SUMMARY
The transition from the juvenile to the adult phase of shoot development in plants is accompanied by changes in vegetative morphology and an increase in reproductive potential. Here we describe the regulatory mechanism of this transition. We show that miR156 is necessary and sufficient for the expression of the juvenile phase, and regulates the timing of the juvenile-to-adult transition by coordinating the expression of several pathways that control different aspects of this process. miR156 acts by repressing the expression of functionally distinct SPL transcription factors. miR172 acts downstream of miR156 to promote adult epidermal identity. miR156 regulates the expression of miR172 via SPL9 which, redundantly with SPL10, directly promotes the transcription of miR172b. Thus, like the larval-to-adult transition in Caenorhabditis elegans, the juvenile-to-adult transition in Arabidopsis is mediated by sequentially operating miRNAs. miR156 and miR172 are positively regulated by the transcription factors they target, suggesting that negative feedback loops contribute to the stability of the juvenile and adult phases.
INTRODUCTION
Genetic analyses of developmental maturation in Caenorhabditis elegans (Moss, 2007; Rougvie, 2005) and plants (Bäurle and Dean, 2006; Chuck and Hake, 2005; Poethig, 2003) have revealed that these phenomena involve several independently regulated processes that must be temporally coordinated for normal development. An important example of this is the coordination between somatic and reproductive maturation, variation in which is the basis for many examples of morphological evolution (Gould, 1977). Each of these maturation processes itself consists of a variety of independently-regulated events that must be temporally coordinated. How this coordination is achieved is a major problem in developmental biology.
In C. elegans, transitions between stages of larval development are mediated by an increase in the expression of two sequentially expressed miRNAs, lin-4 and let-7 (reviewed in Moss, 2007; Pasquinelli and Ruvkun, 2002; Rougvie, 2005). These were the first miRNAs to be discovered, and they have since served as paradigms for the function of this class of regulatory molecules in animals (Lee et al., 1993; Reinhart et al., 2000). Remarkably, miRNAs have a similar function in plants. As a plant grows, it undergoes a transition from a juvenile to an adult stage of vegetative development (vegetative phase change) and then enters a reproductive phase (reproductive phase change or floral induction), during which it produces flowers or other types of reproductive structures. In Arabidopsis, vegetative phase change is marked by changes in the production of trichomes on the abaxial (lower) surface of the leaf, an increase in the length/width (L/W) ratio of the leaf blade, an increase in the degree of serration of the leaf margin and a decrease in cell size (Telfer et al., 1997; Tsukaya et al., 2000; Usami et al., 2009). Recent studies suggest that miR156, and possibly miR172, play pivotal roles in these transitions. In both Arabidopsis and maize, miR156 is highly expressed early in shoot development and decreases with time, while miR172 has the opposite expression pattern (Aukerman and Sakai, 2003; Chuck et al., 2007a; Jung et al., 2007; Lauter et al., 2005; Wu and Poethig, 2006). Over-expression of miR156 in both Arabidopsis and maize prolongs the expression of juvenile vegetative traits and delays flowering (Chuck et al., 2007a; Wu and Poethig, 2006), whereas over-expression of miR172 in Arabidopsis accelerates flowering (Aukerman and Sakai, 2003; Chen, 2004; Jung et al., 2007). These observations suggest that these miRNAs have related, but opposite, functions in shoot maturation.
Although the targets of miR156 and miR172 have been identified, the functions of these targets are still poorly characterized. In Arabidopsis, miR156 targets 10 members of the SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) family of transcription factors (SPL2, SPL3, SPL4, SPL5, SPL6, SPL9, SPL10, SPL11, SPL13, SPL15), while miR172 targets 6 APETALA2-LIKE (AP2-like) transcription factors (AP2, TOE1, TOE2, TOE3, SMZ, SNZ). The gain-of-function or loss-of-function phenotype of single gene mutations reveals a high degree of functional redundancy within these families. Loss-of-function mutations of SPL3 have no obvious phenotype, but constitutive expression of this gene or its closely related paralogs SPL4 and SPL5 produces an early flowering phenotype, accelerates the production of trichomes on the abaxial surface of the leaf (an adult trait) (Gandikota et al., 2007; Wu and Poethig, 2006), and produces changes in cell size and cell number typical of adult leaves (Usami et al., 2009). Over-expression of SPL9 reduces the rate of leaf initiation and increases leaf size (Wang et al., 2008), and a similar phenotype is observed in a gain-of-function mutant of SPL15 (Usami et al., 2009). Loss-of-function mutations in either SPL9 or SPL15 have minor effects on development. Plants doubly mutant for these related genes have a stronger phenotype than the single mutants, which reveals that they promote both vegetative phase change and flowering (Schwarz et al., 2008; Wang et al., 2008). The targets of miR172 have an opposite effect on phase change. Plants lacking TOE1 and TOE2 are early flowering, whereas plants over-expressing TOE1, TOE2, SNZ, or SMZ are late flowering (Aukerman and Sakai, 2003; Jung et al., 2007; Schmid et al., 2003).Although there is still no evidence that these AP2-like genes contribute to vegetative phase change in Arabidopsis, their maize homolog Glossy15 (Gl15) promotes juvenile epidermal identity (Evans et al., 1994; Moose and Sisco, 1994, 1996), suggesting that one or more of the Arabidopsis homologs may do so as well. Whether these SPL and AP2-like genes operate in the same or different pathways is unknown.
We undertook a genetic and molecular analysis of miR156, miR172 and their targets to define their roles in vegetative phase change. Our results indicate that miR156 is both necessary and sufficient for the expression of the juvenile phase, and that it functions as a master regulator of this phase. The targets of miR156 act in several pathways that control both flowering time and different aspects of vegetative development. One of these pathways includes miR172b. We show that miR156 regulates the expression of miR172b via SPL9, which acts as a direct transcriptional regulator of miR172b. Our results suggest a model for the temporal coordination of vegetative phase change and floral induction.
RESULTS
miR156 is a master regulator of the juvenile phase
In Arabidopsis, floral induction affects the development of unexpanded rosette leaves in ways that can make it difficult to observe the juvenile-to-adult transition. This is particularly problematic in genotypes that flower very early in long days, which applies to many of the stocks used in this study. For this reason, all of the experiments reported here were conducted with plants grown in short days to delay flowering.
