Abstract
Sirtuins are NAD+-dependent enzymes that deacetylate a variety of cellular proteins and in some cases catalyze protein ADP-ribosyltransfer. The catalytic mechanism of deacetylation is proposed to involve an ADPR-peptidylimidate, whereas the mechanism of ADP-ribosyltransfer to proteins is undetermined. Herein we characterize a Plasmodium falciparum sirtuin that catalyzes deacetylation of histone peptide sequences. Interestingly, the enzyme can also hydrolyze NAD+. Two mechanisms of hydrolysis were identified and characterized. One is independent of acetyllysine substrate and produces α-stereochemistry as established by reaction of methanol which forms α-1-O-methyl-ADPR. This reaction is insensitive to nicotinamide inhibition. The second solvolytic mechanism is dependent on acetylated peptide and is proposed to involve the imidate to generate β-stereochemistry. Stereochemistry was established by isolation of β-1-O-methyl-ADPR when methanol was added as a co-solvent. This solvolytic reaction was inhibited by nicotinamide, suggesting that nicotinamide and solvent compete for the imidate. These findings establish new reactions of wildtype sirtuins and suggest possible mechanisms for ADP-ribosylation to proteins. These findings also illustrate the potential utility of nicotinamide as a probe for mechanisms of sirtuin catalyzed ADP-ribosyltransfer.
The sirtuins are protein-modifying enzymes broadly found in all phyla of life that utilize NAD+ 1 as a substrate to effect protein modification via deacetylation and/or ADP-ribosyltransfer (1, 2). Sirtuins have been shown to participate in a variety of biological processes, including lifespan regulation, stress response, DNA silencing, transcriptional regulation and metabolic control (3, 4). Sirtuins have phylogenetically highly-conserved active sites, although distinct enzymes catalyze deacetylation (1, 2), ADP-ribosyltransfer to protein nucleophiles (5–7) or both (7). The mechanism for the deacetylation reaction catalyzed by sirtuins is consistent with an imidate mechanism (8) and is supported by a wealth of data (9–16). The protein ADP-ribosyltransfer mechanisms are undetermined and products of ADPR-transfer have been largely uncharacterized (5, 6, 17). Sauve and Schramm argued that sirtuin chemistry could be understood by recognizing that ADP-ribosyltransfer is central to their catalytic function (9). This mechanistic proposal suggests that sirtuin catalysis of protein deacetylation and ADP-ribosyltransfers could be the consequence of the versatile reactivity of a sirtuin-poised NAD+ electrophile activated to react directly with different types of nucleophiles (9). It has also been suggested that ADPribosyl-transfer can derive from the reactivity of an intermediate complex (9, 17). We herein provide evidence for the co-existence of these three different reactive mechanisms (deacetylation, direct-ADPR transfer and intermediate-dependent ADPR-transfer) occurring on the active site of a sirtuin derived from Plasmodium falciparum, called Pf-Sir2.
Our preliminary interest in Pf-Sir2 derived from a proposed role for this enzyme in the persistence of malarial infection (300–500 million cases per year worldwide, 1–2 million deaths per year), in which Plasmodium falciparum is the most responsible pathogen (18). Malaria is difficult to treat, since it typically persists in the host far beyond initial infection or treatment. Persistence of infection is linked to immune avoidance strategies employed by the parasite. The parasite expresses a virulence factor called P. falciparum erythrocyte membrane protein 1 (PfEMP1) on infected erythrocytes. Because it is surface-exposed it could enable clearance of the parasite via an adaptive response of the immune system. However, the parasite swaps the epitope-encoding surface proteins periodically, thereby evading host immunity. Specifically, P falciparum has a repertoire of approximately 60 var genes that encode functionally similar but epitopically variant PfEMP1 proteins of which only a single gene is typically expressed in the host at a time, with the remainder of the genes kept silent by chromatin silencing mechanisms (19). This switching strategy of the parasite is termed antigenic variation (19). Interestingly, deletion of Pf-Sir2 dysregulates silencing of a major subset of var genes (20, 21). It has been proposed that the role of Pf-Sir2 in silencing of var genes is through the enzymatic activity of histone deacetylation, which can promote or maintain heterochromatin (20, 21). In fact, the prototype sirtuin, yeast Sir2 (silencing information regulator 2), is named for its gene silencing functions and it requires intact NAD+ dependent deacetylase activity for these functions (2).
To biochemically characterize the reactivity of Pf-Sir2 we cloned, expressed and purified the enzyme. We confirm it to be a deacetylase of histone H3 and histone H4 and other peptide sequences, as predicted. Unexpectedly, we determined that the enzyme has several additional activities, including the capacity to catalyze NAD+ solvolysis. The solvolysis chemistry features separate stereochemical modalities, dependent and independent of acetylated substrates. The stereochemically distinct solvolytic reactions also exhibit different behaviors with respect to the universal sirtuin inhibitor nicotinamide. Pf-Sir2 provides an instructive and unusual example of several competing reactivities, including deacetylation and two types of ADP-ribosyltransfer, occurring in one sirtuin active site. The respective solvolysis chemistries are first examples of their kind attributed to a wildtype sirtuin. In addition, these reactions illustrate possible mechanisms of protein ADP-ribosyltransfer catalyzed by the sirtuin family.
EXPERIMENTAL PROCEDURES
Reagents and Instrumentation
Synthetic peptides p300: ERSTEL(K-Ac)TEI(KAc)EEEDQPSTS, H3: ARTKQTAR(K-Ac)STGG(K-Ac)APRKQLAS and H4: SGRG(K-Ac)GG(K-Ac)GLG(K-Ac)GGA(K-Ac)RHR were synthesized and characterized by the Proteomics Resource Center at Rockefeller University. They were purified by HPLC before use. All other reagents were purchased from Aldrich or VWR and were of the highest purity commercially available. HPLC analyses were performed on a Hitachi elite LaChrom system equipped with Diode array detector using C18 reverse phase columns. Radiolabeled samples were counted in a Beckman Coulter LS 6500 multi-purpose scintillation counter.
Plasmid Construction and Protein Expression
The gene encoding Sir2 from Plasmodium falciparum (PF13_0152) was cloned from Plasmodium falciparum DNA using PCR by Kirk Deitsch of Weill Medical College of Cornell University. The coding sequence was initially cloned into a pGEM vector and subsequently recloned into Pet28a (Novagen) to obtain protein expressed with an N-terminal poly-histidine tag. The insert was verified by nucleotide sequencing and checked against the published sequence. PetPFSIR2 vector was transfected into Codon-plus® RIPL cells (Stratagene) and protein synthesis was induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at OD600 = 0.25. Cells were grown for 6 hours at 37 °C, pelleted and lysed by freeze-thaw cycles. The protein was purified by Ni-column affinity chromatography, dialyzed overnight in 20 mM potassium phosphate buffer pH 7.0, aliquoted in 20% glycerol and 2 mM DTT and flash-frozen and stored at −80 °C Enzyme concentrations were determined by the method of Bradford (22). The protein molecular weight was determined by MALDI-TOF (Rockefeller Proteomics Resource) and the protein was 95% pure as determined by SDS-polyacrylamide gel electrophoresis. The presence of a catalytically significant histidine residue (position 162 in our construct) was confirmed by LC-MS/MS analysis of a tryptic digest of the purified enzyme performed at Rockefeller University proteomics facility.
Deacetylation assays
Deacetylation reactions were typically performed in 150 mM phosphate buffer pH 7.3 in reaction volumes of 50 ∝L. A typical reaction contained 400 ∝M NAD+, 3 ∝M Pf-Sir2 enzyme and 0–500 ∝M acetylated peptide (H3, H4 or p300). Reactions were initiated by addition of enzyme, incubated for one hour at 30° C and then quenched by addition of trifluoroacetic acid. The reactions were placed on ice for one hour after quench to precipitate protein, centrifuged at 13000 g for 2 min to pellet insolubles then analyzed by HPLC. Peptides were separated using a Waters Xterra RP-18 column running a gradient of 10% to 40 % acetonitrile in 0.1% TFA and chromatograms collected by multi-wavelength diode array with chromatograms analyzed at wavelength of 215 nm. Reactions were quantified by integrating area of peaks corresponding to deacetylated peptides (identities confirmed by mass spectrometry, Proteomics Resource Center, Rockefeller University). Rates were plotted versus peptide concentration and best fit of points to the Michealis-Menton equation was performed by Kaleidagraph®. HPLC observation of nicotinamide and 2′- and 3′-O-AADPR products was accomplished by performing reaction, quench and injection as above, with HPLC elution with 20 mM ammonium acetate pH 7.5. Chromatograms were analyzed at wavelength of 260 nm. To determine nicotinamide inhibition, reactions were performed similarly but nicotinamide at concentrations ranging from 0– 350 ∝M was also added to reaction mixtures. Rates were plotted and points were fit to the equation v = v0 - vinh ([I]/(Ki +[I])) where v is the rate observed for a given concentration of nicotinamide, v0 is the uninhibited rate, vinh is the maximal inhibition, Ki is the apparent inhibition constant and [I] is the concentration of nicotinamide. This equation to fit nicotinamide inhibition of sirtuins has been used previously (11).
14C-Nicotinamide Base Exchange Assay
Reactions containing 400 ∝M NAD+, 500 ∝M H3 or 200 ∝M p300 peptide, and 150 mM phosphate buffer, pH 7.5, with varying concentrations of [carbonyl-14C]nicotinamide (American Radiolabeled Chemicals Inc.) were initiated by addition of Pf-Sir2 enzyme to a concentration of 0.5 ∝M. Reactions were incubated for one hour at 30 °C and quenched by addition of trifluoroacetic acid to pH 2. After centrifugation to remove precipitates, reactions were injected on HPLC (0.5% TFA eluant) to separate nicotinamide and NAD+. Eluant containing nicotinamide and NAD+ was collected and radioactivity determined by scintillation counting. Reactions were run to no more than 10% of the calculated equilibrium position for nicotinamide exchange. Rates were determined as cpm/s incorporated into NAD+, and then converted to a turnover rate (s−1) by adjustment for specific radioactivity of nicotinamide and enzyme concentration. Rates were plotted versus nicotinamide concentration and best fit of plotted data to the Michaelis-Menton curve was performed using Kaleidagraph®. Incubations of less than and greater than 1 hour confirmed that product formation versus time was linear during the course of the assay.
