeta thermophila; Mthe: Methanothermobacter thermautotrophicus; Mthe: Methanothermococcus thermolithotrophicus; Nmar: Nitrosopumilus maritimus; Npha: Natronomonas pharaonis; Nvec: Nematostella vectensis; Oluc: Ostreococcus lucimarinus; Paby: Pyrococcus abyssi; PbCVN : Paramecium bursaria Chlorella virus NY2A; Pfur: Pyrococcus furiosus; Pthe: Pelotomaculum thermopropionicum; Ptor: Picrophilus torridus; Pyae: Pyrobaculum aerophilum; Rbal: Rhodopirellula baltica; Saur: Staphylococcus aureus; Scer: Saccharomyces cerevisiae; Sepi: Staphylococcus epidermidis; Sfum: Syntrophobacter fumaroxidans; Shae: Staphylococcus haemolyticus; Smar: Staphylothermus marinus; Spom: Schizosaccharomyces pombe; Stok: Sulfolobus tokodaii; Susi: Solibacter usitatus; Taci: Thermoplasma acidophilum; Tkod: Thermococcus kodakarensis; Tneu: Thermoproteus neutrophilus; Umet: uncultured methanogen; Usul : uncultured sulfate-reducer.
The C-terminal UFD domain is limited to only three of the active families of eukaryotic E1s (Fig. 1). The Urm1p-activating enzyme UBA4 has a rhodanese domain just like its orthologs from other organism involved in cofactor biosynthesis (MOCS3/ThiF/MoeB). Extensive genetic screens and biochemical characterization to date have not yielded an E2 in urmylation 48. However, in MOCS3 the C-terminal rhodanese domain interacts with the thiolating active site in a manner comparable to the delivery of E2 by the C-terminal UFD of UBA3 18,43. Hence, we predict that the rhodanese domain functions like the E2 with its active cysteine behaving like the E2 catalytic cysteine during urmylation. However, other catalytically active families, namely UBA5 (Ufm activating enzyme) and Agp7/Atg7 (Apg8, Apg12 activating enzyme) which utilize E2s, also lack an UFD domain. We observed that the UBA5 family contains a conserved C-terminal region, which is predicted to form a distinct globular domain apparently unrelated to the UFD domain. Given the C-terminal location of this domain in the UBA5 proteins it is possible that it represents a functional analog of the UFD domain, which independently emerged in this family. In contrast, the Apg7/Atg7 family instead displays a large N-terminal domain which might help it recruit its functional partners, such as the two distinct E2 enzymes, Apg3 and Apg10 (Figure 1 and Table 2). The eukaryotic YKL027W family is closely related to the bacterial YgdL family (see above). YKL027W is fused to the recently reported TRS4-C domain and associates with Rpn6p, the PINT domain subunit of the proteasomal lid 49,50 (Figure 1 and Table 2). The TRS4-C domain is also fused to an Ubl domain in the TRS4 family of proteins 30. These associations suggest that YKL027W might be associated with Ub-conjugation despite the absence of the thiolating cysteine. The TRS4-C domain possibly plays a role analogous to the UFD domain in recruiting a downstream partner after the initial adenylation reaction catalyzed by the E1-like domain.
Evidence from domain architectures: prediction of novel interactions related to secondary metabolite biosynthesis
In the FeeI, MccB, GodD, PaaA and Rv3196 families we identified several domain architectures that predict a close linkage between the E1 catalyzed adenylation and other associated reactions in secondary metabolite biosynthesis (Figure 1 and Table 2; supplementary material). Of particular interest is the frequent fusion to the McbD domain in the GodD family. In the processing of microcin B17, another peptide with heterocyclic modifications, a McbD domain protein forms a complex with a flavin-dependent oxidoreductase (McbC) belonging to the same family as those fused to some FeeI proteins and encoded by predicted operons of the GodD family (Figure 1 and Table 2). These proteins are required for the formation of aromatic heterocyclic thiazole or oxazole rings from cysteine or serine respectively and their adjacent residue 51. Formation of both thiazole and oxazole rings involve a dehydrogenase and a dehydratase reaction 52. The flavin-dependent oxidoreductase McbC is likely to catalyze the former reaction. While McbD was earlier claimed to show similarity to GTPases 53, we found that neither sequence profile searches, nor the conservation pattern, nor alignment-based secondary structure predictions support this relationship. Instead the McbD domain was predicted to adopt an α/β fold that showed a completely different conservation pattern with a glycine-rich loop and other absolutely conserved polar residues suggestive of a distinct enzymatic role. Hence, McbD probably catalyzes the dehydratase reaction required in these modifications. Additionally, in the case of the thiazole formation, there is likely to be an adenylation step catalyzed by the E1-like domain of the GodD-family enzyme prior to carbon-sulfur bond formation. Many McbD domain proteins that lack fusions to the GodD family are instead fused to OsmC-like domains 54 with conserved cysteines that are capable of carrying sulfur atoms. In these cases the OsmC domains might provide an alternative mechanism for sulfur delivery in conjunction with the heterocyclization catalyzed by McbD domains.