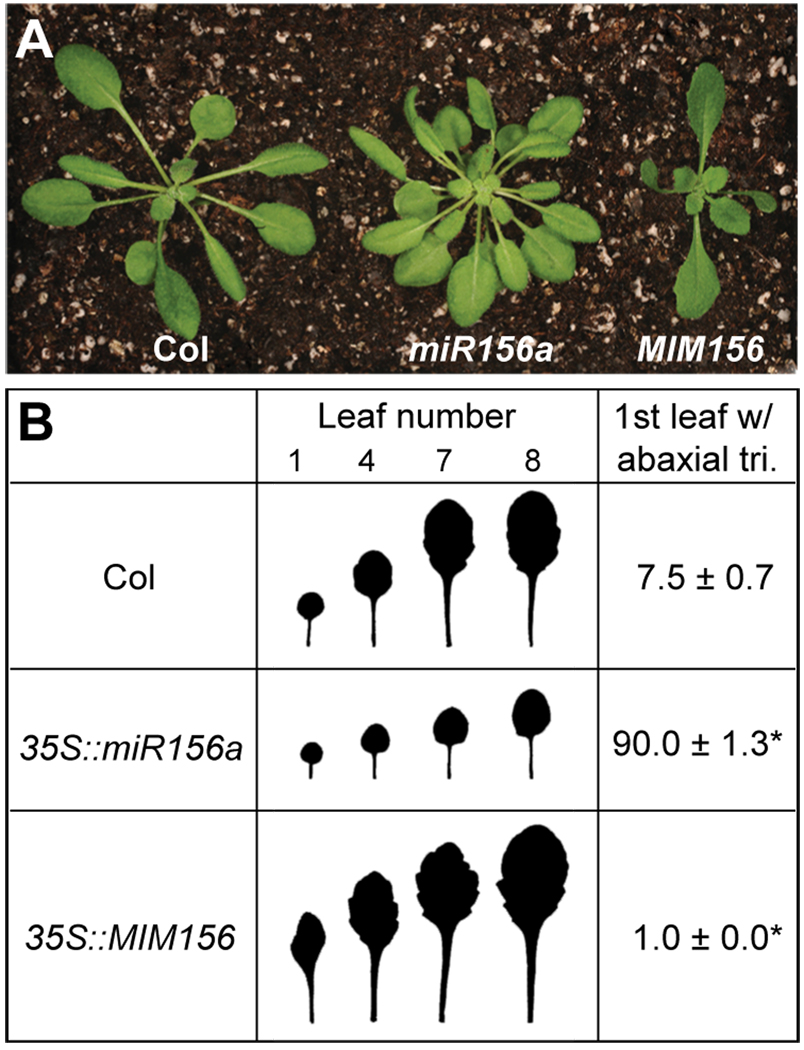
Under short day conditions, plants expressing miR156a under the regulation of the constitutive 35S promoter produced approximately 90 leaves that resembled juvenile leaves in size, shape, and their lack of abaxial trichomes (Figure 1A, B). In contrast, all leaves produced by plants in which the activity of miR156 was suppressed by constitutively expressing a transcript with non-cleavable miR156 target site (a target-site mimic, MIM156) (Franco-Zorrilla et al., 2007) resembled adult leaves. The effect of 35S∷MIM156 on leaf development was particularly striking in the case of the first two rosette leaves. In 35S∷MIM156 plants, the first two rosette leaves were unusually large and elongated, and possessed serrated leaf margins and abaxial trichomes--features of adult leaves. Later-formed leaves were larger, but nearly identical in shape to these first two rosette leaves. Thus, miR156 promotes the expression of all juvenile leaf traits, and is both necessary and sufficient for the expression of these traits.
Figure 1. miR156 is necessary and sufficient for the juvenile vegetative phase.
(A) 25-day-old wild-type, 35S∷miR156a and 35S∷MIMI156 plants grown in short days. (B) The shape and the abaxial trichome phenotypes of fully expanded rosette leaves of wild type, 35S∷miR156a and 35S∷MIMI156 plants. 35S∷miR156a prolongs the duration of the juvenile phase and 35S∷MIM156 eliminates this phase. Asterisks indicate significant difference from wild-type (P < 0.01, n = 18, ± SD).
SPL genes have different roles in vegetative phase change
The SPL genes targeted by miR156 can be grouped into four major clades: SPL3/SPL4/SPL5, SPL2/SPL10/SPL11, SPL9/SPL15, SPL6/SPL13 (Guo et al., 2008). SPL3, SPL9 and SPL10 are representative members from three of these clades. To investigate the function of SPL3, SPL9 and SPL10, we first examined their spatial expression pattern in vegetative shoot apices by RNA in situ hybridization. SPL3 was present uniformly throughout the shoot apex and in expanding leaf primordia and increased in abundance between 15 and 22 days after planting (Figure 2A–C). SPL9 was expressed at a much lower level than SPL3, and was barely visible in young leaf primordia in 22 day-old shoots (Figure 2D–E). To confirm this expression pattern, we examined plants transformed with pSPL9∷rSPL9, a construct that expresses a miR156-resistant SPL9 transcript under the control of its native promoter. Consistent with its wild-type expression pattern, transgenic plants expressed SPL9 in both pre-emergent and expanding leaf primordia (Figure 2F). These results are consistent with previous suggestions (Wang et al., 2008) that miR156 regulates the abundance of SPL9 transcripts, but not their spatial expression pattern. SPL10 transcripts were undetectable in wild-type shoot apices.
Figure 2. SPL3, SPL9 and SPL10 have diverse roles in vegetative development.
(A–C) In situ expression pattern of SPL3.
(A) 22 day-old vegetative shoot apex hybridized with a sense strand control.
(B) 15-day-old vegetative shoot apex hybridized with antisense probe.
(C) 22-day-old vegetative shoot apex hybridized with antisense probe. The abundance of SPL3 mRNA increases with time.
(D–F) In situ expression pattern of SPL9; all samples hybridized with an antisense probe.
(D) 22-day-old vegetative shoot apex of an RNA-null allele of SPL9.
(E) 22-day-old wild-type vegetative shoot apex.
(F) 22-day-old vegetative shoot apex from a plant expressing a miR156-insensitive SPL9 genomic sequence under the control of the SPL9 promoter. SPL9 is expressed in young leaf primordia.