Thionicotinamide Base-Exchange Assay
Reactions containing 400 ∝M NAD+, 400 ∝M H3 peptide, and 150 mM phosphate buffer, pH 7.3, were performed in the presence of concentrations 0 to 2000 ∝M thionicotinamide. Reactions were initiated with addition of Pf-Sir2 enzyme to a final concentration of 1 ∝M. After one hour incubation at 30°C reactions were quenched by addition of trifluoroacetic acid to pH 2. ThioNAD formation was quantified by HPLC on a Waters SymmetryShield RP8 column running 0.1% TFA with a gradient to 25% MeOH starting at 15 minutes and by comparison to an authentic standard for quantitation and identification, prepared via base exchange of thionicotinamide into NAD+ catalyzed by CD38 (23). Product formation rates were plotted versus thionicotinamide concentration and points fit to the Michaelis-Menton equation to obtain the Michealis parameters Km and kcat.
HPLC Assay Measuring Hydrolysis of NAD+
Reaction mixtures containing 400 ∝M NAD+, and one of p300, H3 or H4 peptides (of varying concentrations 0– 800 ∝M) in 150 mM phosphate buffer, pH 7.3, were initiated with the addition of Pf-Sir2 (typically 2–4 ∝M final concentration). Production of ADPR was quantitated by HPLC using a 20 mM ammonium acetate isocratic system on a Waters C-18 column. Comparison to SIRT1 reactions reacted under similar conditions established minimal contribution to apparent hydrolysis from breakdown of AADPR. As stated in the text, Pf-Sir2 added to SIRT1 reactions also did not increase formation of ADPR from AADPR. Moreover, in almost all cases ADPR formation rate exceeded the deacetylation rate that forms AADPR by at least 5 fold. Rate of formation of ADPR was plotted against peptide concentration and fit to a modified curve kobs = kcat(no peptide) + kcat(peptide)[S]/(Km + [S]) where kobs is the observed rate, kcat(no peptide) is the observed rate of hydrolysis in the absence of peptide, kcat(peptide) is the component of the observed rate of reaction that is dependent on peptide, Km is the Michaelis constant for peptide and [S] is the peptide concentration.
To analyze nicotinamide effects on reactions (in the presence or absence of added H3 peptide), varying amounts of nicotinamide, 0–1000 ∝M, were added to reaction mixtures and reactions were assayed as above quantifying rate by observed ADPR. Inhibition curves were of the form v = v0 - vinh ([I]/(Ki +[I])) where v is the rate observed for a given concentration of nicotinamide, v0 is the uninhibited rate, vinh is the maximal inhibition, Ki is the apparent inhibition constant and [I] is the concentration of nicotinamide.
DEAE-Sephadex Ion-Exchange Assay for Measurement of ADPR formation
Reactions were typically performed in 100 mM phosphate and contained 400 ∝M [2-, 8-3H]NAD+ or [8-14C]NAD+ (synthesized by coupling of NMN and [2-, 8-3H]ATP or [8-14C]-ATP (American Radiolabeled Chemicals Inc.) as previously described (24)) with or without peptide. Reactions were initiated by addition of Pf-Sir2 and allowed to incubate for one hour at 37°C. After quenching with TFA, the reactions were centrifuged to remove precipitate and loaded onto columns containing pre-equilibrated DEAE-Sephadex (equilibrated with 5 mM ammonium acetate pH 7). The reaction mixture was then eluted with eight 2 mL fractions of 10 mM ammonium acetate pH 7 and five 2 mL fractions of 100 mM ammonium acetate pH 7. The radioactivity contained in the eluted samples was then quantified by scintillation counter. Radiolabeled NAD+ eluted in the 10 mM washes, while ADPR eluted in the 100 mM fractions. Reactions performed with p300 were corrected for AADPR production by HPLC assay of the eluant of 100 mM ammonium acetate (which contained ADPR, major species and AADPR, minor species) and by independent HPLC determination of deacetylation rate of the peptide under identical reaction conditions. Reactions were corrected with appropriate negative controls.
HPLC Assay for Methanolyses of NAD+ catalyzed by Pf-Sir2
A β-1-methyl-O-ADPR standard was synthesized using the known methanolysis reaction catalyzed by CD-38 (25). This enzyme was incubated with NAD+ in 150 mM phosphate buffer and 30% MeOH, and monitored by HPLC. A single new peak was formed isolated by HPLC, NMR was taken and confirmed to be β-1-O-methyl-ADPR. Mass spectrum by MALDI-TOF confirmed the correct mass (m/z = 574, positive ion). The corresponding α-1-O-methyl-ADPR standard was formed by heating NAD+ in 30 % methanol in 50 mM phosphate pH 7.5 to 80 °C. This procedure is known to generate both methanolysis stereochemistries. The methanolysis product α-1-O-methyl-ADPR was isolated and identified by NMR and by MALDI-MS (m/z = 574, positive ion).
Methanolysis reactions catalyzed by Pf-Sir2 were carried out in the presence of 400 ∝M NAD+, 20% by volume methanol (5.1 M) unless noted otherwise, and varying H3 concentrations in 150 mM phosphate buffer, pH 8.5. The reactions were initiated by addition of Pf-Sir2 (final: 8 ∝M) and then incubated for one hour at 30 °C. After quenching with TFA the solutions were centrifuged to remove precipitate and immediately analyzed by HPLC. Standards of β-1-O-methyl-ADPR and α-1-O-methyl-ADPR were run on the same day and spiked into reaction mixtures to confirm peak identity. To confirm identity of these compounds in Pf-Sir2 reaction mixtures, eluant of peaks were collected, lyophilized and analyzed by MALDI-TOF (positive ion mode, CHCA matrix).
RESULTS
Pf-Sir2 has NAD+ Dependent Protein Deacetylase Activity
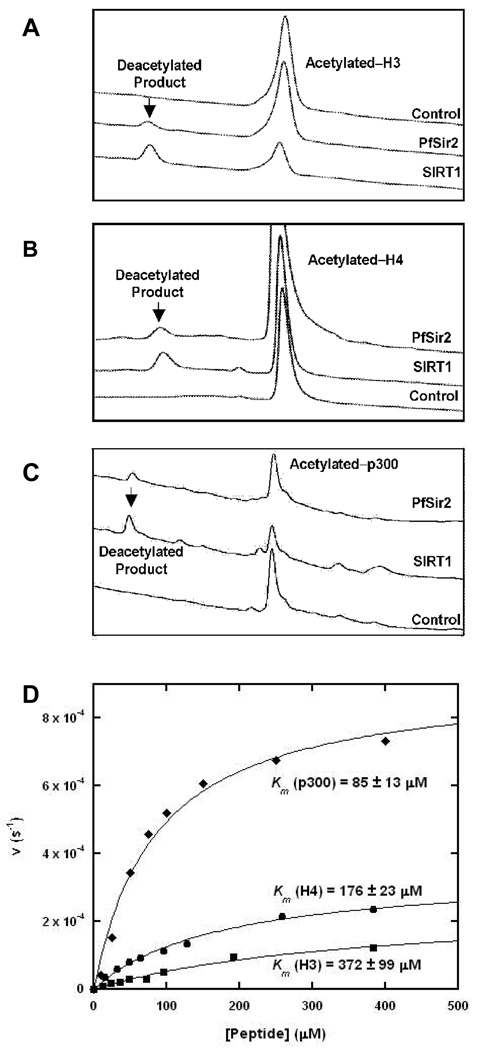
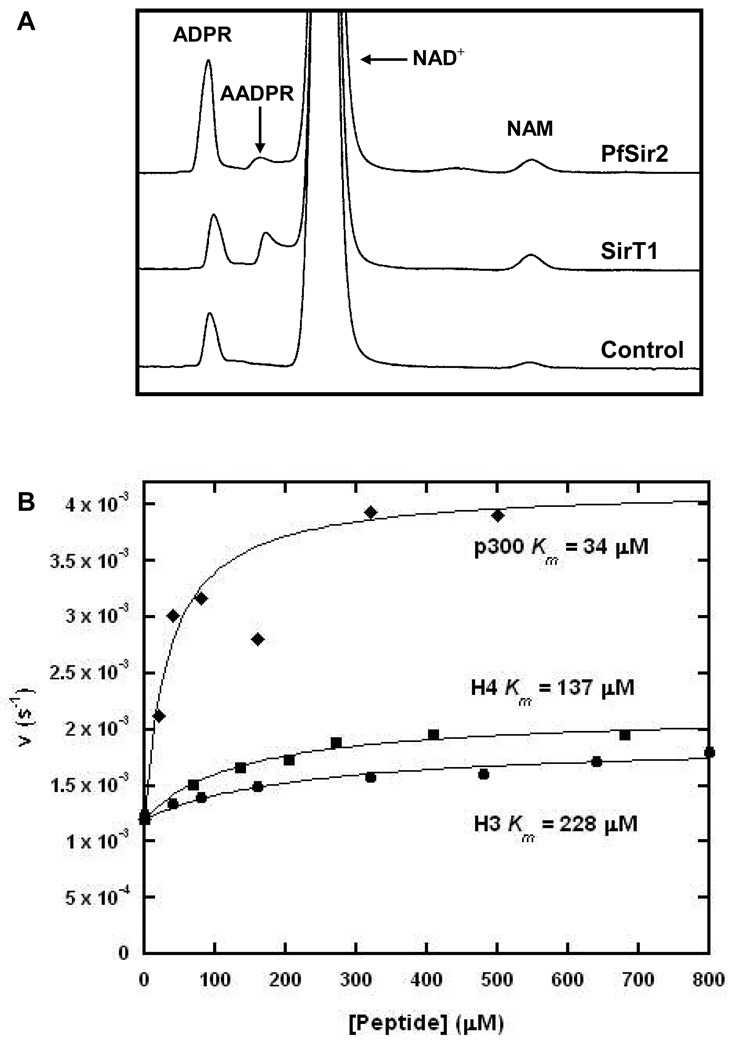
Pf-Sir2 has been found to localize to heterochromatin in P. falciparum and is proposed to contribute to var gene silencing via deacetylation of histones (20, 21). We examined the ability of recombinant Pf-Sir2 to catalyze NAD+ dependent protein deacetylase activity by using peptides homologous to N-terminal histone sequences of H3 and H4 (see experimental for sequences). Deacetylation activity of Pf-Sir2 was compared with SIRT1 as a positive control. Peptide substrates were reacted with NAD+ in the presence of enzyme and peptide deacetylation products were determined by HPLC. SIRT1 and Pf-Sir2 reactions generated similar chromatograms for H4 and H3 reactions (Figure 1A and 1B). It is known that SIRT1 predominantly deacetylates at K16 of tetra-acetylated H4, identical to the sequence used in this study (26). Based on retention time, this same residue is deacetylated by Pf-Sir2 (Figure 1B) and was confirmed by MS analysis (performed by Rockefeller Proteomics Resource). Deacetylation of a di-acetylated H3 peptide by Pf-Sir2 and SIRT1 produced similar HPLC chromatograms as well (Figure 1A). SIRT1 deacetylates positions AcK9 and AcK14 of the N-terminal H3 sequence (26). The single peak was collected in the SIRT1 and Pf-Sir2 HPLC elutions and analyzed by MS (performed by Rockefeller Proteomics Resource). These results confirmed deacetylation of K9 and K14 for both reactions. It was determined that deacetylation depends on the presence of NAD+, and the Km for NAD+ in H3 deacetylation was determined to be 120 ∝M. These results corroborate a recent study showing that Pf-Sir2 is a histone deacetylase for N-terminal tails of whole histones H3 and H4 (27), although Michaelis parameters were not determined and only Westerns were used to identify deacetylated products (27).
