Abstract
Quantitative studies of protein abundance rarely span more than a small number of experimental conditions and replicates. In contrast, quantitative studies of transcript abundance often span hundreds of experimental conditions and replicates. This situation exists, in part, because extracting quantitative data from large proteomics datasets is significantly more difficult than reading quantitative data from a gene expression microarray. To address this problem, we introduce two algorithmic advances in the processing of quantitative proteomics data. First, we use space-partitioning data structures to handle the large size of these datasets. Second, we introduce techniques that combine graph-theoretic algorithms with space-partitioning data structures to collect relative protein abundance data across hundreds of experimental conditions and replicates. We validate these algorithmic techniques by analyzing several datasets and computing both internal and external measures of quantification accuracy. We demonstrate the scalability of these techniques by applying them to a large dataset that comprises a total of 472 experimental conditions and replicates.
Keywords: kd-tree, orthogonal range query, quantitative proteomics, space partitioning data structures, tandem mass spectrometry
One of the driving aims of studies that quantitatively measure gene expression across hundreds of experimental conditions and replicates is the identification of genes and pathways involved in disease. These are identified by a rich set of analytical techniques that include clustering, identification of differential expression, and functional network construction. Despite the many successes of these approaches, there exists a significant underlying problem with studies that measure only gene expression. The identification of pathways is limited by the fact that gene expression provides an incomplete readout of cellular physiology. Pathways involved in disease may show changes in overall protein abundance or in proportions of posttranslationally modified variants of these proteins, possibly without a transcriptional signature. Quantitative proteomics aims to address this problem by providing experimental tools to measure such changes (1–3). The primary measurement tool of quantitative proteomics is liquid chromatography followed by tandem mass spectrometry (LC-MS/MS).
Despite significant advances in LC-MS/MS instrumentation, quantitative proteomics studies have been limited to small numbers of experimental conditions and replicates. Whereas quantitative measurements are collected from gene expression microarrays by simply reading intensity values from an imaging device, extraction of quantitative measurements from LC-MS/MS datasets entails significant computational processing. This processing of the data presents several major challenges.
Background
LC-MS/MS Data.
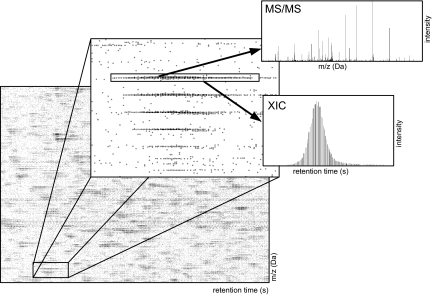
A quantitative proteomics experiment generates a dataset, called an LC-MS/MS scan, that consists of millions of data points that have three values: retention time (t), mass-to-charge ratio (m/z), and intensity (I) (see Fig. 1). In a typical experiment, the data are generated from a sample after protein denaturation, cysteine alkylation, and tryptic digestion of proteins into fragments. The tryptic peptides are separated by using liquid chromatography, ionized, and injected into a hybrid mass spectrometer. High-intensity precursor ions of these tryptic peptides are further fragmented, and the resulting fragments are measured a second time in the instrument (2).
Fig. 1.
A representative LC-MS/MS scan. A scan consists of millions of peaks, each with a specific retention time, m/z, and intensity. The intensity of a peak is indicated in grayscale (darker gray corresponds to higher intensity). The first Inset shows several extracted ion chromatograms (XICs) corresponding to the naturally occurring isotopes of a tryptic peptide. The area under an XIC provides a measure of the relative abundance of a peptide. In this example, one peak in the XIC has an MS/MS fragmentation spectrum from which the peptide's amino acid sequence and, hence, the identity of the protein from which it derives can be determined.
Computational Challenges.
The first computational challenge entails finding sets of data points that correspond to quantitative measurements of specific peptides. These patterns are known as extracted ion chromatograms (XICs) (4). Each XIC is a series of peaks that occur in a narrow m/z range over an interval of time. A typical scan contains many thousands of XICs, each corresponding to a different peptide from a complex mixture of proteins, along with additional data points due to various sources of noise. The area under an XIC measures the relative abundance of the corresponding tryptic peptide (see Fig. 1).