(G) Four-week-old rosettes of wild-type, mutant and transgenic lines of Arabidopsis grown in short days. rSPL3 = 35S∷rSPL3, rSPL9 = pSPL9∷rSPL9, rSPL10 = rSPL10∷rSPL10.
(H) The shape and abaxial trichome phenotypes of fully expanded leaves of wild-type, spl9-4, and transgenic lines expressing miR156-resistant forms of SPL3, SPL9 and SPL10. These genes promote different adult traits. Asterisks indicate significant difference from wild-type (P <0.01, n= 18, ± SD).
To define the roles of these genes in vegetative phase change, we characterized their loss- and gain-of-function phenotypes. spl9-4 delayed abaxial trichome production by 2.8 plastochrons and caused the leaf blade to become rounder (Figure 2G, H). SPL15 is the closest paralog of SPL9. spl15-1 mutants had no obvious effect on abaxial trichome production or leaf shape. Plants doubly mutant for spl9-4 and spl15-1 produced abaxial trichomes 1.6 plastochrons later than spl9-4 but had same leaf shape as spl9-4 (L/W = 1.48 ± 0.08 for spl9 spl15 vs. 1.50 ± 0.11 for spl9). These results imply that SPL15 has overlapping functions with SPL9, but has a less important role in leaf morphogenesis than SPL9. Loss-of-function mutations in SPL3 and SPL10 had no obvious vegetative phenotype, presumably because their function overlaps with other SPL genes. Transgenes expressing miR156-resistant versions of SPL3, SPL9 and SPL10 affected leaf development in different ways. 35S∷rSPL3 accelerated abaxial tricome production by 1.5 plastochrons, but had no significant effect on leaf shape (Figure 2G, H). pSPL9∷rSPL9 and pSPL10∷rSPL10 accelerated the expression of all adult-specific leaf traits, producing leaves with an elongated leaf blade, serrated leaf margin, and abaxial trichomes (Figure 2G, H). However, the leaves of rSPL10 plants were flatter, rounder and more serrated than those of rSPL9, and also had a more distinct petiole (Figure 2G, H). As expected from their high degree of sequence similarity, the phenotype of pSPL11∷rSPL11 was similar to that of pSPL10∷rSPL10 (data not shown). Thus, SPL3, SPL9 and SPL10/SPL11 have overlapping, but distinct functions in vegetative development.
miR172 promotes adult epidermal identity via TOE1 and TOE2
miR172 promotes flowering when over-expressed (Aukerman and Sakai, 2003; Chen, 2004; Jung et al., 2007), but whether it plays a role in Arabidopsis vegetative phase change is unknown. To address this question we examined the vegetative phenotype of plants transformed with a genomic fragment containing the miR172b precursor under the control of 35S promoter (35S∷miR172b). The leaves of plants expressing 35S∷miR172b produced abaxial trichomes two plastochrons earlier than normal, but these leaves were otherwise morphologically normal (Table 1). 35S∷miR172a had essentially the same phenotype as 35S∷miR172b; furthermore, a T-DNA insertion in miR172a (SALK_045787) delayed abaxial trichome production by about three plastochrons while having no obvious effect on leaf morphology (Table 1). Thus, miR172 promotes adult epidermal identity, but has little, if any, role in the regulation of leaf shape.
Table 1.
The effect of miR172, TOE1 and TOE2 on abaxial trichome production and leaf shape
| Genotype | 1st leaf w/ abaxial trichomes | Leaf length/width (leaf 7) |
|---|---|---|
| Wild type | 7.0 ± 0.5 | 1.64 ± 0.09 |
| 35S∷miR172b | 5.0 ± 0.9a | 1.63 ± 0.09 |
| miR172a-1 (SALK_045787) | 9.6 ± 1.0a | 1.61 ± 0.10 |
| Wild type | 6.6 ± 0.5 | 1.64 ± 0.09 |
| toe1-2 | 5.6 ± 0.6a | 1.65 ± 0.11 |
| toe2-1 | 5.5 ± 0.6a | 1.66 ± 0.14 |
| toe1-2, toe2-1 | 3.2 ± 0.4a | 1.68 ± 0.13 |
| 35S∷TOE1 | 10.4 ± 0.9a | 1.51 ± 0.07c |
| Wild type | 7.2 ± 1.0 | |
| toe2-1 | 5.5 ± 0.5 | |
| 35S∷miR156a | 90.0 ± 1.6 | |
| toe2-1, 35S∷miR156a | 9.5 ± 1.4b | |
| Wild type | 7.2 ± 0.7 | |
| spl9-4 | 9.8 ± 1.3 | |
| toe1-2, toe2-1 | 3.0 ± 0.2 | |
| spl9-4, toe1-2, toe2-1 | 3.7 ± 0.7b |
significantly different from wild type (p< 0.01, n = 24, ± SD)
significantly different from all other genotypes (p<0.01, n = 24, ± SD)
significantly different from wild type (p<0.01, n=10, ± SD)
miR172 targets 6 AP2-like genes in Arabidopsis, including TOE1 and TOE2 (Aukerman and Sakai, 2003; Schmid et al., 2003). Gain- and loss-of-function mutations in TOE1 and TOE2 have been shown to affect flowering time (Aukerman and Sakai, 2003; Jung et al., 2007), but their effect on vegetative development is unknown. Abaxial trichome production was accelerated by one plastochron in plants homozygous for toe1–2 or toe2-1 (hereafter referred to as toe1 and toe2), and by three plastochrons in toe1 toe2 double mutants. Neither the single mutants or the double mutant had an effect on leaf shape (Table 1). In contrast, constitutive expression of TOE1 (35S∷TOE1) delayed abaxial trichome production by 4 plastochrons and caused the leaf blade to become slightly rounder than normal (Table 1). We conclude that TOE1 and TOE2 act primarily to promote juvenile epidermal identity, and probably mediate the effect of mR172 on vegetative development.
miR172 acts downstream of miR156
Previous studies have shown that miR156 decreases during shoot development in Arabidopsis (Wu and Poethig, 2006), whereas miR172 increases (Aukerman and Sakai, 2003; Jung et al., 2007); however, the expression of these miRNAs has not been directly compared in the same material. For this purpose, RNA blots of shoot apices harvested 12, 19 and 26 days after planting were hybridized sequentially with probes to these miRNAs. miR156 and miR172 were expressed in inverse patterns: miR156 declined between 12 and 19 days after planting, whereas miR172 increased during this same period (Figure 3A). To determine if these changes are causally related, we examined the level of miR156 and miR172 in plants over-expressing these miRNAs. Plants transformed with 35S∷miR156a had half the normal amount of miR172, whereas plants transformed with 35S∷MIM156 had over twice the normal amount of miR172 (Figure 3B). By contrast, 35S∷miR172b had little or no effect on miR156. Thus, miR156 regulates the expression of miR172, but not the reverse. To determine if this effect is functionally significant, we examined the phenotype of plants homozygous for both 35S∷miR156a and 35S∷miR172b. These double transgenic plants had leaves that were the size and shape of 35S∷miR156a leaves, but initiated abaxial trichome production earlier than 35S∷miR156a plants (Figure 3C). Indeed, their pattern of abaxial trichome production was much closer to that of 35S∷miR172b than to 35S∷miR156a. This result supports the conclusion that miR172 acts downstream of miR156, and provides additional evidence that miR172 primarily regulates epidermal differentiation.