Figure 1. Deacetylation of peptide substrates catalyzed by Pf-Sir2.
(A) HPLC chromatograms showing the deacetylation of acetylated-H3 by Pf-Sir2 and SirT1 over time. (B) Deacetylation of H4. (C) Deacetylation of p300. (D)Saturation curves for steady-state deacetylation rates for the three different peptides.
Interestingly, a human di-acetylated p300 sequence (ERSTEL(K-Ac)TEI(K-Ac)EEEDQPSTS corresponding to amino acid positions 1034–1053 of the full p300 sequence) was determined to be a superior substrate of Pf-Sir2, and the deacetylated peptide product is again similar to that formed by SIRT1 as shown by HPLC (Figure 1C). The preferred site of deacetylation for this sequence by SIRT1 has been characterized (28) and corresponds to the AcLys in the peptide corresponding to amino acid position 1040 of the full p300 sequence (see above). Acetylation modifications in this sequence have been demonstrated to affect p300 functions as a transcriptional co-regulator (28).
Deacetylation reactions were further studied by HPLC in which peptide concentrations were varied in the presence of enzyme and 400 ∝M NAD+. Michaelis-Menton curves fit to determined deacetylation rates versus peptide concentrations are shown in Figure 1D. Km and kcat values are listed for the different peptide substrates in Table 1. Maximal rates for deacetylation of H3 and H4 sequences were similar and remarkably slow as indicated by maximal turnover rates of 2.5 · 10−4 s−1 and 3.5 · 10−4 s−1 respectively (Table 1). The p300 peptide reacted at least 2.5 times faster than either H3 or H4 (9 · 10−4 s−1) with a Km value of 85 ∝M, 2 times smaller than H4 (176 ∝M) and 4 times smaller than the Km of H3 (372 ∝M). Deacetylation reactions of H3, H4 and p300 substrates were determined to form 2′- and 3′- O-acetyl-ADP-ribose (AADPR), known co-products of sirtuin deacetylation reactions (8).
Table 1.
Parameters for Deacetylation, Exchange, Hydrolysis and Inhibition Reactions for Pf-Sir2 with Various Substrates.
| Deacetylation |
Exchangeb |
Hydrolysis |
|||||
|---|---|---|---|---|---|---|---|
| kcat | Km | Ki(NAM)a | kcat | Km | kcat | Km | |
| 10−4 s−1 | ∝M | ∝M | 10−2 s−1 | ∝M | 10−3 s−1 | ∝M | |
| H3c | 2.5 ± 0.4 | 372 ± 99 | 35 ± 2 | 2.5 ± 0.1 | 61 ± 5 | 1.9 ± 0.1 | 228 ± 28 |
| H4c | 3.5 ± 0.2 | 176 ± 23 | NM | NM | NM | 2.2 ± 0.1 | 137 ± 29 |
| p300c | 9.2 ± 0.5 | 85 ± 13 | 91 ± 4 | 6.4 ± 0.2 | 80 ± 7 | 3.4 ± 0.2 | 33 ± 15 |
| NAD+d | NA | NA | NA | ND | NA | 1.2 ± 0.2 | NM |
Conditions for measurements are described in experimental section. All reactions conducted in the presence of 400 ∝M NAD+.
Ki(NAM) is the inhibition constant for nicotinamide inhibition of deacetylation. The values are determined by varying nicotinamide (NAM) concentrations, measuring deacetylation by HPLC and plotting rate of deacetylation versus NAM concentration. Fits of points to the inhibition curve described in experimental determines the value of the parameter.
The values are determined from the saturation curve for 14C-nicotinamide base-exchange.
Peptide primary sequence and acetylation are described in the materials and methods section. NM: Not Measured. ND: Not detected, highest concentration: 5 mM 14C-NAM.
No acetylated peptide is added to these reactions. NA: Not applicable.
Pf-Sir2 Catalyzes Nicotinamide Exchange and Inhibits Deacetylation
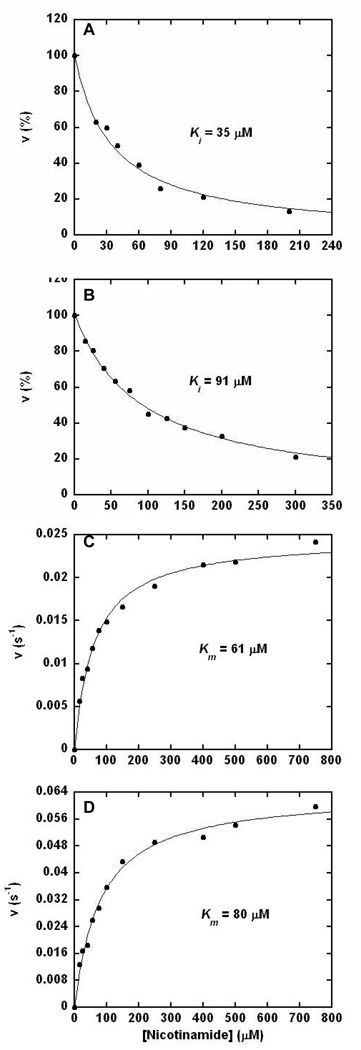
Sirtuin deacetylation reactions are inhibited by the general sirtuin inhibitor nicotinamide, which is a product of sirtuin deacetylation chemistry (11, 12, 29, 30). To interrogate inhibition of H3 and p300 deacetylation as a function of nicotinamide concentration, an HPLC assay was employed (see experimental for details). As shown in Figure 2A and 2B deacetylation was almost completely inhibited by nicotinamide added to reaction mixtures with Ki values for nicotinamide of 35 and 91 ∝M for H3 and p300 respectively. These results indicate that Ki may be dependent upon the substrate peptide sequence. The inhibition constants are potent and suggest that nicotinamide is likely to exert effects on deacetylation activity of Pf-Sir2 enzymes under physiologic conditions. These nicotinamide inhibition constants are similar to those reported for inhibitions of deacetylation reactions catalyzed by yeast, human and archaeal sirtuins (11, 12).
Figure 2. Nicotinamide Base-Exchange Catalyzed by Pf-Sir2.
(A) and (B): Inhibition of the deacetylation reaction by increasing concentrations of nicotinamide. (A) 500 ∝M H3 peptide, 250 ∝M NAD+ and 150 mM phosphate buffer, pH 7.3, quantified by integration of deacetylated product peaks in the HPLC chromatograms. (B) 200 ∝M p300 peptide, 400 ∝M NAD+ and 150 mM phosphate buffer, pH 7.5, and quantified by integration of deacetylated product peaks in the HPLC chromatograms. (C) and (D): Kinetics of the base-exchange chemistry catalyzed by Pf-Sir2 as measured by exchange of [carbonyl-14C]-nicotinamide into unlabeled NAD+. (C) 200 ∝M H3 peptide, 400 ∝M NAD+ and 150 mM phosphate buffer, pH 7.5. (D) 200 ∝M p300 peptide, 400 ∝M NAD+ and 150 mM phosphate buffer, pH 7.5.
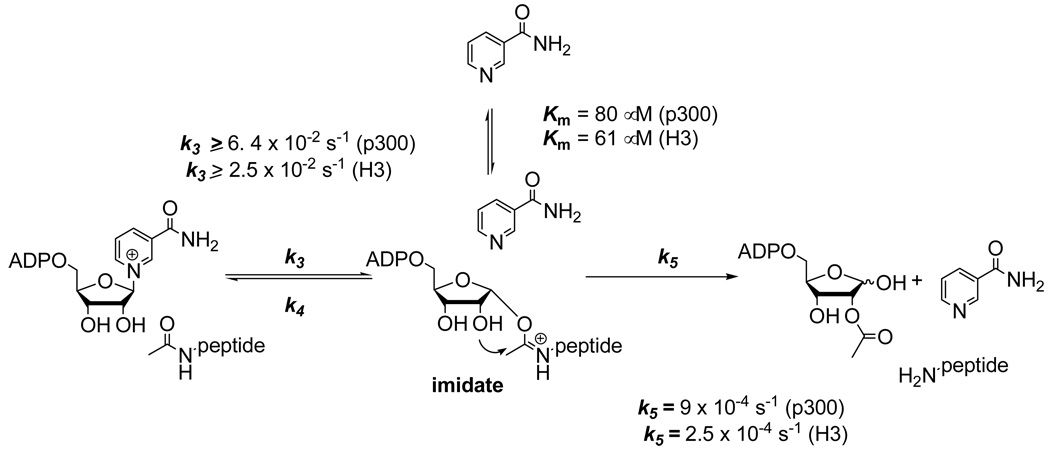
Nicotinamide inhibition of sirtuin catalyzed deacetylation is linked to a sirtuin reaction which catalyzes nicotinamide exchange into NAD+ called “base-exchange”. The base-exchange reaction is thought to compete with the deacetylation reaction for a common reaction intermediate, called the ADPR-peptidyl-imidate (11, 12). Depletion of the imidate by base-exchange inhibits the deacetylation reaction (Scheme 1). Correlation of Km and Ki is expected since base-exchange and inhibition of deacetylation emanate from the same kinetic process (11). We measured rates of base-exchange catalyzed by Pf-Sir2 using [carbonyl-14C]nicotinamide as the exchange base, as described previously (11). Values for rate versus nicotinamide were plotted and the points fit to the Michaelis-Menten equation (Figure 2C and 2D). As expected, Pf-Sir2 catalyzed base-exchange with H3 and p300 as substrates and responded saturably with increasing nicotinamide concentrations. Curve fits determined Km values of 61 ∝M (H3) and 91 ∝M (p300). Corresponding kcat values were 0.025 s−1 (H3) and 0.064 s−1 (p300). These results establish that the kinetic parameter Km corresponds well to the corresponding Ki values as predicted (11). The steady-state rate of base exchange exceeds the corresponding deacetylation rate by 70 fold and 100 fold for p300 and H3 respectively. If deacetylation and base exchange share the imidate as a common intermediate, then the slow deacetylation rate is caused by a rate-limiting step downstream of the imidate, possibly attack of the 2'-OH on the imidate (11). The reactivity of the NAD+ and peptide substrate to form the imidate (Scheme 1) must be at least as fast as the steady state base-exchange rate.