The second computational challenge entails determining the identity of the peptide associated with an XIC. Specifically, if one of the peaks in this series has a fragmentation spectrum, the spectrum can be used to search a database of predicted spectra based on the genome sequence of the species under study (5–7). From a high-scoring match to the database, one can determine the sequence of the tryptic fragment and the protein from which the fragment originated. We note that such database search is in itself a difficult computational challenge, one that is often addressed independently of quantification. We rely on existing algorithms for database search (5, 6).
The third computational challenge in quantification entails handling hundreds of LC-MS/MS scans that correspond to replicates and experimental conditions. To obtain relative protein abundance across the scans, XICs corresponding to the same peptides must be identified in each scan. Nonlinear variation in retention time of these XICs due to slight differences in chromatography, along with measurement error in m/z, complicates this process. A solution requires a multiscan alignment step to correct for these sources of variation. As the number of scans increases, it becomes significantly more difficult to assure correctness of the global alignment (8).
Previous Work.
Existing methods do not adequately address the computational challenges of XIC detection and multiscan alignment and are limited to a small number of scans. Several of these methods rely on image processing to collect quantitative data from LC-MS/MS scans (9–12). Because image processing typically requires computation on all individual pixels, these methods require a large amount of computation per LC-MS/MS scan. Furthermore, they require expensive image-correlation computations to nonlinearly align LC-MS/MS scans (13). Our algorithmic techniques avoid pixel-level image representations (see below).
To address the limitations of image-based methods, more recent approaches have relied on complex algorithms that process individual spectra one retention time point at a time and combine those results (14–16). We skip this individual scan-processing step, representing the data in two dimensions: retention time and m/z. Our algorithm is much simpler because it exploits structure in the data across both dimensions. Furthermore, existing methods rely on iterative optimization techniques to align scans. These iterative techniques become increasingly less reliable and computationally more expensive as problem dimensionality increases with the introduction of additional scans (8). In this work, we present a technique that is not susceptible to these dimensionality problems and demonstrably scales to hundreds of LC-MS/MS scans.
Results
Validation by Using Spiked-In Proteins.
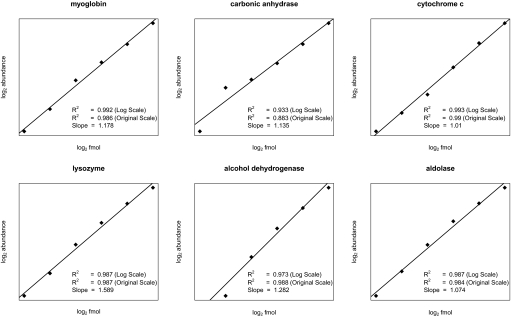
We tested whether our algorithm could accurately track the known abundance of several proteins across multiple LC-MS/MS scans by analyzing the dataset generated by Mueller et al. (16). The bulk human serum background was sufficiently complex to provide a challenge for our algorithm. Specifically, we observed 18,702 XIC groups that occurred in at least two of the three replicates of each dilution in three or more of the six dilution datasets. The large number of grouped XICs indicated that many different tryptic fragments were present, requiring our algorithm to accurately discern XICs from the spiked-in proteins. Next, we correlated the relative protein-abundance data computed by our algorithm from the LC-MS/MS data with the known concentrations of the spiked-in proteins. We observed nearly perfect correlations, with R2 values between 0.97 and 0.99 (see Fig. 2). These results indicate accurate quantification of the relative abundance of specific proteins in a complex mixture. The entire analysis required only 2 min and 4 s of time on a CPU with 4 cores, compared with the 105 min of time reported by Mueller et al. (16) for their method on a CPU with a single core. Accounting for differences in computer configurations, we estimate a processing time improvement of >12-fold.
Fig. 2.
Comparison of known protein concentration with relative abundance measured by mass spectrometry. Measured protein abundance is plotted on a log–log scale against known femtomole concentration for each of six nonhuman proteins spiked in to human serum by Mueller et al. The lines show best fit by regression, and the corresponding log–log scale slopes and correlation values (R2) for both the log–log scale and the original scale are shown below each plot.
Validation on a Large Dataset.
To test whether our algorithm could quantify large numbers of proteins across hundreds of experimental conditions, we analyzed the dataset generated by Foss et al. (8). This dataset comprises 472 LC-MS/MS scans of total yeast protein. Four hundred twenty eight of the scans represent two biological and two technical replicates for each of 107 progeny from a cross between two yeast strains.