Figure 3. miR172 acts downstream of miR156.
(A) Blot of small RNA from the shoot apex of wild-type plants of different ages hybridized sequentially with probes to miR156 and miR172. The levels of these miRNAs change in a complementary fashion. U6 served as a loading control.
(B) Blots of small RNA from 35S∷miR156a 35S∷miR172b (14-day-old) and 35S∷MIM156 (20-day-old) plants hybridized sequentially with probes to miR156 and miR172. miR156 represses miR172. U6 was used as loading control.
(C) Leaf shape and abaxial trichome phenotypes of fully expanded rosette leaves of wild-type, 35S∷miR156a 35S∷miR172b and 35S∷miR156a 35S∷miR172b double transgenic plants. 35S∷miR172b partially rescues the 35S∷miR156a over-expression phenotype.
Numbers indicate fold change relative to wild-type. Asterisks indicate significant difference from wild-type (P <0.01, n=18 plants, ± SD).
If miR172 mediates the effect of miR156 on epidermal identity by repressing the expression of TOE1 and TOE2, then the early abaxial trichome phenotype of toe1 and/or toe2 should be epistatic to the late abaxial trichome phenotype of 35S∷miR156. Consistent with this prediction, toe2 nearly completely rescued the abaxial trichome phenotype of 35S∷miR156a in double mutants (Table 1). We were unable to examine the genetic interaction between toe1 and 35S∷miR156a because the miR156a transgene was silenced in toe1–2 35S∷miR156a plants, probably because toe1–2 is a T-DNA induced mutation and shares sequences with the 35S∷miR56a construct (Daxinger et al., 2008). Although toe2-1 is also a T-DNA induced mutation, it does not silence 35S∷miR56a.
SPL9 and SPL10 promote the transcription of miR172
miR156 represses 10 members of the SPL gene family (Rhoades et al., 2002; Schwab et al., 2005). To identify the SPL genes that mediate the effect of miR156 on miR172, we analyzed the expression of miR172 in plants expressing miR156-resistant versions of SPL3, SPL4, SPL5, SPL9 and SPL10. Although the phenotypes of 35S∷rSPL3, 35S∷rSPL4 and 35S∷rSPL5 are similar to that of 35S∷miR172b and toe1 toe2 (Table 1), these transgenes had no effect on the abundance of miR172 (Figure 4A) or the abundance of the TOE1 and TOE2 transcripts (Figure 4B). To determine if SPL3, SPL4 and SPL5 act downstream of TOE1 and TOE2 we examined the their expression in toe1 toe2 mutants and 35S∷TOE1 plants. SPL3 was slightly (1.5 to 2-fold) but consistently over-expressed in toe1 toe2, but was either slightly down-regulated or unaffected by 35S∷TOE1 (Figure 4C). SPL4 and SPL5 were expressed much more variably than SPL3 in toe1 toe2. Although in some cases we observed a slight increase in their expression, in other experiments there was no significant difference between their expression level in toe1 toe2 and wild-type plants (Figure 4D). These results suggest the effect of toe1 toe2 on SPL3, SPL4 and SPL5 expression is indirect.
Figure 4. SPL3, SPL4 and SPL5 act independently of miR172 and TOE1, TOE2.
(A) RNA blots of small RNA from 20-day-old wild-type, 35S∷rSPL3, 35S∷rSPL4 and 35S∷rSPL5 rosettes hybridized with a probe to miR172 reveals that these transgenes have no effect on the expression of miR172. U6 served as a loading control. Numbers indicate fold change relative to wild-type.
(B) qRT-PCR analysis of TOE1 and TOE2 mRNA from 14-day-old wild-type, 35S∷rSPL3 35S∷rSPL4 and 35S∷rSPL5 rosettes indicates that these transgenes have no effect on the expression of TOE1 and TOE2.
(C) qRT-PCR analysis of SPL3 mRNA in wild-type, toe1 toe2, and 35S∷TOE1 rosettes reveals that the expression of SPL3 is increased by toe1 toe2, but unaffected by 35S∷TOE1.
(D) qRT-PCR analysis of SPL4 and SPL5 mRNA in 2-week-old wild-type and toe1 toe2 rosettes reveals no consistent change in the expression of these genes; the results of two experiments are shown.
qRT-PCR data represent the average of three technical replicates; samples were normalized to wild-type at each time point; ± SD.