Scheme 1.
Reactions of acetylysine peptides in base exchange and deacetylation pathways. Rate constants for deacetylation and base exchange are shown for the respective steps.
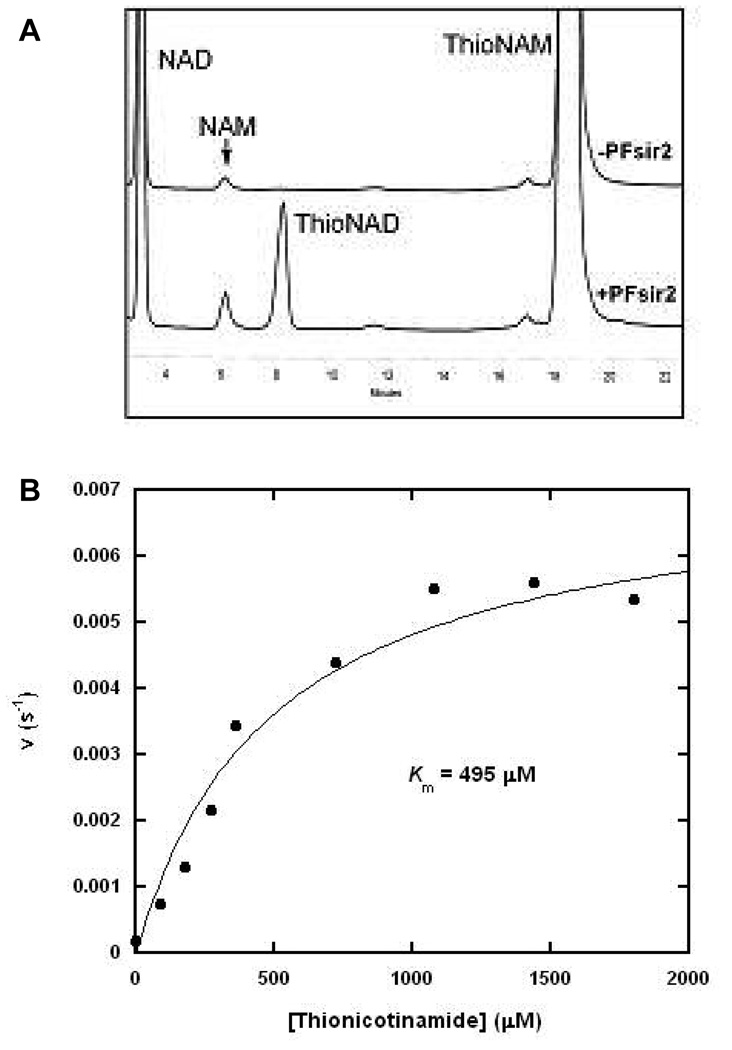
To further probe for substrate specificity in the base-exchange reaction, we employed thionicotinamide as the base-exchange substrate. Thionicotinamide has previously been shown to be competent as a base-exchange substrate of the sirtuin HST2 (12). Thionicotinamide is proposed to form thioNAD+ upon reaction with an imidate or an ADPR-intermediate complex (12). HPLC chromatograms show that thioNAD+ formation is catalyzed by Pf-Sir2 in the presence of thionicotinamide (Figure 3A). To characterize thionicotinamide exchange, thionicotinamide concentrations were varied and the rates of formation of thioNAD+ were determined using H3 as a substrate. Fit of the individual data points to the Michaelis-Menton equation is shown in Figure 3B. The Km of thionicotinamide was 495 ∝M, and kcat was 6 · 10−3 s−1. Under similar reaction conditions thionicotinamide reacts approximately 4 times slower than nicotinamide and has an 8 times higher Km. The basis for the slower reaction of thionicotinamide is unlikely to reflect intrinsic reactivity differences, since nicotinamide and thionicotinamide pKa values are reportedly nearly identical (12). The weaker binding of thionicotinamide and slower reaction suggests that substrate binding interactions and geometry are less optimal than for reaction of nicotinamide on the active site. These results provide an example of how reactivity differences can emerge with even slight perturbation of substrate structure, not necessarily related to intrinsic reactivity, requiring care in using rate differences in “isosteric substrates” for addressing questions of mechanism.
Figure 3. Thionicotinamide Base-Exchange Catalyzed by Pf-Sir2.
(A) HPLC chromatograms showing the thionicotinamide base-exchange reaction and negative control. (B) Steady-state saturation kinetics of the thionicotinamide base-exchange reaction as catalyzed by Pf-Sir2 in the presence of 400 ∝M NAD+ and 400 ∝M H3 peptide and increasing concentrations of thionicotinamide.
Pf-Sir2 Catalyzes Hydrolysis of NAD+
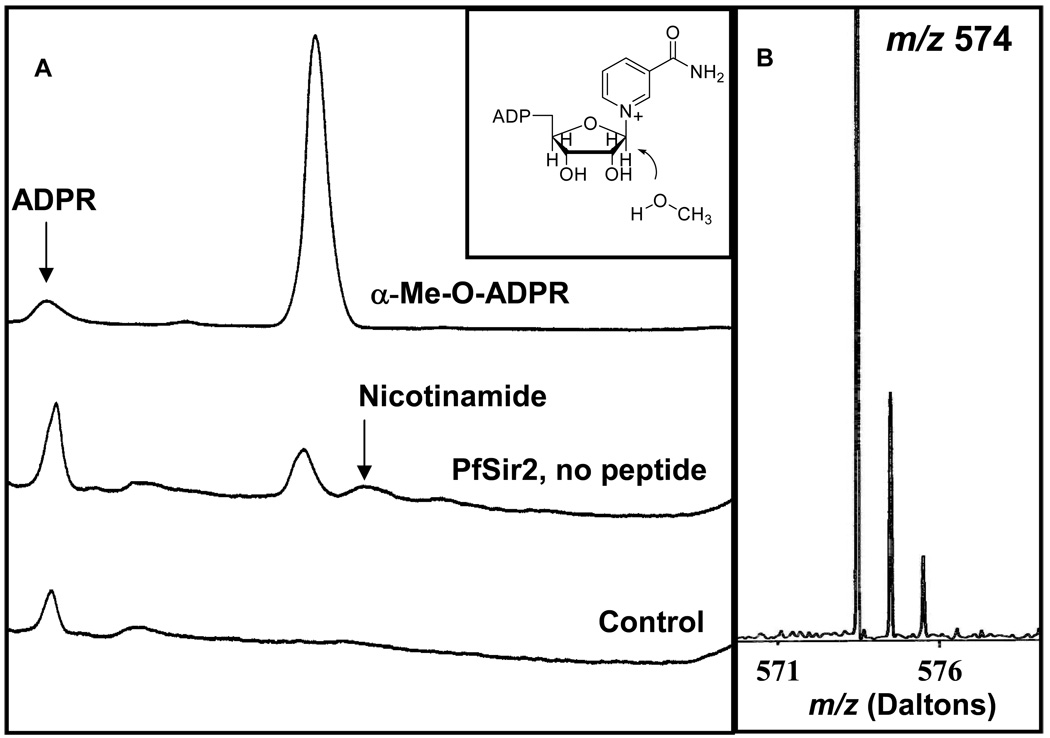
The NAD+ hydrolysis product ADPR is often detected in NAD+ dependent deacetylation reactions. ADPR is typically formed by slow non-enzymatic hydrolysis of NAD+, which is the minor pathway, and by the uncatalyzed decomposition of AADPR, the product of deacetylation chemistry (8), which is the major pathway. HPLC chromatograms of deacetylation reactions containing NAD+ and Pf-Sir2 showed that amounts of ADPR were much higher than expected in comparison with reaction of SIRT1 under similar conditions of pH, substrates and rate of NAD+ consumption (Figure 4A). We considered the possibility that Pf-Sir2 catalyzed decomposition of AADPR. However, purified AADPR in the presence of Pf-Sir2 was not decomposed faster relative to a control (data not shown). Moreover, AADPR was not decomposed when Pf-Sir2 was added to a SIRT1 reaction mixture (data not shown). This suggested to us that Pf-Sir2 might possess an independent activity capable of catalyzing solvolysis of NAD+. Although hydrolysis is catalyzed by a mutated sirtuin in which the conserved catalytic histidine in the active site is replaced by alanine (H135A, HST2) (10), there are no examples of wildtype sirtuins that have an established NAD+ glycohydrolase activity. We did check for the presence of the active site histidine in recombinant Pf-Sir2 protein by trypsin digestion. We observed the tryptic peptide containing the catalytic histidine by MS analysis (Rockefeller Proteomics Resource). We also sequenced the vector. Finally, the predicted MW of the protein was confirmed by MALDI (predicted 32507.6 found 32504.9). Thus, a mutated histidine on Pf-Sir2 could not explain formation of ADPR.
Figure 4. Hydrolysis of NAD+ Catalyzed by Pf-Sir2.
(A) HPLC chromatograms showing the production of ADPR by SirT1 and Pf-Sir2 under similar conditions. (B) the kinetics of the hydrolysis reaction for varying concentrations of peptide carried out in the presence of 400 ∝M NAD+ and 150 mM phosphate buffer, pH 7.3.