Our algorithm detected 14,208 XICs that grouped across 40 or more progeny. 11,888 of these XIC groups had at least one associated MS/MS spectrum. For 3,692 XIC groups, a peptide was identified by database search (see Methods). These peptides represented 635 unique proteins. The ability to quantify 635 proteins across 40 or more progeny represents a ≈3-fold improvement in proteome coverage over the previous analysis of this dataset (see supporting information (SI) Fig. S1).
We next examined the quality of protein quantification. First, we observed a median correlation of 0.98 between technical replicates and 0.95 between biological replicates (see Fig. S2), showing that quantification was highly reproducible. Second, for nine proteins previously measured in the parent strains by quantitative Western blot analysis, the relative abundance was in agreement with the results computed by our algorithm from the LC-MS/MS data (see Fig. S2). Third, we took advantage of genotyping data for the progeny strains to carry out linkage analysis of protein abundance for the 635 proteins measured in at least 40 strains. Linkage analysis tests for significant differences in abundance between groups of strains divided by genotype at a given genetic marker. Errors in XIC detection and grouping would result in poor quantitative profiling of protein abundance, degrading our ability to detect such group differences. We detected linkage for 154 of the 635 proteins at a false-discovery rate of 5%, showing that measurement noise is small relative to real genetic effects. The fraction of proteins with significant linkages was ≈2-fold higher than that obtained at the same false-discovery rate in the previously published analysis, indicating improvement in both the accuracy of quantification and the number of segregants in which a typical protein is measured. The entire analysis of 472 scans required 27 min of CPU time and 13 GB of memory.
Isotope-Labeled Data Validation.
To test whether our algorithm could also quantify isotope-labeled data, in addition to the label-free data examined above, we analyzed an isotope-labeled dataset by Baek et al. (17). This dataset contained a complex mixture of labeled and unlabeled peptides. Our algorithm detected 119,140 paired XICs for which the light or the heavy XIC had fragmentation spectra. 29,062 of these paired XICs were assigned a peptide by database search. We grouped all independent measurements of peptides from a given protein and computed a median ratio between heavy and light forms for 1,816 individual proteins that had three or more measurements. One thousand six hundred two unique proteins were measured both by us and Baek et al., and we observed a Spearman's correlation of r2 = 0.69 between our protein abundance ratios and those reported by Baek et al. (see Fig. S3).
This correlation exceeded the Spearman's correlation of r2 = 0.59 observed in the Baek et al. study between technical replicates, indicating reasonably accurate quantification of relative protein abundance by both algorithms. We observed this high correlation despite two main sources of variation: (i) we used a different algorithm for MS/MS database search; (ii) we used our own algorithm for quantification. Processing isotope-labeled data was no slower than processing label-free data. The algorithm required 4 min and 14 s of time and 2.2 GB of memory to quantify 29,062 peptides on a CPU with four cores. For comparison, the quantification algorithm used by Baek et al. required 57 min on a similar isotope-labeled dataset to quantify 3,445 peptides by using a CPU with two cores (18). Accounting for differences in computer configurations, we estimate our approach is >56-fold faster per peptide quantified.
To assess the accuracy with which XICs were paired, we used a smaller, more manageable stable isotope-labeled dataset. The dataset was collected from an isotopically labeled sample containing only histone H3 from mouse embryonic fibroblasts (MEFs) and mouse embryonic stem cells (mESCs). It was hand-curated to quantify the extent of gene-activating or silencing marks in the mESCs, especially acetylated or methylated H3 histones. In this less-complex dataset, we used precision and recall as measures of XIC pairing accuracy. We observed a recall of 100%; that is, the algorithm found 22 of the 22 hand-curated D5 pairs. We observed a precision of 91.6%; that is, of the 24 pairs found by the algorithm, 22 were hand-curated pairs. The high precision and recall values indicated that XIC pairing was accurate. All of the hand-curated pairs that showed differences between MEFs and mESCs were also detected as different by our algorithm, indicating that the algorithm could detect quantitative differences in posttranslational modifications. As expected, gene-silencing marks such as H3K9me3 or H3K27me3 were depleted in mESCs, whereas gene-activating marks (H3K14ac) were enriched in the mESCs.