In contrast to 35S∷rSPL3, 35S∷rSPL4 and 35S∷rSPL5, plants expressing pSPL9∷rSPL9 and pSPL10∷rSPL10 had more than 4-fold higher levels of miR172 (Figure 5A). Furthermore, qRT-PCR revealed that the primary transcript of miR172b—one of 5 loci encoding miR172—is expressed at uniformly high level in pSPL9∷rSPL9 and pSPL10∷rSPL10 throughout shoot development (Figure 5B). To determine if miR172b is a direct target of SPL9, we took advantage of an inducible expression system based on the posttranscriptional activation of the rat glucocorticoid receptor (GR) (Lloyd et al., 1994). GR was fused to the 5' end of rSPL9, and this fusion gene was expressed in transgenic plants under the regulation of the 35S promoter. Transgenic seeds were plated on MS medium, and treated with the synthetic ligand dexamethasone (DEX) in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX) for 4 hours; RNA was then extracted and the abundance of miR172b was assessed by qRT-PCR. CHX was used to block the translation of mRNAs regulated by SPL9, and thus prevent secondary effects. In the presence of DEX, this line has a phenotype similar to that of the pSPL9∷rSPL line illustrated in Figure 2G. The miR172b primary transcript was elevated about 3-fold in samples treated with DEX and with DEX+CHX, strongly suggesting that miR172b is a direct transcriptional target of SPL9 (Figure 5C). We then tested if SPL9 binds to miR172b by examining the chromatin fragments that immunoprecipitate with a FLAG-tagged SPL9 protein; this epitope-tagged protein was expressed in transgenic plants under the regulation of the SPL9 promoter and produced the phenotype illustrated in Figure 2G, demonstrating that the protein is functional. As a control, we used plants expressing cMyc-tagged SPL9 under the regulation of the same promoter. The abundance of several 200-bp regions containing the core SPL binding sequence GTAC (Figure 5D) was measured in immunoprecipitated material using qPCR (Figure 5E). One site (S1) located approximately 1.2 kb upstream of the transcriptional start site of miR172b was enriched approximately 5-fold in FLAG-SPL9 plants compared to SPL9-cMyc controls; no significant enrichment was observed for the other sites we examined (Figure 5E). To determine if SPL9 is required for the transcription of miR172b we examined the effect of spl9-4 and spl9-4 spl15-1 on the abundance of the miR172b primary transcript. Although spl9-4 had no significant effect on the miR172b transcript, the level of this transcript was slightly reduced in spl9 spl15 double mutants (Figure 5F), consistent with the observation that this double mutant has a more severe phenotype than either single mutant. These results indicate that SPL9 directly promotes the transcription of miR172b.
Figure 5. Regulation of miR172b by SPL9, SPL10, TOE1, and TOE2.
(A) Northern blot of small RNA from 20-day-old wild-type, pSPL9∷rSPL9 and pSPL10∷rSPL10 rosettes. U6 was used as a loading control. Numbers indicate the fold change relative to wild-type. pSPL9∷rSPL9 and pSPL10∷rSPL10 increase the expression of miR172.
(B) qRT-PCR analysis of the miR17b precursor in 12, 16, 19 and 24-day-old wild type, pSPL9∷rSPL9 pSPL10∷rSPL10 rosettes. The fold change relative to the 12-day-old wild type sample is shown; SD bars are obscured by symbols. These transgenes increase the expression of miR172b and eliminate its temporal expression pattern.
(C) qRT-PCR analysis of the miR17b precursor in 20-day-old 35S∷GR-rSPL9 seedlings treated with DEX in the absence or presence of CHX. GR-SPL9 promotes the expression of miR172b in the absence of protein synthesis.
(D) The location of three putative SPL9 binding sites in the miR172 locus that were tested by ChIP analysis. Open box indicates the miR172b transcript.
(E) qPCR analysis of putative SPL9 binding sites in the chromatin of 14-day-old pSPL9∷SPL9r–cMyc and pSPL9∷3XFLAG-SPL9r rosettes immunoprecipitated with an antibody to FLAG. The immunoprecipitated values were first normalized to the input values then divided by the pSPL9∷SPL9r–cMyc value to get a fold enrichment. The numbers represent the fold difference relative to pSPL9∷SPL9r–cMyc sample. Values are the average of two biological replicates. eIF4A was used as a negative control.
(F) qRT-PCR analysis of the miR172b precursor in wild-type and spl9-4 spl15-1 rosettes at different stages of vegetative development. miR172b is slightly reduced at all of these stages.
(G) Blot of small RNA from the rosette of 14-day-old wild-type and toe1 toe2 plants hybridized with a probe to miR172. U6 served as a loading control. Numbers indicate the fold change relative to the wild-type sample.
(H) qRT-PCR analysis of the miR172b precursor in wild-type, toe1 toe2 and 35S∷TOE1 rosettes.
(I) qRT-PCR analysis of the miR156a precursor in 12- and 16-day-old pSPL9∷rSPL9 and pSPL10∷rSPL10 plants.
qRT-PCR data represent the average of three technical replicates; samples were normalized to wild-type at each time point; ± SD.
If SPL9 promotes adult epidermal identity by up-regulating the transcription of miR172b and thereby repressing TOE1 and TOE2, these AP2-like genes should be required for the juvenilized epidermal phenotype of spl9-4. To test this prediction, we examined the timing of abaxial trichome production in spl9-4, toe1 toe2, and the spl9-4 toe1 toe2 triple mutant. The timing of abaxial trichome production in the triple mutant was slightly but significantly different from toe1 toe2 (Table 1). This result suggests that TOE1 and TOE2 contribute to the epidermal phenotype of spl9-4, but are not solely responsible for this phenotype.
Feedback loops in which a miRNA-regulated transcription factor regulates the transcription of its cognate miRNA have been described in a number of animals (Fazi et al., 2005; Johnson et al., 2005; Kim et al., 2007; Varghese and Cohen, 2007). To determine if miR172 is regulated in this fashion, we examined the effect of TOE1 and TOE2 on mature miR172 and the miR172b precursor. Both of these molecules were reduced about 50% in toe1 toe2 double mutants; conversely, the miR172b precursor was slightly elevated in plants over-expressing TOE1 (Figure 5G, H). We then examined if SPL genes have a similar effect on the expression of the miR156a precursor. This transcript was elevated about 1.5 fold in 12-day-old, and 2.5-to-3 fold in 16-day-old pSPL9∷rSPL9 and pSPL10∷rSPL10 plants (Figure 5I). Plants over-expressing SPL3, SPL4 and SPL5 also had elevated levels of miR156a, but this effect was only observed around 2 weeks after planting (data not shown). The evidence that miR172b and miR156a are positively regulated by the transcription factors they target suggests that the expression of these targets is modulated by a negative feedback loop that buffers against small changes in the level of their mRNA.
DISCUSSION
Shoot maturation depends on the coordinated activity of multiple interacting pathways. The pathways that mediate the transformation of an adult vegetative shoot into a flower-bearing shoot (floral induction) have been intensively studied, and are now well understood (Bäurle and Dean, 2006; Parcy, 2005). Although much less is known about the regulation of vegetative phase change, decades of research on woody plants (Hackett and Murray, 1997), and more recent studies of maize and Arabidopsis (Chuck and Hake, 2005; Kerstetter and Poethig, 1998; Poethig, 2003) suggest that this transition also involves the activity of multiple pathways. The results presented here provide new insight into the structure of these pathways in Arabidopsis, and suggest the mechanism by which their expression is temporally coordinated.