To quantitate ADPR formation we used HPLC conditions similar to those in Figure 4 using saturating NAD+ and different acetylated peptides (see experimental for details). Determined rates of formation of ADPR are shown in Table 1. Strikingly, the ADPR formation rate was 3–8 times higher than the maximal deacetylation rate for all peptide substrates (H3, H4 and p300). ADPR formation increased as a function of the peptide concentration and was dependent on substrate identity (H3, H4 or p300) as shown in Figure 4. Quantitating rate of ADPR formation as a function of peptide concentration led to the unusual finding that ADPR formation was observable even in the absence of a peptide substrate and occurred at a rate of 1 · 10−3 s−1 (Figure 4B). We fit the rate of ADPR formation to a modified Michealis-Menton expression that is described in the experimental section (see Figure 4B). The Km values for peptide substrates in deacetylation and in hydrolysis were in good correspondence for all three peptides (Figure 1, Figure 4B and Table 1). The best hydrolysis stimulating substrate was p300. Hydrolysis in the absence of acetylated peptide was intriguing since peptide-independent NAD+ hydrolysis has not been reported for any sirtuin, mutant or wildtype. In addition, the observation of a peptide-stimulated hydrolysis reaction suggested to us that Pf-Sir2 might catalyze two different types of hydrolysis, which we sought to further elucidate.
In the Absence of Peptide, Pf-Sir2 Catalyzes α–face Methanolysis
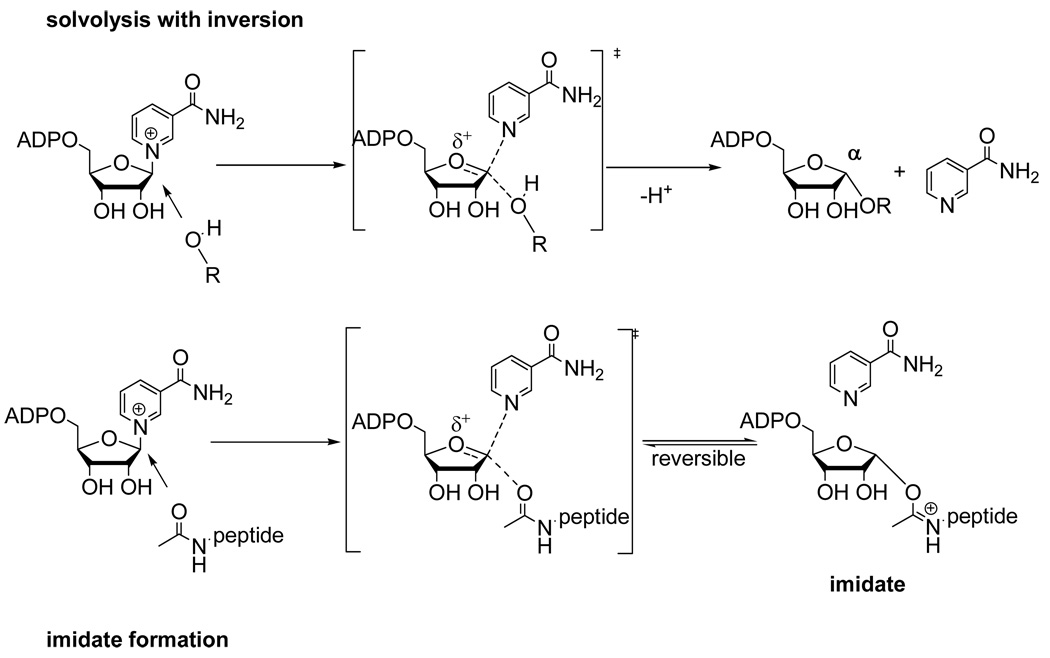
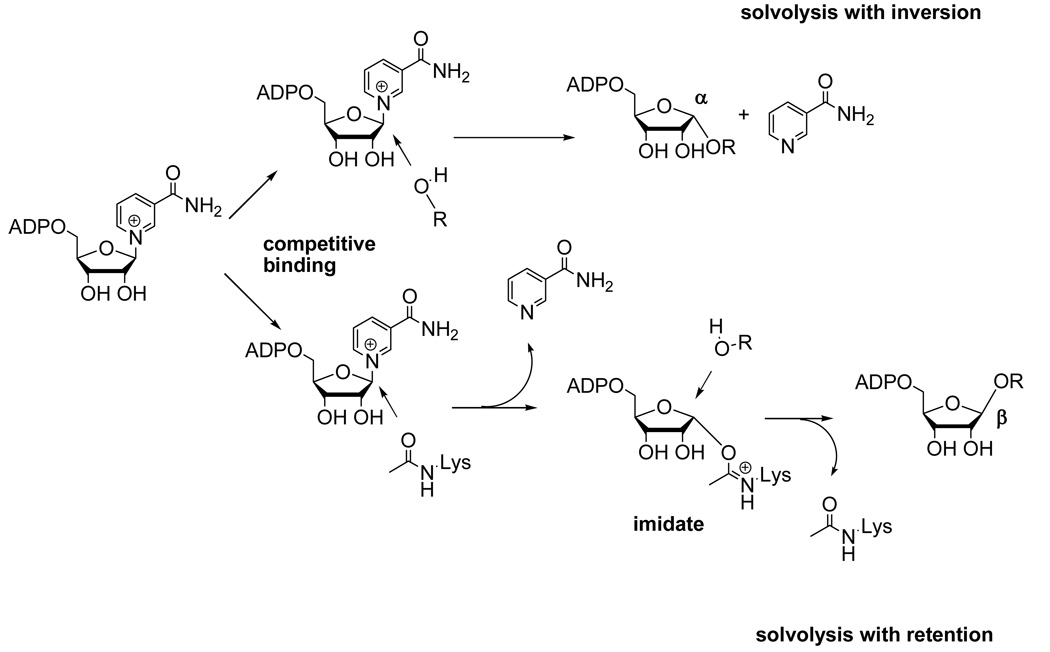
We probed the mechanism of peptide independent solvolysis by employing methanol as a co-solvent. Methanol has previously been used to determine the stereochemistry of solvolysis reactions of NAD+ catalyzed by CD38/NAD+ glycohydrolase (25, 31). Methanol is superior to water for determining stereochemical outcome, because while the hydrolysis product rapidly undergoes mutarotation which randomizes stereochemistry at the anomeric carbon, the methanolysis product retains its original stereochemistry and thereby reports on the mechanism of its formation. The relevant α-1-O-methyl-APDR and β-1-O-methyl-ADPR standards were synthesized by known methods (see Methods section), and Pf-Sir2 solvolysis reactions were carried out in the absence of a peptide substrate but in the presence of methanol (30%). HPLC analysis of the reaction revealed a new reaction product that eluted at the same time as the α-1-O-methyl-ADPR standard (Figure 5A). The product was collected and analyzed by MALDI-MS and it behaved identically to the standard, with a positively charged parent ion M/z = 574 (Figure 5B). Furthermore, when the reaction was carried out in deuterated methanol, the isolated product gave a positively charged ion of m/z = 577, the expected value for the deuterated compound α-1-O-CD3-ADPR (data not shown). Thus, the reaction that generates solvolysis in the absence of a peptide substrate generates a product with inverted stereochemistry versus NAD+. This result confirmed a new mode of chemistry for a sirtuin, namely a single-displacement ADP-ribosyltransfer to solvent independent of acetylated substrate. The solvolysis is envisioned to occur via binding of solvent to the active site pocket typically occupied by an acetylated substrate. The acetylated substrate in the presence of NAD+ processes forward to form an imidate (Scheme 2), whereas in the absence of the acetylated substrate the poised NAD+ could capture the coordinated solvent to produce a product with inverted stereochemistry (Scheme 2).
Figure 5. Production of α-face Methanolysis Product.
(A) HPLC chromatograms showing the α-MeOADPR standard, a Pf-Sir2 reaction run without peptide and in the presence of 30% MeOH, and a control reaction containing no enzyme run in the presence of 30% MeOH. (B) MALDI-MS data showing the m/z values for the methanolysis products collected from the Pf-Sir2 catalyzed reactions in the presence of 30% CH3OH and 30% CD3OH respectively.
Scheme 2.
Proposed reaction of NAD+ in active site of Pf-Sir2 with either solvent of acetyllysine.
In the Presence of Peptide, Pf-Sir2 Catalyzes both α– and β–face Methanolysis
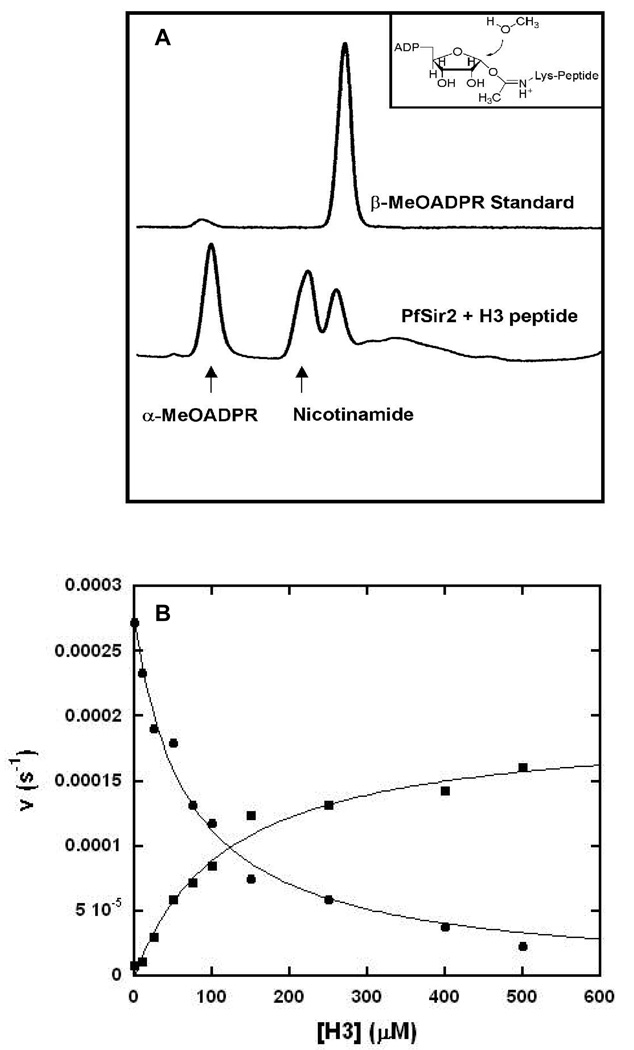
To investigate the mechanisms of solvolysis reactions stimulated by the presence of acetylated peptide substrate, we conducted solvolysis reactions in which NAD+ and H3 were reacted in the presence of Pf-Sir2, in which 30% methanol was present as a co-solvent. An HPLC chromatogram of a reaction mixture showed that the α-methanolysis product appeared again, along with an additional new peak (Figure 6A). The new product had the same elution time as an authentic β-1′-MeO-ADPR standard (Figure 6A). We collected this new product and a MALDI-MS spectrum determined the mass of the compound (m/z =574) identical to β-1-O-methyl-ADPR. As before, the corresponding reaction run in the presence of d4-deuterated methanol gave a mass shifted species (m/z = 577) corresponding to the tri-deuterated product. Thus, we conclude that in the presence of peptide Pf-Sir2 produces a new solvolysis product generated with overall retention of stereochemistry versus NAD+.