Discussion
We have devised and implemented algorithms for the analysis of large-scale proteomics data that differ from previous work in their ability to scale to hundreds of LC-MS/MS scans. The algorithm we present scales to very large datasets because of its reliance on a space-partitioning data structure that accelerates planar orthogonal range queries. Space-partitioning data structures and planar orthogonal range queries are well-studied ideas in computer science. They are of tremendous practical utility and are ubiquitous in computational geometry, computer graphics, and geographic information systems. Furthermore, there exists a significant body of work on practical aspects of their implementation, engineering, and use (19). Graph theoretic approaches, machine learning, signal processing, and statistics have made significant inroads into proteomics, and we expect the same of computational geometry techniques.
We note that the techniques presented here are not limited to measurements of protein-abundance data and posttranslational modifications, but they also apply to quantitative analysis of other biomolecules with mass spectrometry. We expect that they can be readily adapted to quantitative measurement of metabolite abundance and lipid abundance. Adapting these techniques to these other types of molecules will be the subject of future work. Combined with advances in experimental mass spectrometry techniques, we expect that the algorithms developed here will play a key role in providing a more complete picture of cellular physiology, thereby enabling sensitive and accurate identification of genes and pathways involved in disease.
Methods
Additional details are given in SI Text.
Data Structure.
Central to all of the algorithms below is a data structure that indexes points based on two of their dimensions: retention time (t) and m/z. The data structure must support the following interface: RangeQuery (D, t1, t2, m1, m2) uses the data structure D to conduct a planar orthogonal range query, returning objects (t, m/z) in a rectangular region defined by t1 < t < t2 and m1 < m/z < m2.
In computer science, there are many data structures that support this interface, each with its own space, speed, simplicity, and efficiency tradeoffs. Examples include kd-trees, range trees, quad-trees, binary space partitioning trees, and all of their many variants (19, 20). For this work, we use a kd-tree because it occupies linear space with respect to the number of data points and requires O() time to conduct a single range query, where N is the number of (t,m/z) objects (millions in a typical scan).
Ion Chromatogram Extraction.
The first step of processing an LC-MS/MS scan is to remove peaks that are caused by noise (see Fig. S4A). Given a kd-tree on all of the data points, the algorithm iterates through each peak and performs a planar orthogonal range query with specified width in retention time and height in m/z around the peak. If the number of peaks returned by the query exceeds a threshold, and the peak is above a nominal absolute intensity threshold, the peak is labeled as signal. Otherwise, the peak is labeled as noise. For the datasets analyzed here, we selected an absolute intensity threshold of 100, which we observed by visual inspection to remove only low-intensity noise peaks. After this process, the peaks labeled as noise are removed from further processing.
To find XICs, the algorithm uses planar orthogonal range queries and an undirected graph structure. First, the algorithm makes each peak a node in the graph. Second, the algorithm constructs a kd-tree on all of the signal points. Next, the algorithm iterates through each signal peak and connects the current node to any signal peak nodes returned in a query with specified width in retention time dimension and height in m/z. Last, the algorithm finds XICs by computing the connected components of the constructed graph. An individual connected component corresponds to an XIC (see Fig. S4B).
An XIC has a start retention time and an end retention time. Furthermore, an XIC may have one or more peaks with fragmentation spectra. These can be used to determine the sequence of the corresponding tryptic peptide. An XIC also has an m/z value that corresponds to the average m/z value of all of the peaks grouped into the XIC, and a retention time that corresponds to the retention time of the most-intense peak in the XIC. Because XICs have a center point, we also index them by using a kd-tree data structure.
Extracted Ion Chromatogram Alignment and Grouping.
Before grouping the XICs in multiple LC-MS/MS scans, the algorithm first aligns each scan to a reference scan via simple translation. This alignment compensates for any differences in when the samples began to elute out of the LC column. The algorithm iterates through each XIC in the current scan and finds the nearest XIC in the reference scan that is within a rectangular window with specified width in retention time and height in m/z. For this nearest XIC in the reference scan, the algorithm also computes the reciprocal nearest XIC in the current scan. If this reciprocal XIC is the same as the current XIC, the difference in retention time between the current XIC and the nearest reference XIC is stored in a list. After all of the XICs in the current scan are processed, the median of these differences in retention time is used to translate each XIC in the current scan to the reference scan.