The MccB, GodD, PaaA and Rv3196 families are united by the presence of an N-terminal winged HTH domain (Figure 1 and Table 2), which we established by means of sequence profile analysis (PSI-BLAST e-value 10−3). Based on available E1 structures we predict that the N-terminal wHTH domain in these families probably forms a cap over the active site of the adjacent monomer. Thus, it could potentially provide an additional nucleotide-binding interface and also a means to guide the peptide substrate to the active site. Such a role played by the wHTH domain might explain some unusual features observed in these families, such as loss of the arginine finger, the N-terminal divergence and loss of the nucleotide-binding loop between S2 and H2 (Table 1).
EVOLUTIONARY HISTORY OF THE E1 SUPERFAMILY
Early evolution and prokaryotic adaptations of E1-like proteins
The presence of at least one representative of the E1-like superfamily in the three superkingdoms of life suggests that it was present in the last universal common ancestor (LUCA). Based on extant versions we can infer that this ancestral version resembled ThiF/MoeB proteins and functioned as a dimer with a symmetric pair of adenylating and thiolating active sites. Earlier studies on Rossmannoid superfamilies have shown that S-AdoMet dependent methyltransferases and FAD/NAD(P) dependent dehydrogenases had already diversified to spawn multiple lineages by the time of LUCA 22,23. Thus, the evolutionary divergence of E1-like domains and these related Rossmannoid superfamilies occurred prior to LUCA. In contrast to the E1-like domain, the ThiS and MoaD families of Ubls appear to have been distinct in LUCA 30 itself, probably providing the primary determinants of pathway specificity. Hence, it is likely that LUCA possessed a single multi-functional E1-like domain that interacted with both ThiS and MoaD-like proteins. Subsequently, by the time of divergence of the bacterial superkingdom functional associations between E1-like domains and sulfur-carrying rhodanese and JAB peptidase domains appear to have emerged. Superposition of domain architectures and gene neighborhoods on the phylogenetic tree of the prokaryotic ThiF/MoeB proteins suggests that at least in bacteria the rhodanese domain was ancestrally fused to the E1 or associated as a neighboring gene in an operon.
In some bacteria the rhodanese domain was also displaced by the non-homologous CCTBP domain (Figure 3). The TBP domain is universally conserved in the archaeo-eukaryotic lineage as a component of the basal transcription apparatus. Other than the CCTBP domain, the only TBP-like domain found in bacteria is also predominantly present in low GC Gram-positive bacteria and is found fused to the RNAse domain in RNase HIII proteins 55,56. These phyletic patterns suggest that the CCTBP- and RNAse HIII- associated TBP-like domains were laterally transferred to low GC Gram-positive bacteria from archaea. However, the CCTBP domain appears to have acquired a role in mediating sulfur transfer/redox reactions prior to the transfer. The lack of concordance between the protein tree and the prokaryotic species tree (Figure 3) 44 suggests rampant lateral transfer of the E1 domain between distant lineages in the post-LUCA evolution of the superfamily. Moreover, on multiple occasions the E1 enzyme duplicated to give rise to separate paralogs dedicated to MoCo and thiamine biosynthesis (e.g. independently in γ-proteobacteria and in the low GC Gram-positive bacteria) or cysteine biosynthesis (e.g. in mycobacteria). The independence of these events is also supported by the distinct domain architectures of the E1-like proteins associated with the thiamine and MoCo pathways in each of these lineages (Figure 3).
