Abstract
Genome-wide association studies (GWAS) have been widely used to identify genetic effects on complex diseases or traits. Most currently used methods are based on separate single-nucleotide polymorphism (SNP) analyses. Because this approach requires correction for multiple testing to avoid excessive false positive results, it suffers from reduced power to detect weak genetic effects under limited sample size. To increase the power to detect multiple weak genetic factors and reduce false positive results caused by multiple tests and dependence among test statistics, a modified forward multiple regression (MFMR) approach is proposed. Simulation studies show that MFMR has higher power than the Bonferroni and false discovery rate (FDR) procedures for detecting moderate and weak genetic effects, and MFMR retains an acceptable false positive rate even if causal SNPs are correlated with many SNPs due to population stratification or other unknown reasons.
Keywords: GWAS, SNP, MFMR, separate SNP analysis, multiple regression analysis
INTRODUCTION
With the development of genotyping and computer technology, genome-wide association studies (GWAS) have become feasible because currently available genotyping platforms for association mapping can accommodate up to 1 million single-nucleotide polymorphisms (SNPs), with a density of genotyping average 1 SNP per 3 kb, across the whole genome. However, large numbers of false positive results become a major challenge in genome-wide association studies due to multiple testing and correlations among SNPs located in different genomic regions. In GWAS for a 500K SNP data, if one controls Type I error at 5% in single SNP tests, there will still be about 25,000 positive SNPs even if none of the SNPs are truly associated with disease or trait. In addition, if there are some causal SNPs affecting the disease or trait of interest, and if these SNPs are correlated with other SNPs not in the same linkage disequilibrium (LD) region because of population stratification or other unknown reasons, we may observe additional false-positive SNPs (here we define the positive SNPs in the same LD region of a causal SNP as true positives, and the positive SNPs not in the same LD region of a causal SNP as false positives). So, identifying which, if any, of these results is a true positive versus the many expected false positives presents a major statistical problem to localize the true genes that relate to the complex disease or trait under study. To solve this problem, a Bonferroni correction based on the significance level of an individual test divided by the number of total tests has been used to control the Family-Wise Error Rate (FWER). Alternatively, Bonferroni Step-Down [Holm, 1979], Westfall and Young Permutation [1993], Benjamini and Hochberg’s false discovery rate (FDR) correction [1995], a Monte Carlo approach [D.Y. Lin, 2005], two-stage GWAS design [Satagopan and Elston, 2003], principal components approach [Price at al, 2006] and many other methods have been proposed to control for multiple tests in order to yield an acceptable FWER or FDR. Some of these methods such as principle components analysis also allow for population stratification.
Among the above mentioned methods, the Bonferroni correction is the most stringent approach. Practical application shows it is adequate for controlling FWER and detecting relatively strong genetic effects, but it is too stringent to detect weak or moderate genetic effects. FDR (defined as the expected proportion of false rejections among the total number of rejected hypotheses) correction tolerates more false positives than many other methods and has thus been found to yield more power to detect weak genetic effects than other approaches. However, when the number of tests is very large, such as in GWAS, the FDR approach still lacks power to find weak genetic effects. For example, in the study of genome-wide association for lung cancer at 15q25.1 [Amos et al, 2008], the risk allele of SNP rs1051730 conveyed an odds ratio 1.31 versus the wild allele in Chi-square test, and gave a generalized R2 = 0.009 in logistic regression [Nagelkerke, 1991], showing a rather weak effect of this SNP in influencing lung cancer risk. Even though 2291 individuals (1154 cases and 1137 controls) were studied, the p-value was only 1.14 × 10−5 from Chi-square test, and 5.38 × 10−5 from logistic regression. Because there were 315,824 SNPs, a test employing the Bonferroni correction did not have enough power to detect this risk SNP, and the FDR procedure (q = 0.05) also failed to detect this weak genetic factor.
In general, for a genome-wide screen to detect multiple genetic factors for a complex trait, methods based on separate SNP analyses have required criteria that are either too strict to detect weak genetic effects or yield too many false positive results if the criteria are relaxed to α > 10−4. Therefore, the conventional methods based on separate SNP analysis including the Bonferroni and FDR methods may not effectively control both type I error and provide adequate power unless a large enough sample size is used. To increase the power to detect weak genetic effects and at the same time control the number of false positives under limited sample size, we propose a modified forward multiple regression (MFMR) approach based on maximum order statistics.