Once each scan is aligned to the reference scan via translation, XICs are grouped across these scans. First, each XIC belonging to a scan is labeled by using the LC-MS/MS scan's identifier. Then, each XIC from that scan is combined into a larger set of labeled XICs from all scans (see Fig. S5A). Last, we use the same technique applied to find XICs from peaks to group XICs across scans. Each XIC starts out as a node in a graph. Planar orthogonal range queries between the start and end of an XIC in retention time and XIC width in m/z are used to connect XICs in a graph. Connected components in this graph correspond to grouped XICs (see Fig. S5B). The planar orthogonal range queries used in this step automatically compensate for any nonlinear differences in the positions of XICs. Furthermore, the range queries can be expanded far beyond the start and end of the XICs to compensate for more-severe differences in XIC retention time.
Collecting Relative Abundance Data.
Once these XICs are grouped across scans, relative abundance values are computed, and an amino acid sequence and protein identity is assigned to the group. Relative abundance values across scans are computed as the areas under each XIC in the group. These relative protein abundance values are normalized by using median of medians normalization to adjust for differences in overall scan intensity (21). Then, the entire XIC group is assigned an amino acid sequence for a particular tryptic peptide by database search. Database search is conducted by using all or a subset of fragmentation spectra within an XIC group.
Once the XIC group is assigned an amino acid sequence and protein identity, relative abundance data are available for a known tryptic peptide from a known protein. Because individual proteins are digested into several tryptic fragments, there may be many different XIC groups for a single protein, but in most applications, only the relative abundance of a protein is required. Because each tryptic fragment from a protein ionizes with varying efficiency, the quantitative values in each XIC group cannot be combined by simple averaging. Instead, a representative XIC group is selected for each protein. Specifically, we select the XIC group with highest signal-to-noise ratio. We note that other criteria, tailored to specific applications, can be used to select representative XIC groups.
Stable Isotope-Labeled Data Processing.
An alternative to label-free quantification is isotope-labeled quantification. In this experimental technique, one of two experimental samples is labeled with a heavier isotope tag (22). Because both the unlabeled and isotope-labeled tryptic fragments are subject to the same chromatographic conditions, they elute out of a liquid chromatography column at approximately the same retention time. Isotope-labeled quantification reduces to finding pairs of XICs within a single scan at the same retention time that are spaced according to the charge of the tryptic fragment and the weight difference of the heavier isotope tag. The ratio of the areas of these XICs measures the relative abundance of the corresponding tryptic fragment between the experimental samples.
Instead of aligning and grouping XICs across scans, the algorithm handles isotope-labeled data by grouping XICs within a scan. This is accomplished by reciprocal planar orthogonal range queries between XICs. XICs are processed in increasing m/z order, starting with XICs corresponding to the lighter variant of the peptide. XICs that have already been paired are skipped. Based on the charge of the XIC, the expected isotopic spacing of the heavier variant is computed. An orthogonal range query between the start and end of the current XIC and an m/z width creates a putative paired XIC. The longest of the returned XICs is used to conduct a reciprocal planar orthogonal range query between the start and end of the putative paired XIC. If the current XIC is returned by this reciprocal query, the two XICs are paired.
Fragmentation spectra assigned to the paired XICs are used to determine the amino acid sequence and protein identity associated with the pair. Because proteins are digested into several tryptic fragments, a dataset may contain several paired XICs per protein. Unlike label-free data, the abundance ratios computed by these pairs are comparable. The median ratio from all of these pairs can be used to compute a robust estimate of the abundance ratio for an individual protein.
Datasets.
We used four datasets to test and validate our algorithms. The first dataset is a spike-in dataset used in a previous study by Mueller et al. (16). The dataset contains six nonhuman proteins added at six different known concentrations to a background sample of bulk human serum (see Table S1). Three replicates of each dilution were collected by using an FT-LTQ ThermoElectron mass spectrometer and an Agilent 1100 chromatographic separation system. In total, this dataset consists of 18 LC-MS/MS scans and is 15 GB in size. We downloaded the data from http://prottools.ethz.ch/muellelu/web/Latin_Square_Data.php.
The second dataset, described by Foss et al. (8), measured total unfractionated cellular proteins from 107 genotyped segregants from a cross between two parental strains of yeast (BY4716 and RM11–1a). Four replicate LC-MS/MS scans were carried out for each segregant. The data also include 10 replicates each of parent strain and 2 replicates of each of six gas-phase fractions from each parent strain. In total, this dataset includes 472 LC-MS/MS scans and is 42 GB in size. The dataset was generated by using a ThermoElectron Corp. LTQ-FT mass spectrometer and a Michrom Bioresources Paradigm MS4B MDLC nanoflow liquid chromatography system. We obtained the data from the authors.