Figure 6. Production of β-face Methanolysis Product.
(A) HPLC chromatograms showing the β-MeOADPR standard, and a Pf-Sir2 reaction run in the presence of H3 peptide and 30% MeOH. (B) Bottom: Pf-Sir2 catalyzed methanolysis with different concentrations of H3 Productions of α-MeOADPR (circle) and β-MeOADPR (square) with different concentrations of H3 are shown. The reactions contained 400 ∝M of NAD+ in 100 mM phosphate buffer with 0, 10, 25, 50, 75, 100, 150, 250, 400 and 500 ∝M of H3 all at pH 8.5. The following Michaelis parameters were determined by fitting the curves with KaleidaGraph: Ki (α-MeO-ADPR) = 70 ∝M, k0 = 2.7 × 10−4 s−1 Km (β-MeO-ADPR) = 119 ∝M, k500 = 1.6 × 10−4 s−1 ksolvolysis ([H3] = 0) = 1.38 × 10−3 s−1, ksolvolysis ([H3] = 500 ∝M) = 1.40 × 10−3 s−1
In light of this finding, we considered the possibility that two solvolytic mechanisms were occurring on the enzyme, one that happens by one-step single displacement mechanism to produce inverted stereochemistry. The second reaction was hypothesized to be a peptide dependent double-displacement mechanism, probably imidate-dependent, which generates products exhibiting retention of stereochemistry versus NAD+ (Scheme 3). If these are the operative mechanisms, then these two mechanisms should be competitive in nature, given that the solvent and acetylated peptide are proposed to require the same pocket on the enzyme to generate the α-solvolysis products or imidate to generate β-solvolysis products. If this idea is correct, increasing concentration of peptide should increase β–product formation and inhibit α-product formation concomitantly. Thus, a set of reactions were performed varying H3 concentrations and analyzed by HPLC to quantitate ADPR, and both stereochemical methanolysis products. As shown β–methanolysis product increased with increasing peptide concentrations, eventually saturating with Km of 120 ∝M (Figure 6B), while the α– product was inhibited to near zero over the same concentration range with a inhibition constant for peptide of Ki = 70 ∝M. (The lower apparent Km (and thus Ki) for H3 under these conditions (as compared to the Km values in Table 1) may be a consequence of methanol effects on H3 binding). Interestingly, the overall rate of solvolytic turnover of NAD+ in the absence and presence of peptide under these conditions are virtually identical but the solvolytic mechanism completely changes (See Figure 6 legend for rates of combined ADPR and 1-O-methyl-ADPR for these conditions). The result supports the model that acetylated substrate competes for the α-face of C1' of NAD+, displacing solvent from the active site thus preventing the formation of solvolysis products with inversion of stereochemistry. Correspondingly, the acetylated substrate can react forward to form the imidate complex, which is proposed to undergo its own solvolysis reaction (Scheme 3) to generate overall stereochemical retention and the observed β-1-O-methyl ADPR (Scheme 3).
Scheme 3.
Reaction choices of solvolysis with inversion of stereochemistry or competition reaction of acetyllysine to form imidate which can react with solvent to form product with overall retention of stereochemistry.
Differential Nicotinamide Inhibition of Solvolytic Mechanisms
Observation of two separate solvolytic products, formed through distinct and independent pathways suggested to us that nicotinamide might affect solvolysis differently under conditions in which peptide is present or absent from reaction mixtures. As already discussed, nicotinamide is a general sirtuin inhibitor and inhibits deacetylation of Pf-Sir2 by virtue of its ability to capture the imidate intermediate before it decomposes to products. Nicotinamide does not appear to inhibit Michaelis complex formation, since increasing nicotinamide concentration does not inhibit base-exchange chemistry, even when the base-exchange rate is fully saturated (Figure 2, bottom panels). Thus, we would predict that nicotinamide cannot inhibit peptide independent solvolysis, because it does not inhibit NAD+ binding. Conversely, since peptide dependent chemistry is likely imidate-dependent, we predict that nicotinamide should inhibit peptide-dependent solvolytic reactions.
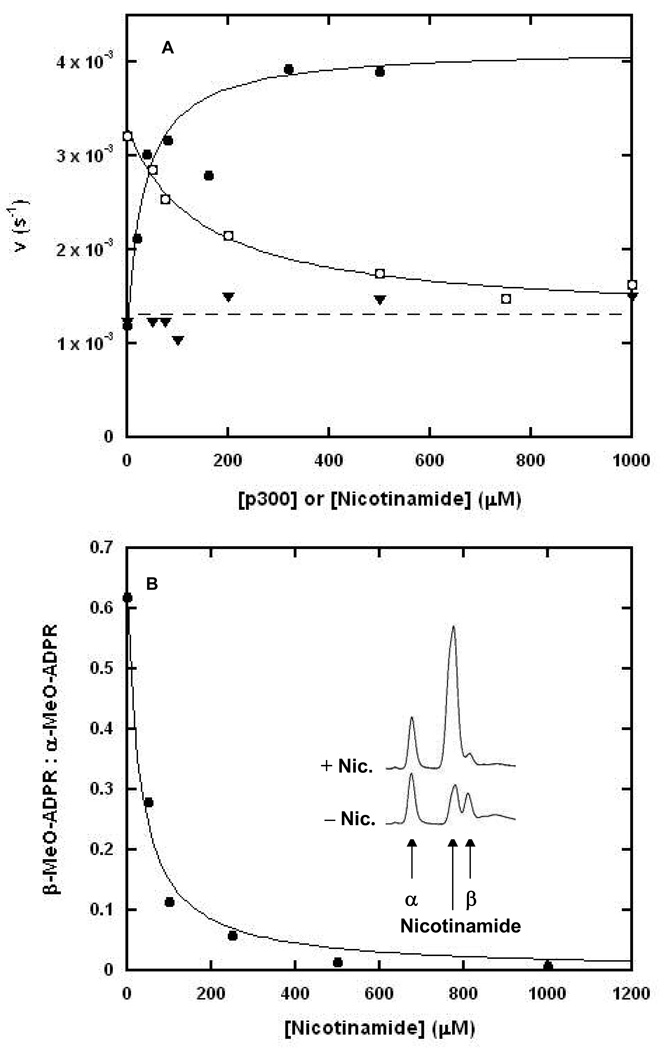
To examine the effect of nicotinamide on hydrolytic rate in the presence and absence of peptide, we conducted reactions in the presence of NAD+ with and without p300 added to solution. In the absence of added peptide, increasing concentrations of nicotinamide have no effect on the rate of hydrolysis, as shown by the line of nearly zero slope in Figure 7A. When p300 is added to reaction, it can stimulate the rate of hydrolysis several fold over the rate in which no peptide is present (overlaid curve Figure 7A). Increasing concentrations of nicotinamide inhibit this increase of hydrolysis in the presence of sub-saturating p300 (subsaturating conditions allow both peptide-dependent and peptide-independent mechanisms to occur simultaneously) but does not fully inhibit hydrolysis, as shown by the fit of the inhibition data to a curve for non-linear inhibition (Ki = 75 ∝M see experimental for details). The uninhibited solvolysis rate is in reasonable agreement with the solvolytic rate when no peptide is present, consistent with the idea that only peptide-dependent hydrolysis is inhibited.
Figure 7. Hydrolysis is Incompletely Inhibited by Nicotinamide.
(A) The effect of nicotinamide on the hydrolytic activity of Pf-Sir2 in the presence and absence of peptide. The curve fit to the data for hydrolysis in increasing P300 concentrations (solid circles) is shown alongside the hydrolysis data obtained by running the reactions in 400 ∝M P300 and increasing nicotinamide concentrations (circles in squares). The hydrolysis rate is incompletely inhibited and plateaus at a level equal to the rate of peptide independent hydrolysis (dashed line). The data for peptide independent hydrolysis in varying nicotinamide concentrations is shown (triangles) and is fit by a straight line (dashed line). (B) The plot of the ratio of β to α stereochemistry of methanolysis products with increasing nicotinamide concentration as determined by HPLC. The plotted points were fit to a curve of form r = r0 – rmax ([I]/(Ki +[I])) where r is the ratio of stereochemistry observed for a given concentration of nicotinamide, r0 is the observed stereochemical ratio when no nicotinamide is present, rmax is the maximal suppression of the ratio, Ki is the apparent inhibition constant of nicotinamide and [I] is the concentration of nicotinamide. The HPLC chromatograms (inset) illustrate that when nicotinamide is added there is a decrease in the β–1-O-methylADPR peak, but there is little change in the production of α–1-O-methylADPR. The nicotinamide visible in the lower trace was produced by the normal hydrolysis and methanolysis of NAD+ catalyzed by Pf-Sir2.
To further probe the idea that only the peptide-dependent solvolysis is sensitive to nicotinamide concentrations, we examined the β/α stereochemistry ratio of methanolysis products as a function of increasing nicotinamide concentrations under experimental conditions in which both methanolysis stereoisomers are generated. As shown in the inset of Figure 7B, HPLC chromatograms reveal that formation of the β product is inhibited by nicotinamide, whereas, formation of the α product is not inhibited. The effect of increasing nicotinamide concentrations on β/α product ratio was determined by HPLC and plotted, with points fit to a predicted curve as described in the figure legend. The ratio decreases to near 0 as a function of increasing nicotinamide concentrations, as predicted if nicotinamide selectively inhibits hydrolysis from the imidate complex, without inhibiting peptide-independent solvolysis of NAD+ (Figure 7B). The peptide-independent hydrolysis of NAD+ by Pf-Sir2 establishes the first catalytic activity of a sirtuin that is insensitive to nicotinamide inhibition--other than base-exchange--at concentrations of nicotinamide exceeding the mM range.