Our results indicate that miR156 is necessary and sufficient for the expression of the juvenile phase and demonstrate that it operates by repressing the expression of SPL genes that act in pathways with different developmental functions (Figure 6). These results are consistent with a recent study indicating that the precocious phase change phenotype of the squint mutation in Arabidopsis is attributable to a defect in the activity of miR156 (Smith et al., 2009), and with previous descriptions of the phenotype of plants expressing miR156-resistant versions of SPL genes (Gandikota et al., 2007; Usami et al., 2009; Wang et al., 2008; Wu and Poethig, 2006). Remarkably, over-expression of miR156 in maize produces a phenotype similar to that produced by over-expression of miR156 in Arabidopsis (Chuck et al., 2007a). Along with the evidence that miR156 is one of the most highly conserved miRNAs in the plant kingdom (Axtell and Bowman, 2008), these results suggest that miR156 is a master regulator of the juvenile phase in plants.
Figure 6. A model for the regulation of vegetative phase change by miR156 and miR172.
Temporal changes in the level of miR156 and SPL proteins are illustrated by the shaded bars; time increases from left to right. We propose that miR156 coordinates the expression of several pathways by repressing the expression of SPL genes that act in these pathways. Each of these pathways controls different phase-specific traits, but have components in common (e.g. SPL9, SPL10) and may also share downstream targets. The relationship between TOE1 and TOE2 and SPL3, SPL4, SPL5 is unclear.
The identity of all the SPL genes that mediate the effect of miR156 on vegetative phase change is difficult to establish because of the high degree of functional redundancy within this family; furthermore, the genomic organization of some of these genes makes it difficult to generate the combination of mutations necessary for this analysis. The genes described in this paper are important components of this mechanism, however. The loss- and gain-of-function phenotypes of SPL9 demonstrate that it promotes most, if not all, of the traits associated with the adult phase. SPL10 also regulates all aspects of vegetative phase change, but appears to have different functions than SPL9 because its over-expression phenotype differs in several respects from that of SPL9. SPL3, SPL4, and SPL5 have more limited roles in leaf development, acting primarily, although perhaps not exclusively (Usami et al., 2009; Wu and Poethig, 2006), to promote adult patterns of epidermal differentiation. In addition to their roles in vegetative development, all of these genes promote flowering under long day conditions (Cardon et al., 1997; Schwarz et al., 2008; Wu and Poethig, 2006). Indeed, the early flowering phenotype of plants over-expressing SPL3, SPL4, and SPL5 raises questions about the previously reported effects of these genes on leaf shape (Wu and Poethig, 2006) and cell size and number (Usami et al., 2009) because floral induction has major effects on leaf development and these previous studies were conducted under floral inductive conditions.
miR172 has been implicated in the regulation of flowering time and floral organ identity in both maize and Arabidopsis (Aukerman and Sakai, 2003; Chen, 2004; Chuck et al., 2007b; Zhao et al., 2007). In maize, miR172 targets Gl15, a gene that promotes juvenile epidermal identity (Lauter et al., 2005). These genes have complementary expression patterns, so it is reasonable to hypothesize that miR172 plays a role in vegetative phase change. However, there is still no evidence that miR172 is actually important for this process. For example, mutations in ts4, which encodes miR172e, have no effect on vegetative phase change in maize (Chuck et al., 2007b). Our results indicate that in Arabidopsis miR172 promotes adult epidermal identity, and that this is its primary function during vegetative development. This function is mediated by two of its six targets, TOE1 and TOE2, as demonstrated by the observation that loss-of-function mutations in these genes actually have a more severe effect on abaxial trichome production than the 35S∷miR172b transgene used in these studies. The difference in the severity of these phenotypes can probably be attributed to the relatively small increase of miR172 in this transgenic line. The observation that miR172 levels are affected by changes in the level of miR156, as well as the observation that 35S∷miR172b and toe2 nearly completely correct the epidermal phenotype of 35S∷miR156a, provide convincing evidence that miR172 acts downstream of miR156, and mediates the effect of miR156 on epidermal identity.
We were intrigued by the possibility that SPL3, SPL4 and SPL5 might mediate the interaction between miR156 and miR172 because over-expression of these SPL genes produces a vegetative phenotype very similar to that of toe1 toe2 mutants or plants over-expressing miR172 (Wu and Poethig, 2006). Furthermore, all of these genotypes are early flowering under long days (Aukerman and Sakai, 2003; Cardon et al., 1997; Chen, 2004; Gandikota et al., 2007; Wu and Poethig, 2006). However, over-expression of SPL3, SPL4 or SPL5 had no effect on the abundance of miR172, indicating that these genes cannot be responsible for the effect of miR156 on miR172. An alternative possibility is that these SPL genes act downstream of TOE1 and TOE2. Although SPL3 transcripts are elevated in toe1 toe2, over-expressing TOE1 did not produce a corresponding decrease in SPL3 mRNA; furthermore, SPL4 and SPL5 were largely unaffected in toe1 toe2. Consequently, we suspect that the effect of toe1 toe2 on SPL3 expression is indirect. Our results are more consistent with the hypothesis that SPL3, SPL4, SPL5 regulate the same downstream targets as TOE1 and TOE2, but operate largely independently of these genes (Figure 6).
How does miR156 regulate the expression of miR172? Our results indicate that SPL9 is a direct transcriptional activator of miR172b, and probably acts redundantly in this process with SPL10 and several other SPL genes. The obvious candidates are SPL15--the closest homolog of SPL9 in Arabidopsis--and SPL11, the closest homolog of SPL10. Indeed, the observation that spl9 spl15 double mutants have slightly reduced levels of miR172b supports the conclusion that SPL15 cooperates with SPL9 in the regulation of this miRNA. SPL11 is adjacent to SPL10 in the genome and is nearly identical in sequence to SPL10, so it reasonable to assume that these genes have overlapping, if not identical, functions. This conclusion is supported by the observation that T-DNA insertions in these genes have no obvious phenotype (Wu and Poethig, unpublished observations). A rigorous test of the role of various SPL genes in the regulation of miR172b will require generating plants lacking combinations of SPL genes, and this is particularly difficult in the case of SPL10 and SPL1l because of their proximity.