Another major facet of the post-LUCA evolution of E1-like domains in the bacteria was the emergence of several novel lineage-specific paralogs associated with the innovation of novel metabolic capabilities. For example, E1-like enzymes involved in biosynthesis of siderophores and related protective compounds were derived from the MoeB/ThiF proteins (Figure 3). They were recruited to perform biochemically similar reactions as the latter in these new secondary metabolism pathways. The most dramatic adaptation of this type was the origin and radiation of the monophyletic group of FeeI, MccB, PaaA, Rv319, and GodD families (Figure 1 and Table 3). These families display extraordinary sequence divergence relative to E1-like domains involved in the more conserved primary metabolic systems. Hence, they are possibly under strong selection due to the need to recognize a rapidly diversifying set of secondary metabolite substrates that range from fatty acids to several small peptides with no detectable sequence similarity.
Origin and evolution of Eukaryotic E1 enzymes
New insights regarding the origin of the E1s of Ub/Ubl conjugation systems had emerged from our earlier discovery of potential bacterial cognates of eukaryote-type Ub/Ubl conjugation systems 36. E1-like domains of these systems belong to a cluster of five related families (6A–E in Fig. 1), which are consistently found in operons or fused to E2 domains. Gene-neighborhoods encoding these E1-like proteins never contain any genes for cofactor, cysteine or secondary metabolite biosynthesis. This strongly supports the conjecture that these E1-like domains function in association their cognate E2s in primitive Ub/Ubl conjugation-type systems. A version of these bacterial systems probably spawned the ancestral E1–E2 pair of all eukaryotic E1s functioning in conjunction with an E2 in the first eukaryotic common ancestor. The abundance of these systems in α-proteobacteria 36, from which the mitochondrial endosymbiont emerged, makes it a plausible source for this ancestral E1–E2 pair. By the time of the last eukaryotic common ancestor (LECA), E1s had radiated into 7 distinct families, which appears to have occurred concomitantly with an even more extensive radiation of Ubls, resulting in a wide range of protein modifiers 30. Prior fixation of robust de-ubiquitination and degradation systems in the form of the proteasome and its lid complex in the eukaryotic progenitor possibly favored this proliferation of modifications by Ub/Ubls.
The first divergence in the pre-LECA evolution of E1s appears to have given rise to the Apg7/Atg7 family that conjugates Apg12/Apg8 to protein and lipid substrates 25. The next divergence resulted in formation of the respective ancestors of the active and inactive subunits of all extant Ub/Ubl-conjugating enzymes. By LECA the ancestral active and inactive monomers had concomitantly diversified into 3 families each (Fig. 1), of which an active and inactive pair fused to give rise to the UBA1 family. These 3 pairs of families constituted the activating enzymes of Ub (UBA1 N- and C-terminal domains), SUMO (UBA2 and AOS1/SAE1 families) and Nedd8 (UBA3 and APPBP1 families). Subsequently the UBA5 family appears to have emerged just prior to the divergence of kinetoplastid-heterolobosean lineage, and acquired specificity for Ufm1, a pre-existing Ubl. Further, throughout eukaryotic evolution there were several lineage-specific duplications of E1-like domains. This was most rampant in the UBA1 family, where a duplication in the common ancestor of amoebozoa, fungi and metazoa resulted in UBA6, which might activate Fat10 57. In vertebrates, another duplication in the UBA1 family resulted in UBE1L, the activating enzyme for ISG15 involved in interferon response 58. Similarly, a lineage-specific duplication of the UBA1 family occurred in ciliates along with a fusion to an N-terminal E2 domain of the BRUCE family 59. The UBA1 family also underwent sporadic lineage-specific duplications in stramenopiles and kinetoplastids suggesting their possible diversification into different functional contexts.