METHODS
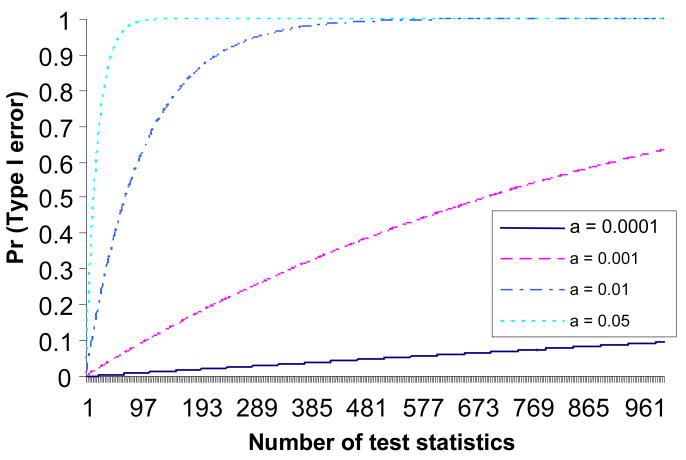
Let X1, X2, …, Xn be independent random variables with respective cumulative distribution function Fi(x) (i = 1, 2, …, n), and let Yn = X(n) be the maximum order statistic of X1, X2, …, Xn. Then Yn has cumulative distribution function Gn(yn) = P(Yn ≤yn) = P(Xi ≤ yn; i = 1, 2, …, n) = . Considering a GWAS using general linear regression, let Xi = MSR/MSE be the mean square for regression divided by mean square for error for the ith SNP (i = 1, 2, …, n). Let us treat Xi (i = 1, 2, …, n) independently by assuming each SNP is from a different LD block and no population stratification or population substructure has been controlled in genome data. The null hypotheses indicate no additive mean trait effect among the genotypes (AA, AB and BB) of the ith SNP. The alternative hypotheses indicate that there is additive mean trait effect among the genotypes of the ith SNP. If null hypotheses (i = 1, 2, …, n) are true, Xi (i = 1, 2, …, n) are independently and identically distributed from the same F distribution F(X; 1, N−2) where N is sample size. If the alternative hypotheses (i = 1, 2, …, n) are true then Xi (i = 1, 2, …, n) are independent and follow non-central F distributions Fi(X; 1, N−2,λi) [Hocking, 2003]. In approaches of separate SNP analyses, we set a critical value c0 such that P(Xi > c0 | ; i = 1, 2, …, n) = α, where α is the probability of Type I error in each test, and the corresponding power for each test is πi = P(Xi > c0 | ). Now we use the same critical value c0 for the maximum order statistic Yn, and let be the joint null hypothesis that there is no true positive SNP in whole genome, then the probability of Type I error for Yn is P(Yn > c0 | ) = 1 − P(Yn ≤ c0 | ) = 1 − P(Xi ≤ c0 | ; i = 1, 2, …, n) = 1 −[P(X ≤ c0 | H0)]n = 1 − [1 − α]n ≈ nα, which is just the FWER in an approach of separate SNP analysis assuming all tests are independent. When we use Bonferroni approach, the α is about 0.05/n if we control FWER at 0.05 level, which is 5.0×10−7 for a GWAS with 100,000 SNPs. From the above formula, the Type I error rate of a maximum order statistic tends to 1 when α > 0 and n increases. Supplementary Figure 1 shows the Type I error rate of a maximum order statistic for α = 0.0001, 0.001, 0.01 and 0.05.
Let the joint alternative hypothesis for the maximum order statistic Yn be = {the SNP with the maximum order statistic has additive genotype effect and there is at least one SNP with additive genotype effect in genome.}, then the power for Yn is P(Yn> c0 | ) = 1 − P(Yn ≤c0 | ) = 1 − P(Xi ≤ c0 | ; i = 1, 2, …, n). Assume the Jth test statistic XJ is the largest statistic among X1, X2, …, Xn. Because the alternative hypothesis for the maximum order statistic includes the alternative hypothesis for the Jth test statistic , and the hypothesis implies , they are actually equivalent. Also because the hypothesis for Jth SNP has nothing to do with other SNPs by independent assumption, we have P(Xi ≤ c0 | ) = P(Xi ≤ c0) for i ≠ J, and P(Yn>c0 | ) = 1 − P(Xi ≤ c0 | ; i = 1, 2, …, n) = 1 − P(Xi ≤ c0 | ; i = 1, 2, …, n) = , where P(XJ ≤ c0 | ) is the probability of Type II error for the largest test statistic in separate SNP test approach. In the separate SNP test approach, because we do not have (or do not use) the condition that XJ is the maximum order statistic, XJ follows univariate distribution.