Although SPL9 promotes the transcription of miR172b, this is not the only way by which it regulates epidermal identity. This conclusion is supported by the observation that spl9-4 produces a small but significant delay in abaxial trichome production without having an obvious effect on the abundance of miR172. That is, the effect of spl9-4 on epidermal identity cannot be explained by a decrease in the level of miR172. Moreover, pSPL9∷rSPL9 has a stronger effect on abaxial trichome production than 35S∷miR172. The simplest interpretation of these observations is that SPL9 has multiple targets involved in epidermal differentiation.
The genes that mediate the effects of SPL9 and SPL10 on leaf development are unknown. Many leaf shape mutations have been identified in Arabidopsis. However, to our knowledge, the only mutations that have major effects on phase-specific traits are mutations in genes required for the expression or function of miR156, miR172 and their direct targets. The morphological differences between juvenile and adult leaves are relatively subtle in Arabidopsis, so genes that act downstream of specific SPL genes or TOE1 and TOE2 may have been missed in mutant screens. It is also possible that phase-specific aspects of leaf morphology are regulated by genes that individually have only a small effect on these traits. Identifying these downstream genes is an important goal for future research.
There is growing evidence from animal systems that miRNA-regulated transcription factors frequently regulate the transcription of their cognate miRNAs (Fazi et al., 2005; Johnson et al., 2005; Kim et al., 2007; Varghese and Cohen, 2007). A genome-wide survey identified 23 such feedback loops in C. elegans (Martinez et al., 2008). The evidence that TOE1 and TOE2 positively regulate their repressor, miR172b, suggests that the expression of TOE1 and TOE2 may be modulated by a negative feedback loop involving miR172, and our data suggest that miR156 and its targets have a similar relationship. Interestingly, the miR172 target AP2 negatively regulates its own expression (Schwab et al., 2005), and may represent another example of this phenomenon. Negative feedback loops typically act to buffer small changes in the expression of proteins with important regulatory functions (Martinez et al., 2008). Negative feedback regulation is an attractive mechanism for stabilizing the expression of genes involved in vegetative phase change, and may be responsible for the remarkable stability of the juvenile and adult phases in some species (Hackett, 1985).
Finally, it is important to emphasize the potential significance of these results for understanding the mechanism of floral induction. Although it has long been known that floral induction depends on the transition to the adult vegetative phase (Hackett, 1985; Zimmerman et al., 1985) the relationship between vegetative phase change and floral induction is still unclear because many factors that affect flowering time (e.g. photoperiod, flowering time mutations) do not affect the timing of vegetative phase change, or have a relatively modest effect on this transition (Telfer et al., 1997). This raises the question of how these processes interact. Several of the targets of miR156 affect flowering time as well as vegetative phase change (Cardon et al., 1997; Schwarz et al., 2008; Wu and Poethig, 2006), and it is reasonable to propose that these genes act as licensing factors for the transition to flowering. If so, this will provide a solution to the long-standing question of how changes in the vegetative morphology of the shoot are coordinated with changes in its reproductive potential.
EXPERIMENTAL PROCEDURES
Genetic stocks and growth conditions
All of the genetic stocks used in this paper were in a Columbia background. 35S∷rSPL3, 35S∷miR156a, pSPL9∷rSPL9, pSPL10∷rSPL10 and pMIM156 have been described previously (Franco-Zorrilla et al., 2007; Wang et al., 2008; Wu and Poethig, 2006). Additional pSPL9∷rSPL9 and pSPL10∷rSPL10 lines generated in our laboratory were also used for some experiments. Milo Aukerman (DuPont) provided toe1–2 toe2-1. miR172a-1 (SALK_045787), spl9-4 (SAIL_150_B05) and spl15-1 (SALK_074426) were obtained from the Arabidopsis Biological Resource Center. Seeds were grown on Metromix 200 (Scotts) or Fafard #2 soil and left at 4°C for 2 days. Plant age was measured from the time seeds were transferred to the growth chamber. For phenotypic analysis, plants were grown in Conviron E7/2 chambers in short days (10h light:14h dark, 23°C), under a 3:1 combination of cool white (F032/841/Eco,Sylvania) and wide spectrum (Gro Lite WS, Interlectric Corp.) fluorescent lights, at light intensity of 300 µmol/m2/sec. Abaxial trichomes were scored 2–3 weeks after planting with a stereomicroscope. For leaf shape analysis, fully expanded leaves were removed, attached to cardboard with double-sided tape and flattened with transparent tape, and then scanned in a digital scanner. Rips in the leaf blade produced during this process were filled in using Photoshop.
Transgenic plants
The TOE1 and SPL9 coding sequence and a 1018 bp genomic sequence harboring the precursor of miR172b were PCR-amplified with pfu TURBO using cDNA (TOE1, SPL9) or genomic DNA (miR172b) as a template. The glucocorticoid receptor (GR) sequence was amplified from pBIΔGR and fused to a miR156-insensitive SPL9 cDNA. All of these constructs were cloned downstream of the CaMV 35S promoter in pEZR-CL. The intergenic regions containing the SPL9 or SPL10 promoter and open reading frame were PCR-amplified and mutations were introduced into the miR156 binding sequence. These sequences were cloned into pEG302 and pEG303 gateway vectors with an FLAG and cMyc epitope tag, respectively (Earley et al., 2006). The whole SPL9 intergenic region was also PCR-amplified and 3 copies of the FLAG epitope tag were fused to the N terminus of SPL9 protein. The fused sequence was then cloned into the SmaI and NcoI sites in pCambia3301. Plants were transformed using the floral dip method, and transformants were selected on Kanamycin or BASTA. Lines containing single insertions were selected on the basis of the segregation ratio of the resistant or susceptible plants to Kanamycin or BASTA in the progeny of these primary transformants, and homozygous stocks were established from these lines.
GR induction and RNA quantitation
35S∷GR-rSPL9 seeds were plated onto half strength MS medium containing 50 mg/L Kanamycin. Plates were moved to short days after 2 days a 4°C. On day 20, the plates were flooded with 0.1% ethanol (mock), 10 µM DEX in 0.1% ethanol, 10 µM CHX in 0.1% ethanol, and 10 µM DEX plus 10 µM CHX. After 4 hours, seedlings were harvested, frozen in liquid nitrogen, and stored at −80 °C.