Interestingly, there appear to have been additional independent transitions of other eukaryotic E1-like families to Ubl-conjugation-related roles. Eukaryotic MoeB/ThiF orthologs (e.g. MOCS3) have been shown to function like their prokaryotic counterparts in MoCo biosynthesis along with their Ubl partners 44. However, the yeast ThiS/MoaD ortholog, Urm1p is conjugated to protein targets by its cognate E1-like enzyme (UBA4, the fungal MOCS3 ortholog). Genetic studies have also implicated a distinct complex of proteins (Table 2), which are additionally required for synthesis of 2-thiouridine at the wobble position of the tRNA 48, in conjugation of Urm1p to some target proteins. Of these Ncs2p and Ncs6p are PP-loop ATPases, which catalyze a adenylating reactions similar to the E1-like enzymes 23,60. Hence, Ncs2p and Ncs6p could have independently acquired an E1-like function required for some of the Urm1 conjugation events, possibly functioning in conjunction with Elp2p a WD40-type β-propeller and Elp6p, an unusual RecA superfamily P-loop NTPase. In this light it would be of interest to investigate if Urm1p-like ThiS/MoaD orthologs are also involved in tRNA thiobase synthesis as sulfur carriers in conjunction with the above protein complex. The YKL027W family appears to have emerged from an independent lateral transfer of the bacterial YgdL family into eukaryotes after the divergence of diplomonads and parabasalids. It also appears to have independently acquired an Ub-related function in eukaryotes (Table 2; see above).
CONCLUSIONS AND GENERAL OBSERVATIONS
Here we present a synthetic view of the natural history of the E1-like superfamily by combining all available sequence, structure, biochemical and contextual information. Consequently, we were able to develop a natural classification of the superfamily that allowed us to reconstruct its structural and biochemical diversification. We also clarify the multiple origins and subsequent evolution of different Ub/Ubl-activating versions in eukaryotes. The observations reported here have generated several hypotheses (e.g. Table 2) testable by experimental biochemical studies. We hope that this synthesis provides a resource (see supplementary material) that spurs new directed investigations on the less-studied E1-like families and their functions.
MATERIALS AND METHODS
The FSSP program was used for structure similarity searches 61, and the MUSTANG program to generate structural alignments 62. Protein structures were visualized and manipulated using Swiss-PDB 63 and PyMol (http://pymol.sourceforge.net/) programs. Sequence profile searches were performed against the NCBI non-redundant (NR) protein database (National Center for Biotechnology Information, NIH, Bethesda, MD), and a locally compiled database of proteins from completely or near-completely sequenced genomes. PSI-BLAST searches were performed using an expectation value (E-value) of 0.01 used as the threshold for inclusion into the profile 64; searches were iterated until convergence. Alignment-derived HMM searches were performed using the HMMer package 65. Multiple alignments were constructed using the MUSCLE 66 and Kalign 67 programs, followed by manual correction based on PSI-BLAST high-scoring pairs, secondary structure predictions, and available crystal structures. Protein secondary structure was predicted using a multiple alignment as the input for the JPRED2 program, which uses information extracted from a PSSM, HMM, and residue frequencies in alignment columns68. Pairwise comparisons of HMMs, using a single sequence or multiple alignment as query, against profiles of proteins in the PDB database were performed with the HHPRED program 69. Similarity-based clustering was performed using the BLASTCLUST program [ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html] with empirically determined length and score threshold parameters. Gene neighborhoods in prokaryotes were obtained by isolating conserved genes immediately upstream and downstream of the gene in question showing separation of less than 70 nucleotides between gene termini. Neighborhoods were determined by searching NCBI PTT tables (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Genome) with a custom PERL script. Phylogenetic analysis was carried out using neighborhood-joining and minimum evolution-based methods with gamma distributed rates and a JTT substitution matrix as implemented in the MEGA4 program 70. The shape parameter α was estimated empirically through a series of trials. Maximum likelihood trees were also obtained by first generating the least-squares tree (FITCH program of the PHYLIP package 71) with subsequent local rearrangement using the PROTML program (MOLPHY package 72). The reliability of the tree topology was assessed using the RELL bootstrap method of MOLPHY, with 10 000 replicate 72. All large-scale procedures were carried out using the TASS software package (Anantharaman V, Balaji S, Aravind L, unpublished).
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the Intramural research program of the National Library of Medicine, National Institutes of Health, USA for funding their research.