DISCUSSION
Deacetylase activity
Protein deacetylase activity had been previously predicted for Pf-Sir2 based upon biological data implicating this sirtuin in regulation of chromatin structure and in silencing of epitope and virulence associated genes encoded by the var family (20, 21). Patterns of var gene expression are involved in antigenic variation, a process by which P. falciparum avoids the immune system of the host by swapping expression of these genes, while keeping the remainder silent (20, 21). We determined that Pf-Sir2 in the presence of NAD+ catalyzes deacetylation of N-terminal histone sequences of H3 and H4 identical to N-terminal histone sequences. Pf-Sir2 deacetylates H3 peptides at sequences lysine 9 and 14, and also deacetylates H4 at lysine 16. Pf-Sir2 deacetylation generates AAPDR as a product. Two other recent reports have confirmed deacetylase activity for this enzyme (27, 32). In the process of examining other substrates for Pf-Sir2 we identified a human p300 sequence to be a superior substrate for Pf-Sir2 as compared with peptide H3 and H4 sequences. Although we currently do not understand the biochemical or functional basis for Pf-Sir2 substrate preference of p300 over histone sequences, we speculate that context-dependent factors such as post-translational modifications of histone sequences or macromolecular structures of histones and Pf-Sir2 localization to histones may affect catalytic efficiency of Pf-Sir2 for histone sequences in a native context. We do not currently know if p300 is a substrate for Pf2-Sir2 in vivo, although P falciparum is an intracellular parasite of human cells and could theoretically act on human protein sequences.
The deacetylation reaction is slow, even relative to sirtuin standards, which are typically slow turnover enzymes and typically have rates of deacetylation of 0.1 – 0.01 s−1. In the case of H3 and H4 sequences, we determined kcat values of 2.5 and 3.5 ×10−4 s−1 respectively. For a p300 sequence deacetylation rate was only slightly faster with a maximal rate 9 × 10−4 s−1. A recent study of recombinant Pf-Sir2 with a non-physiologic 8-mer acetylated substrate determined a similarly slow deacetylation rate (32). We propose that the slow rate of deacetylation under steady-state conditions is caused by a stalled imidate complex (Scheme 1) which is supported in part by the determination that the nicotinamide cleavage step, which precedes imidate formation, occurs approximately 50–100 times faster (Scheme 1). The rate constants for nicotinamide cleavage are no slower than the observed rate of steady-state base-exchange, which for H3 and p300 are 2.5 × 10−2 s−1 and 6.5 × 10−2 s−1 respectively (Scheme 1 and Figure 2).
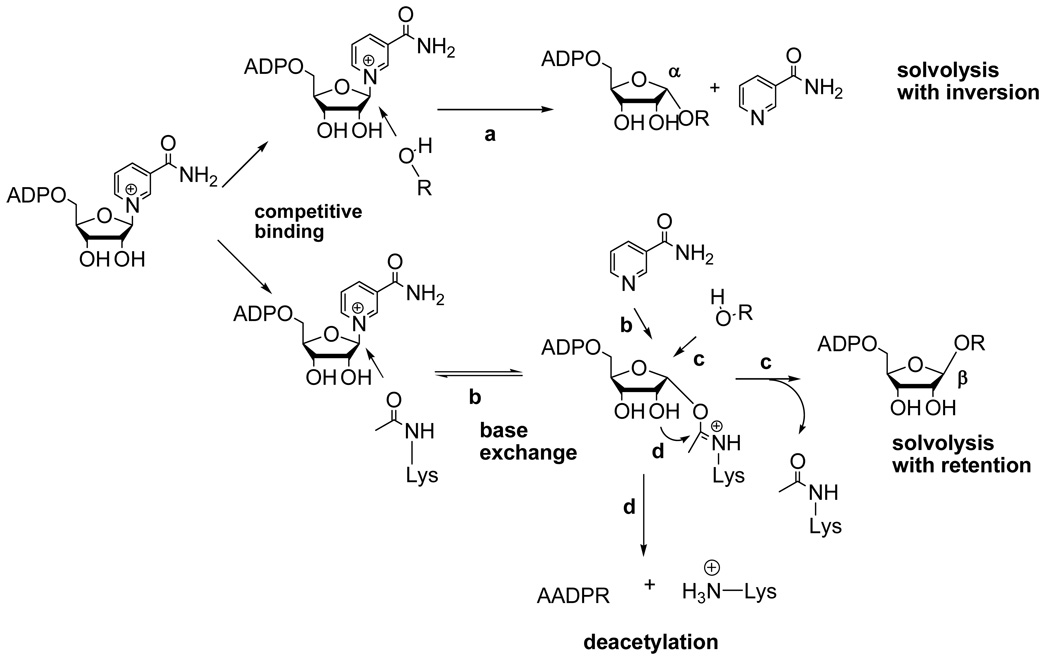
Slow reaction of the imidate and observation of hydrolysis from the imidate
Evidence that imidate forward reaction is rate-limiting for deacetylation includes the observation that the imidate decomposes via hydrolysis at rates that are 4–10 times faster than deacetylation (Table 1), suggesting that the 2'-OH attack of the imidate, which commits the imidate to deacetylation chemistry, is slower. Consistently, the imidate hydrolysis reaction is interpreted to be a direct consequence of the slow rate of deacetylation chemistry (Scheme 4, step d). Reaction of solvent at the anomeric carbon of the Pf-Sir2 imidate is predicted to give overall retention of stereochemistry, which was observed in reactions performed in the presence of both acetyllysine peptide and methanol. A literature precedent is available for such a reaction, in which the conserved active site histidine of the sirtuin HST2 was mutated, causing decreased deacetylation rate. In consequence of slowed deacetylation chemistry, imidate hydrolysis was observed, a reaction not observed on the wildtype HST2 enzyme (10).
Scheme 4.
Overall reaction scheme of Pf-Sir2 catalyzed reactions. Top scheme depicts solvolytic chemistry in the absence of peptide which gives inversion of stereochemistry (reaction a). Bottom scheme depicts acetyllysine dependent chemistries which occur via the imidate including base exchange (reaction b), solvolysis from the imidate (reaction c) and deacetylation (reaction d)
Nicotinamide inhibition of imidate-dependent reactions
Nicotinamide inhibition occurs mainly through the reaction of nicotinamide through the imidate complex. Competitive nicotinamide capture of the imidate (Scheme 4 reaction b) is predicted to inhibit the deacetylation (11, 12) and imidate solvolysis (Scheme 4, reactions d and c respectively). This was observed. Interestingly, the steady-state nicotinamide exchange reaction occurs substantially faster than deacetylation or solvolysis, implying that the imidate and Michaelis complex (NAD+ and acetylated peptide) pseudo-equilibrate when nicotinamide concentrations saturate base-exchange (Scheme 4). Analogous pseudo-equilibration effects of base-exchange with an active site ADPR-intermediate have been observed with the enzyme CD38 (33). We conclude that the imidate is thermodynamically destabilized with respect to the Michaelis complex when nicotinamide saturates the base-exchange reaction, otherwise the imidate concentration would not be depleted and inhibition of deacetylation would not be observed (11). In contrast, in the absence of nicotinamide re-binding to the active site, the imidate is kinetically stable with respect to return to the Michaelis complex, and partitions between deacetylation and hydrolysis (β-stereochemistry).
Peptide-independent solvolysis and insensitivity to nicotinamide
The Pf-Sir2 enzyme also displayed an unusual capacity to catalyze NAD+ hydrolysis in the absence of any added acetylated substrate. In the absence of acetylated substrate, we identified only α-1-O-methyl-ADPR as a product (no β) in addition to ADPR when methanol was added as a probe for stereochemistry of reaction. This finding confirmed that the solvolytic reaction in the absence of peptide occurs with inversion of stereochemistry at C1′ (Scheme 4, reaction a). This type of direct displacement solvolysis has not been reported for any other sirtuin, although cholera and diphtheria toxins, which are NAD+ dependent protein ADP-ribosyltransferases, hydrolyze NAD+ in the absence of cognate substrates, presumably via direct displacement mechanisms, to give inversion in hydrolysis products (34–36). Interestingly, methanolysis appears to be non-reactive in the cholera toxin solvolysis reaction, making confirmation of this prediction problematic (37). The Pf-Sir2 peptide-independent solvolytic reaction occurs at a rate that exceeds the deacetylation rate for H3, but is inhibited by this peptide, as shown by inhibition of α-stereochemistry methanolysis with increasing concentrations of acetylated peptide. This suggests to us that acetyllysine and water (or methanol) occupy the same site on the enzyme leading to imidate formation or α-hydrolysis, with the ratio determined by the relative occupancy of the site. We also determined that increased H3 peptide concentration, while inhibiting the α-methanolysis concomitantly increases β-methanolysis, implying that H3 peptide provides a mechanism for imidate formation and the intermediate solvolysis pathway. The peptide-independent solvolysis pathway is not sensitive to inhibition by nicotinamide, since it does not require imidate formation.