The third dataset, used to test performance on isotope-labeled data, derives from a study by Baek et al. (17) on the effect of microRNAs on protein levels. In this dataset, the unlabeled sample originated from the cytosolic fraction of mouse cultured cells. The isotope-labeled sample originated from the cytosolic fraction of mouse cultured cells with a miR-223 knockout. The dataset was generated with a ThermoElectron LTQ-OrbitrapXL and consists of 16 LC-MS/MS scans from 16 gel fractions. The sample was labeled with Arg-6 and Lys-6. We obtained the data from the authors.
The fourth dataset used a deuterated isotope label (5-Da shift) (23). To generate these data, samples of histone H3 were extracted from MEF and mESC. The MEF sample was derivatized with propionic anhydride (D0, 56 Da), whereas the mESC sample was derivatized with an isotopically labeled propionic anhydride (D5, 61 Da). The data were collected with a ThermoElectron LTQ-Orbitrap and consisted of single LC-MS/MS scan. This dataset was generated as part of this study.
Protein Identification.
Amino acid sequence and protein identity were assigned by using the X! Tandem database search algorithm (version 08-02-01-3 (6), e-value <0.1). For the Foss et al. data, we additionally used the OMMSA algorithm (version 2.1.1 (5), e-value <0.1).
CPU and Availability.
Algorithm timing and peak memory usage was collected on a machine with two dual-core AMD Opteron 2220 2.8 Ghz Processors and 32 GB of RAM (four CPU cores total). All of the algorithms described in these methods were implemented in the Princeton LC-MS/MS Data Viewer. Complete source code is available at http://compbio.cs.princeton.edu/pview.
Supplementary Material
Footnotes
Conflict of interest statement: A patent for related technology has been filed with the U.S. Patent and Trademark office by Princeton University. Z.K., M.S., and L.K. are coinventors.
This article is a PNAS Direct Submission. A.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904100106/DCSupplemental.
References
- 1.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 4.Ong S, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 5.Geer LY, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 6.Craig R, Beavis RC. A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 7.Eng JK, McCormack AL, Yates JR. An approach to correlate MS/MS data to amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 8.Foss E, et al. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39:1369–1375. doi: 10.1038/ng.2007.22. [DOI] [PubMed] [Google Scholar]
- 9.Jaffe JD, et al. PEPPeR, a platform for experimental proteomic pattern recognition. Mol Cell Proteomics. 2006;5:1927–1941. doi: 10.1074/mcp.M600222-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kohlbacher O, et al. TOPP—The OpenMS proteomics pipeline. Bioinformatics. 2007;23:191–197. doi: 10.1093/bioinformatics/btl299. [DOI] [PubMed] [Google Scholar]
- 11.Bellew M, et al. A suite of algorithms for the comprehensive analysis of complex protein mixtures using high-resolution LC-MS. Bioinformatics. 2006;22:1902–1909. doi: 10.1093/bioinformatics/btl276. [DOI] [PubMed] [Google Scholar]
- 12.Palagi PM, et al. MSight: An image analysis software for liquid chromatography–mass spectrometry. Proteomics. 2005;5:2381–2384. doi: 10.1002/pmic.200401244. [DOI] [PubMed] [Google Scholar]
- 13.Listgarten J, Emili A. Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry. Mol Cell Proteomics. 2005;4:419–434. doi: 10.1074/mcp.R500005-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Park SK, Venable JD, Xu T, Yates JR. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finney GL, et al. Label-free comparative analysis of proteomics mixtures using chromatographic alignment of high-resolution μLC-MS data. Anal Chem. 2008;80:961–971. doi: 10.1021/ac701649e. [DOI] [PubMed] [Google Scholar]
- 16.Mueller LN, et al. SuperHirn—A novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics. 2007;7:3470–3480. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 17.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakalarski CE, et al. The impact of peptide abundance and dynamic range on stable-isotope-based quantitative proteomic analyses. J Proteome Res. 2008;7:4756–4765. doi: 10.1021/pr800333e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samet H. Foundations of Multidimensional and Metric Data Structures. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 20.Bentley JL. Multidimensional binary search trees used for associative searching. Commun ACM. 1975;18:509–517. [Google Scholar]
- 21.Callister SJ, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res. 2006;5:277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 23.Gazzar ME, et al. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29:1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.