Footnotes
SUPPLEMENTARY INFORMATION
A comprehensive alignment containing all E1-like families, alignments of different domains fused to E1-like domains, conserved gene neighborhoods and a comprehensive list of gis are provided at ftp://ftp.ncbi.nih.gov/pub/aravind/temp/supplementary_material_E1.html
REFERENCES
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York, NY: Garland Science Publishing; 2002. [Google Scholar]
- 2.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. Bioessays. 2000;22(5):442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.McGrath JP, Jentsch S, Varshavsky A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. Embo J. 1991;10(1):227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duda DM, Walden H, Sfondouris J, Schulman BA. Structural analysis of Escherichia coli ThiF. J Mol Biol. 2005;349(4):774–786. doi: 10.1016/j.jmb.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann C, Begley TP, Ealick SE. Structure of the Escherichia coli ThiS-ThiF complex, a key component of the sulfur transfer system in thiamin biosynthesis. Biochemistry. 2006;45(1):11–19. doi: 10.1021/bi051502y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leimkuhler S, Wuebbens MM, Rajagopalan KV. Characterization of Escherichia coli MoeB and its involvement in the activation of molybdopterin synthase for the biosynthesis of the molybdenum cofactor. J Biol Chem. 2001;276(37):34695–34701. doi: 10.1074/jbc.M102787200. [DOI] [PubMed] [Google Scholar]
- 7.Lake MW, Wuebbens MM, Rajagopalan KV, Schindelin H. Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex. Nature. 2001;414(6861):325–329. doi: 10.1038/35104586. [DOI] [PubMed] [Google Scholar]
- 8.Rudolph MJ, Wuebbens MM, Rajagopalan KV, Schindelin H. Crystal structure of molybdopterin synthase and its evolutionary relationship to ubiquitin activation. Nat Struct Biol. 2001;8(1):42–46. doi: 10.1038/83034. [DOI] [PubMed] [Google Scholar]
- 9.Burns KE, Baumgart S, Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Reconstitution of a new cysteine biosynthetic pathway in Mycobacterium tuberculosis. J Am Chem Soc. 2005;127(33):11602–11603. doi: 10.1021/ja053476x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortese MS, Caplan AB, Crawford RL. Structural, functional, and evolutionary analysis of moeZ, a gene encoding an enzyme required for the synthesis of the Pseudomonas metabolite, pyridine-2,6-bis(thiocarboxylic acid) BMC Evol Biol. 2002;2:8. doi: 10.1186/1471-2148-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis TA, Cortese MS, Sebat JL, Green TL, Lee CH, Crawford RL. A Pseudomonas stutzeri gene cluster encoding the biosynthesis of the CCl4-dechlorination agent pyridine-2,6-bis(thiocarboxylic acid) Environ Microbiol. 2000;2(4):407–416. doi: 10.1046/j.1462-2920.2000.00122.x. [DOI] [PubMed] [Google Scholar]
- 12.Godert AM, Jin M, McLafferty FW, Begley TP. Biosynthesis of the thioquinolobactin siderophore: an interesting variation on sulfur transfer. J Bacteriol. 2007;189(7):2941–2944. doi: 10.1128/JB.01200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onaka H, Nakaho M, Hayashi K, Igarashi Y, Furumai T. Cloning and characterization of the goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiology. 2005;151(Pt 12):3923–3933. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Pastor JE, San Millan JL, Castilla MA, Moreno F. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J Bacteriol. 1995;177(24):7131–7140. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N-P bond formation in microcin C7. J Am Chem Soc. 2008;130(11):3603–3609. doi: 10.1021/ja7101949. [DOI] [PubMed] [Google Scholar]
- 16.Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA. Basis for a ubiquitin-like protein thioester switch toggling E1–E2 affinity. Nature. 2007;445(7126):394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 18.Matthies A, Nimtz M, Leimkuhler S. Molybdenum cofactor biosynthesis in humans: identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry. 2005;44(21):7912–7920. doi: 10.1021/bi0503448. [DOI] [PubMed] [Google Scholar]
- 19.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361(5):1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol. 1997;4(9):686–689. doi: 10.1038/nsb0997-686. [DOI] [PubMed] [Google Scholar]