RNA blots were processed as described previously (Wu and Poethig, 2006). Total RNA was isolated using Trizol (Invitrogen), purified with Qiagen RNeasy, and treated with RNase-free DNAase (Qiagen). qRT-PCR was performed using SuperScript™ II reverse transcriptase and Power SYBR Green PCR master mix (Applied Biosystems), and normalized using eIF4A as a standard. The primers used for qRT-PCR are described in Supplemental Table 1.
Chromatin Immunoprecipitation
pSPL9∷rSPL9-cMyc and pSPL9∷3XFLAG-rSPL9 transgenic seedlings were harvested in 1x PBS and cross-linked with 1% formaldehyde in 1x PBS for 12 minutes using vacuum infiltration. The cross-linking was stopped in 0.1M glycine. Nuclear extracts were prepared and immunoprecipitation was performed as described (William et al., 2004). After chromatin shearing, about 5-µl of anti-FLAG polyclonal anibody (Sigma, F7425) was added to the samples and incubated at 4 °C overnight. Beads were then washed and eluted with the lysis buffer (0.1M NaHCO3, 1%SDS). After reversing the cross-linking, DNA was purified using the QIAquick PCR purification Kit (Qiagen), and resuspended in 80 µl water. 3 µl of diluted DNA was used for real-time qPCR. The sequence of the primers used to amplify different regions in the promoter and coding sequence of miR172b are listed in Supplemental Table 1. PCR conditions were 42 cycles at 94°C for 10s, 57°C for 20s, and 72°C for 30s. Values for the FLAG ChIP samples were first normalized to the input and then were divided by the normalized cMyc signal to obtain a fold enrichment.
In situ hybridization
In situ hybridization was performed using a protocol obtained from Jeff Long (www.its.caltech.edu/~plantlab/protocols/insitu.pdf), with the following minor modifications: the probe was hybridized to slides at a temperature of 60–65°C overnight, and the blocking and antibody dilution solutions were produced using maleic acid instead of Tris-HCl. SPL3 and SPL9 probes were amplified and transcribed using the primers listed in Supplemental Table 1.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to members of the Poethig laboratory for comments on this manuscript and helpful discussions throughout the course of these experiments. We are also grateful to Milo Aukerman for supplying seeds of toe1 toe2. This research was supported by a grant from NIH (R01 GM051893).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. doi: 10.1016/j.tplants.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Bäurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Cardon GH, Hohmann S, Nettesheim K, Saedler H, Huijser P. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: a novel gene involved in the floral transition. Plant J. 1997;12:367–377. doi: 10.1046/j.1365-313x.1997.12020367.x. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007a;39:544–549. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- Chuck G, Hake S. Regulation of developmental transitions. Curr Opin Plant Biol. 2005;8:67–70. doi: 10.1016/j.pbi.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat Genet. 2007b;39:1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I. Unexpected silencing effects from T-DNA tags in Arabidopsis. Trends Plant Sci. 2008;13:4–6. doi: 10.1016/j.tplants.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Evans MM, Passas HJ, Poethig RS. Heterochronic effects of glossy15 mutations on epidermal cell identity in maize. Development. 1994;120:1971–1981. doi: 10.1242/dev.120.7.1971. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3' UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Ontogeny and Phylogeny. Harvard, MA: Belknap Press; 1977. [Google Scholar]
- Guo AY, Zhu QH, Gu X, Ge S, Yang J, Luo J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene. 2008;418:1–8. doi: 10.1016/j.gene.2008.03.016. [DOI] [PubMed] [Google Scholar]
- Hackett WP. Juvenility, maturation and rejuvenation in woody plants. Hort Rev. 1985;7:109–155. [Google Scholar]
- Hackett WP, Murray JR. Approaches to understanding maturation or phase change. In: Geneve RL, Preece JE, Merkle SA, editors. Biotechnology of Ornamental Plants. New York: CAB International; 1997. pp. 73–86. [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19:2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter RA, Poethig RS. The specification of leaf identity during shoot development. Annu Rev Cell Dev Biol. 1998;14:373–398. doi: 10.1146/annurev.cellbio.14.1.373. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter N, Kampani A, Carlson S, Goebel M, Moose SP. microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci U S A. 2005;102:9412–9417. doi: 10.1073/pnas.0503927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lloyd AM, Schena M, Walbot V, Davis RW. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Martinez NJ, Ow MC, Barrasa MI, Hammell M, Sequerra R, Doucette-Stamm L, Roth FP, Ambros VR, Walhout AJ. A C. elegans genome-scale microRNA network contains composite feedback motifs with high flux capacity. Genes Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH. Glossy15 controls the epidermal juvenile-to-adult phase transition in maize. Plant Cell. 1994;6:1343–1355. doi: 10.1105/tpc.6.10.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 1996;10:3018–3027. doi: 10.1101/gad.10.23.3018. [DOI] [PubMed] [Google Scholar]
- Moss EG. Heterochronic genes and the nature of developmental time. Curr Biol. 2007;17:R425–R434. doi: 10.1016/j.cub.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Parcy F. Flowering: a time for integration. Int J Dev Biol. 2005;49:585–593. doi: 10.1387/ijdb.041930fp. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Rougvie AE. Intrinsic and extrinsic regulators of developmental timing: from miRNAs to nutritional cues. Development. 2005;132:3787–3798. doi: 10.1242/dev.01972. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–527. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67:183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MR, Willmann MR, Wu G, Berardini TZ, Möller B, Weiers D, Poethig RS. Cyclophilin40 is required for miRNA activity in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106:5424–5429. doi: 10.1073/pnas.0812729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Tsukaya H, Shoda K, Kim GT, Uchimiya H. Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta. 2000;210:536–542. doi: 10.1007/s004250050042. [DOI] [PubMed] [Google Scholar]
- Usami T, Horiguchi G, Yano S, Tsukaya H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development. 2009;136:955–964. doi: 10.1242/dev.028613. [DOI] [PubMed] [Google Scholar]
- Varghese J, Cohen SM. microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev. 2007;21:2277–2282. doi: 10.1101/gad.439807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William DA, Su Y, Smith MR, Lu M, Baldwin DA, Wagner D. Genomic identification of direct target genes of LEAFY. Proc Natl Acad Sci U S A. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Poethig RS. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Kim Y, Dinh TT, Chen X. miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 2007;51:840–849. doi: 10.1111/j.1365-313X.2007.03181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman RH, Hackett WP, Pharis RP. Hormonal aspects of phase change and precocious flowering. Encycl Plant Physiol. 1985;11:79–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.