Implications of this work for understanding mechanisms of ADP-ribosyl transfer
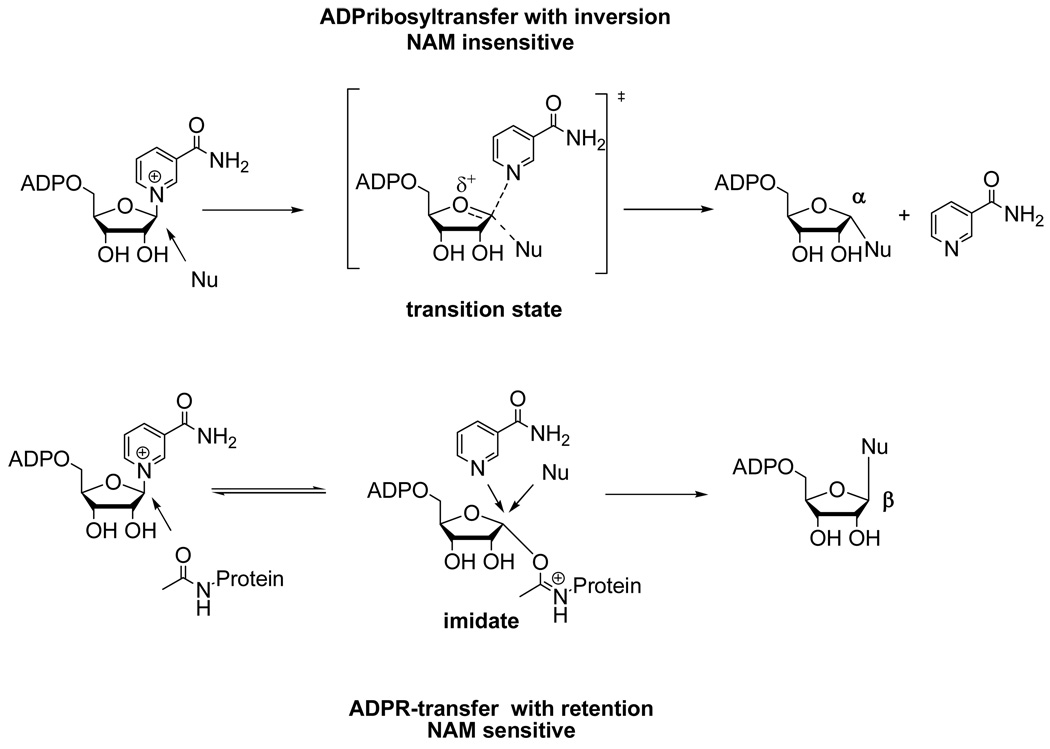
Our findings on Pf-Sir2 are relevant to reports of stable ADP-ribosyltransfer to proteins catalyzed by sirtuins, which have thus far remained mechanistically uncharacterized. From a general chemical perspective the two different types of solvolysis that occur on Pf-Sir2 define mechanistic examples of two different types of stable ADP-ribosyltransfer to nucleophiles. The first of these examples involves direct displacement of NAD+ by a nucleophile to furnish an ADP-ribosyltransfer product with α–stereochemistry (inverted) at C1' of ADPR (Scheme 5, top). This type of reaction is analogous to that observed for both the ADP-ribosylating toxins and the protein modifying ADPribosyltransfer chemistry proposed for the poly-ADP-ribosylpolymerases. Interestingly, this reaction chemistry occurs independently of an acetylated peptide, although these reactions presumably occur by nucleophile occupation of the “acetyllysine pocket”, followed by reaction with enzyme-bound NAD+. Interestingly, we found that nicotinamide cannot inhibit this reaction (Scheme 5), suggesting that nicotinamide only inhibits sirtuin reactions through an imidate complex. We propose that a lack of sensitivity of ADP-ribosyltransfer to nicotinamide can be used to infer the lack of an imidate complex in a sirtuin-catalyzed ADP-ribosyltransfer mechanism.
Scheme 5.
Proposed general mechanisms of ADP-ribosyltransfer catalyzed by sirtuins as determined by chemistries reported in this study. Top chemistry is acetyllysine independent, gives inversion of stereochemistry versus NAD+ and is insensitive to nicotinamide inhibition. Bottom chemistry scheme shows ADP-ribosyltransfer from the imidate complex, to produce ADP-riboysltransfer with retention of stereochemistry versus NAD+. This reaction is predicted to be sensitive to nicotinamide inhibition.
The second type of ADP-ribosyltransfer mechanism determined in this study is reaction of the imidate complex to produce a product with β-stereochemistry (retention) at C1' of ADPR (Scheme 5). This reaction is inherently sensitive to nicotinamide inhibition since nicotinamide can compete for the nucleophile site and for reaction with the imidate (Scheme 5). Thus, imidate-dependent ADP-ribosyltransfer can be distinguished by sensitivity to nicotinamide. We propose that nicotinamide sensitivity can help to determine mechanisms of ADP-ribosyltransfer catalyzed by sirtuins. For example, SIRT4 has been demonstrated to ADP-ribosylate glutamate dehydrogenase, and this reaction is inhibited by nicotinamide (6). Based on our findings, this could imply SIRT4 catalyzes imidate dependent ADP-ribosylation, and we are currently investigating this possibility in our laboratory.
Conclusions
Plasmodium falciparum Pf-Sir2 is a chromatin associated enzyme implicated in silencing of var genes. We found that it can deacetylate acetyllysine peptide substrates such as histone H3 and H4 N-terminal sequences. Pf-Sir2 was also found to catalyze two stereochemically distinct types of solvolysis, one sensitive to nicotinamide inhibition, the other insensitive to nicotinamide inhibition. These are the first reported NAD+ glycohydrolase reactions catalyzed by a wildtype sirtuin. The biologic role of these hydrolytic reactions, which would generate ADPR in the nucleus of the parasite, is not known. We speculate that the solvolyses may be occurring in competition with other kinds of Pf-Sir2 ADP-ribosyltransfer reactions in cells, including protein ADP-ribosyl transfer. A recent paper suggests that Pf-Sir2 is able to catalyze protein ADP-ribosyltransfer (27) although the mechanism of ADP-ribosyltransfer remains undetermined. Investigations to elucidate mechanisms of sirtuin-catalyzed protein ADP-ribosyltransfer reactions are underway in our laboratory.
Abbreviations
- ADPR
adenosine diphosphoribose
- ADPribosyl
adenosine diphosphoribosyl
- AADPR
O-acetyl-adenosine diphosphoribose
- CD38
cell developmental protein 38
- NAD+
nicotinamide adenine dinucleotide oxidized form
- NAM
nicotinamide
- Pf-Sir2
Plasmodium falciparum silencing information regulator 2
- Sir2
Silencing information regulator-2
Footnotes
Research supported in part by assistance from NIH grant R01 DK 73466.
REFERENCES
- 1.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 2.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 3.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 4.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 5.Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- 6.Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Salcedo JA, Gijon P, Nolan DP, Tebabi P, Pays E. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. Embo J. 2003;22:5851–5862. doi: 10.1093/emboj/cdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sauve AA, Celic I, Avalos J, Deng H, Boeke JD, Schramm VL. Chemistry of gene silencing: the mechanism of NAD+-dependent deacetylation reactions. Biochemistry. 2001;40:15456–15463. doi: 10.1021/bi011858j. [DOI] [PubMed] [Google Scholar]
- 9.Sauve AA, Schramm VL. SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates. Curr Med Chem. 2004;11:807–826. doi: 10.2174/0929867043455675. [DOI] [PubMed] [Google Scholar]
- 10.Smith BC, Denu JM. Sir2 protein deacetylases: evidence for chemical intermediates and functions of a conserved histidine. Biochemistry. 2006;45:272–282. doi: 10.1021/bi052014t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauve AA, Schramm VL. Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry. Biochemistry. 2003;42:9249–9256. doi: 10.1021/bi034959l. [DOI] [PubMed] [Google Scholar]
- 12.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem. 2003;278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 13.Zhao K, Chai X, Marmorstein R. Structure of the yeast Hst2 protein deacetylase in ternary complex with 2'-O-acetyl ADP ribose and histone peptide. Structure. 2003;11:1403–1411. doi: 10.1016/j.str.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 14.Marmorstein R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem Soc Trans. 2004;32:904–909. doi: 10.1042/BST0320904. [DOI] [PubMed] [Google Scholar]
- 15.Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. doi: 10.1016/j.molcel.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 17.Kowieski TM, Lee S, Denu JM. Acetylation-dependent ADP-ribosylation by Trypanosoma brucei Sir2. J Biol Chem. 2008;283:5317–5326. doi: 10.1074/jbc.M707613200. [DOI] [PubMed] [Google Scholar]
- 18.Guerra CA, Snow RW, Hay SI. Mapping the global extent of malaria in 2005. Trends Parasitol. 2006;22:353–358. doi: 10.1016/j.pt.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyes SA, Kraemer SM, Smith JD. Antigenic variation in Plasmodium falciparum: gene organization and regulation of the var multigene family. Eukaryot Cell. 2007;6:1511–1520. doi: 10.1128/EC.00173-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 21.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 22.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 23.Yost DA, Anderson BM. Adenosine diphosphoribose transfer reactions catalyzed by Bungarus fasciatus venom NAD glycohydrolase. J Biol Chem. 1983;258:3075–3080. [PubMed] [Google Scholar]
- 24.Scheuring J, Berti PJ, Schramm VL. Transition-state structure for the ADP-ribosylation of recombinant Gialpha1 subunits by pertussis toxin. Biochemistry. 1998;37:2748–2758. doi: 10.1021/bi972594x. [DOI] [PubMed] [Google Scholar]
- 25.Sauve AA, Munshi C, Lee HC, Schramm VL. The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry. 1998;37:13239–13249. doi: 10.1021/bi981248s. [DOI] [PubMed] [Google Scholar]
- 26.Imai S, Johnson FB, Marciniak RA, McVey M, Park PU, Guarente L. Sir2: an NAD-dependent histone deacetylase that connects chromatin silencing, metabolism, and aging. Cold Spring Harb Symp Quant Biol. 2000;65:297–302. doi: 10.1101/sqb.2000.65.297. [DOI] [PubMed] [Google Scholar]
- 27.Merrick CJ, Duraisingh MT. Plasmodium falciparum Sir2: an unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot Cell. 2007;6:2081–2091. doi: 10.1128/EC.00114-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouras T, Fu M, Sauve AA, Wang F, Quong AA, Perkins ND, Hay RT, Gu W, Pestell RG. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–10276. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 29.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 30.Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 31.Berthelier V, Tixier JM, Muller-Steffner H, Schuber F, Deterre P. Human CD38 is an authentic NAD(P)+ glycohydrolase. Biochem J. 1998;330(Pt 3):1383–1390. doi: 10.1042/bj3301383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarty SP, Saikumari YK, Bopanna MP, Balaram H. Biochemical characterization of Plasmodium falciparum Sir2, a NAD(+)-dependent deacetylase. Mol Biochem Parasitol. 2008;158:139–151. doi: 10.1016/j.molbiopara.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Sauve AA, Deng HT, Angeletti RH, Schramm VL. A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. Journal of the American Chemical Society. 2000;122:7855–7859. [Google Scholar]
- 34.Moss J, Osborne JC, Jr, Fishman PH, Brewer HB, Jr, Vaughan M, Brady RO. Effect of gangliosides and substrate analogues on the hydrolysis of nicotinamide adenine dinucleotide by choleragen. Proc Natl Acad Sci U S A. 1977;74:74–78. doi: 10.1073/pnas.74.1.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kandel J, Collier RJ, Chung DW. Interaction of fragment A from diphtheria toxin with nicotinamide adenine dinucleotide. J Biol Chem. 1974;249:2088–2097. [PubMed] [Google Scholar]
- 36.Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35:1137–1149. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 37.Oppenheimer NJ. Structural determination and stereospecificity of the choleragen-catalyzed reaction of NAD+ with guanidines. J Biol Chem. 1978;253:4907–4910. [PubMed] [Google Scholar]