- 21.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. Embo J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aravind L, Mazumder R, Vasudevan S, Koonin EV. Trends in protein evolution inferred from sequence and structure analysis. Curr Opin Struct Biol. 2002;12(3):392–399. doi: 10.1016/s0959-440x(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 23.Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48(1):1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 24.Huang DT, Walden H, Duda D, Schulman BA. Ubiquitin-like protein activation. Oncogene. 2004;23(11):1958–1971. doi: 10.1038/sj.onc.1207393. [DOI] [PubMed] [Google Scholar]
- 25.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395(6700):395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 26.Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32(17):5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burroughs AM, Iyer LM, Aravind L. In: Comparative Genomics and Evolutionary Trajectories of Viral ATP Dependent DNA-Packaging Systems. J-N V, editor. Basel: S. Karger AG; 2007. [DOI] [PubMed] [Google Scholar]
- 28.Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Jr, Holton JM, Schulman BA. The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell. 2003;12(6):1427–1437. doi: 10.1016/s1097-2765(03)00452-0. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Hu W, Cai S, Lee B, Song J, Chen Y. The intrinsic affinity between E2 and the Cys domain of E1 in ubiquitin-like modifications. Mol Cell. 2007;27(2):228–237. doi: 10.1016/j.molcel.2007.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leimkuhler S, Rajagopalan KV. A sulfurtransferase is required in the transfer of cysteine sulfur in the in vitro synthesis of molybdopterin from precursor Z in Escherichia coli. J Biol Chem. 2001;276(25):22024–22031. doi: 10.1074/jbc.M102072200. [DOI] [PubMed] [Google Scholar]
- 32.Brady SF, Chao CJ, Clardy J. Long-chain N-acyltyrosine synthases from environmental DNA. Appl Environ Microbiol. 2004;70(11):6865–6870. doi: 10.1128/AEM.70.11.6865-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady SF, Chao CJ, Clardy J. New natural product families from an environmental DNA (eDNA) gene cluster. J Am Chem Soc. 2002;124(34):9968–9969. doi: 10.1021/ja0268985. [DOI] [PubMed] [Google Scholar]
- 34.Wolf YI, Rogozin IB, Kondrashov AS, Koonin EV. Genome alignment, evolution of prokaryotic genome organization and prediction of gene function using genomic context. Genome Res. 2001;11:356–372. doi: 10.1101/gr.gr-1619r. [DOI] [PubMed] [Google Scholar]
- 35.Huynen M, Snel B, Lathe W, 3rd, Bork P. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 2000;10(8):1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer LM, Burroughs AM, Aravind L. The prokaryotic antecedents of the ubiquitin-signaling system and the early evolution of ubiquitin-like beta-grasp domains. Genome Biol. 2006;7(7):R60. doi: 10.1186/gb-2006-7-7-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, 3rd, Ibba M, Soll D. RNA-dependent cysteine biosynthesis in archaea. Science. 2005;307(5717):1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 38.Liu G, Li Z, Chiang Y, Acton T, Montelione GT, Murray D, Szyperski T. High-quality homology models derived from NMR and X-ray structures of E. coli proteins YgdK and Suf E suggest that all members of the YgdK/Suf E protein family are enhancers of cysteine desulfurases. Protein Sci. 2005;14(6):1597–1608. doi: 10.1110/ps.041322705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Layer G, Gaddam SA, Ayala-Castro CN, Ollagnier-de Choudens S, Lascoux D, Fontecave M, Outten FW. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J Biol Chem. 2007;282(18):13342–13350. doi: 10.1074/jbc.M608555200. [DOI] [PubMed] [Google Scholar]
- 40.Ye H, Abdel-Ghany SE, Anderson TD, Pilon-Smits EA, Pilon M. CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J Biol Chem. 2006;281(13):8958–8969. doi: 10.1074/jbc.M512737200. [DOI] [PubMed] [Google Scholar]
- 41.Einsle O, Tezcan FA, Andrade SL, Schmid B, Yoshida M, Howard JB, Rees DC. Nitrogenase MoFe-protein at 1.16 A resolution: a central ligand in the FeMo-cofactor. Science. 2002;297(5587):1696–1700. doi: 10.1126/science.1073877. [DOI] [PubMed] [Google Scholar]
- 42.Curatti L, Hernandez JA, Igarashi RY, Soboh B, Zhao D, Rubio LM. In vitro synthesis of the iron-molybdenum cofactor of nitrogenase from iron, sulfur, molybdenum, and homocitrate using purified proteins. Proc Natl Acad Sci U S A. 2007;104(45):17626–17631. doi: 10.1073/pnas.0703050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthies A, Rajagopalan KV, Mendel RR, Leimkuhler S. Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc Natl Acad Sci U S A. 2004;101(16):5946–5951. doi: 10.1073/pnas.0308191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krepinsky K, Leimkuhler S. Site-directed mutagenesis of the active site loop of the rhodanese-like domain of the human molybdopterin synthase sulfurase MOCS3. Major differences in substrate specificity between eukaryotic and bacterial homologs. Febs J. 2007;274(11):2778–2787. doi: 10.1111/j.1742-4658.2007.05811.x. [DOI] [PubMed] [Google Scholar]
- 45.Iyer LM, Koonin EV, Aravind L. Adaptations of the helix-grip fold for ligand binding and catalysis in the START domain superfamily. Proteins. 2001;43(2):134–144. doi: 10.1002/1097-0134(20010501)43:2<134::aid-prot1025>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 46.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 47.Aravind L, Koonin EV. THUMP--a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends in biochemical sciences. 2001;26(4):215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- 48.Goehring AS, Rivers DM, Sprague GF., Jr Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell. 2003;14(11):4329–4341. doi: 10.1091/mbc.E03-02-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends in biochemical sciences. 1998;23(6):204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 50.Aravind L, Ponting CP. Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci. 1998;7(5):1250–1254. doi: 10.1002/pro.5560070521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science. 1996;274(5290):1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 52.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. Low-molecular-weight post-translationally modified microcins. Mol Microbiol. 2007;65(6):1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 53.Milne JC, Eliot AC, Kelleher NL, Walsh CT. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry. 1998;37(38):13250–13261. doi: 10.1021/bi980996e. [DOI] [PubMed] [Google Scholar]
- 54.Koonin EV, Aravind L, Galperin MY. In: A comparative-genomic view of the microbial stress response. Storz G, Hengge-Aronis R, editors. Washington DC: ASM Press; 2000. pp. 417–444. [Google Scholar]
- 55.Aravind L, Koonin EV. A natural classification of ribonucleases. Methods Enzymol. 2001;341:3–28. doi: 10.1016/s0076-6879(01)41142-6. [DOI] [PubMed] [Google Scholar]
- 56.Chon H, Matsumura H, Koga Y, Takano K, Kanaya S. Crystal structure and structure-based mutational analyses of RNase HIII from Bacillus stearothermophilus: a new type 2 RNase H with TBP-like substrate-binding domain at the N terminus. J Mol Biol. 2006;356(1):165–178. doi: 10.1016/j.jmb.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 57.Chiu YH, Sun Q, Chen ZJ. E1-L2 activates both ubiquitin and FAT10. Mol Cell. 2007;27(6):1014–1023. doi: 10.1016/j.molcel.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 58.Ritchie KJ, Zhang DE. ISG15: the immunological kin of ubiquitin. Semin Cell Dev Biol. 2004;15(2):237–246. doi: 10.1016/j.semcdb.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14(6):801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 60.Duquesne S, Destoumieux-Garzon D, Zirah S, Goulard C, Peduzzi J, Rebuffat S. Two enzymes catalyze the maturation of a lasso peptide in Escherichia coli. Chem Biol. 2007;14(7):793–803. doi: 10.1016/j.chembiol.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 61.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26(1):316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Konagurthu AS, Whisstock JC, Stuckey PJ, Lesk AM. MUSTANG: a multiple structural alignment algorithm. Proteins. 2006;64(3):559–574. doi: 10.1002/prot.20921. [DOI] [PubMed] [Google Scholar]
- 63.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 64.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lassmann T, Sonnhammer EL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40(3):502–511. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 69.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33(Web Server issue):W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 71.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 72.Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.