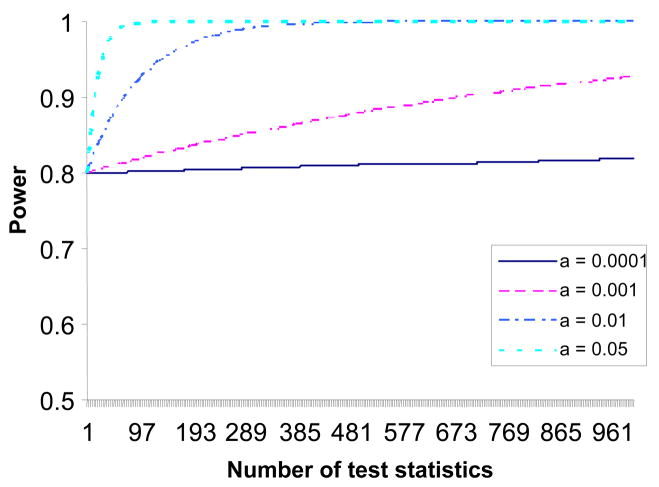
In hypothesis , if there are other m true positive SNPs, then we have (actually if m ≠ 0 and n > m + 1). For example, Assuming the power in an approach of separate SNP analyses for the Jth SNP is 80% at given effect size, allele frequency, α level, and sample size, then the power for the same SNP given the SNP has the maximum order statistic is P(Yn > c0 | ) ≥ 1 − [1−0.8]×[1−α]n−1, which is always greater than the power from separate SNP analyses. Supplementary Figure 2 shows the minimum power of a maximum order statistic for α = 0.0001, 0.001, 0.01 and 0.05 assuming 80% power from the same test statistic in separate SNP test approach.
The above formulas and Figures 1 and 2 suggest that tests based on maximum order statistics have more power than the tests based on separate SNP analyses. When n is large, the power from the maximum order statistic tends to 1. When n decreases, the power is still higher than the power from separate SNP analysis, especially when there are multiple true positive SNPs because the power mainly depends on the term when n is not large. Although a test based on maximum order statistic increases the probability of Type I error, the Type I error rate will reduce quickly when n decreases. Based on these characteristics of Yn, at the first step of forward multiple regression, we pick the SNP with the maximum order statistic in the regression model if its p-value is less than a pre-specified α level, then we chose a number c1 such that P(X > c1 | H0) = 0.05 and drop all SNPs with observed test statistics xi ≤ c1 (i = 1, 2, …, n). We only retain SNPs with the largest 5% of order statistics for the next step of forward multiple regression. Repeating this procedure in the following steps of forward multiple regression, the remaining number of SNPs, ni, at ith forward regression will reduce quickly, so the Type I error rate of the maximum order statistic at the ith forward regression will tend to α quickly. The forward multiple regressions will stop when ni becomes small, because no H0 will be rejected at a small α level if null hypothesis is true. By using this approach, the power to detect weak genetic effect will increase and the number of tests will decrease greatly, so that we can use a less stringent α level to increase power without increasing the number of false positives due to a large number of tests. For example, in the study of genome-wide association for lung cancer by Amos et al in 2008, when we used MFMR and took α = 0.0001, we detected rs1051730 and 26 other positive SNPs out of total 315,824 SNPs. Because the properties of the maximum order statistic do not limit the distributions of the test statistics, here we have extended MFMR from general linear regression to logistic regression where the test statistics follow χ2 distributions. This approach can also be extended to a GWAS using any other test statistics.
Figure 1.
Type I error rate of the maximum order statistic.
Figure 2.
Minimum power of the maximum order statistic assuming 80% power for the same test statistic in single SNP test approach.
SIMULATION STUDIES
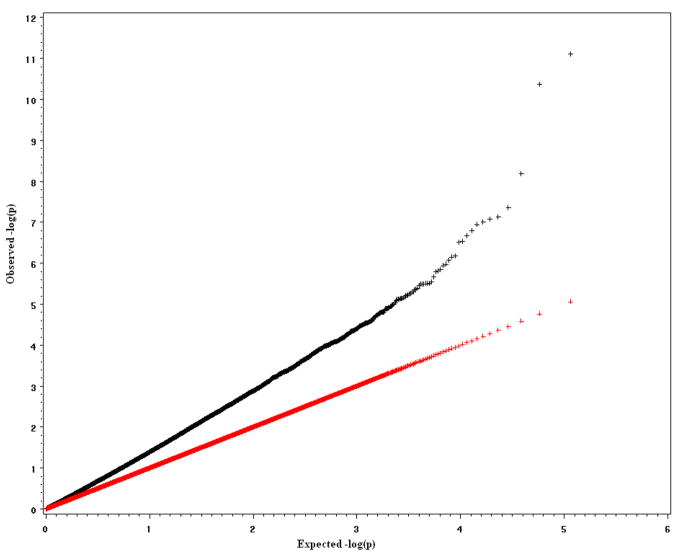
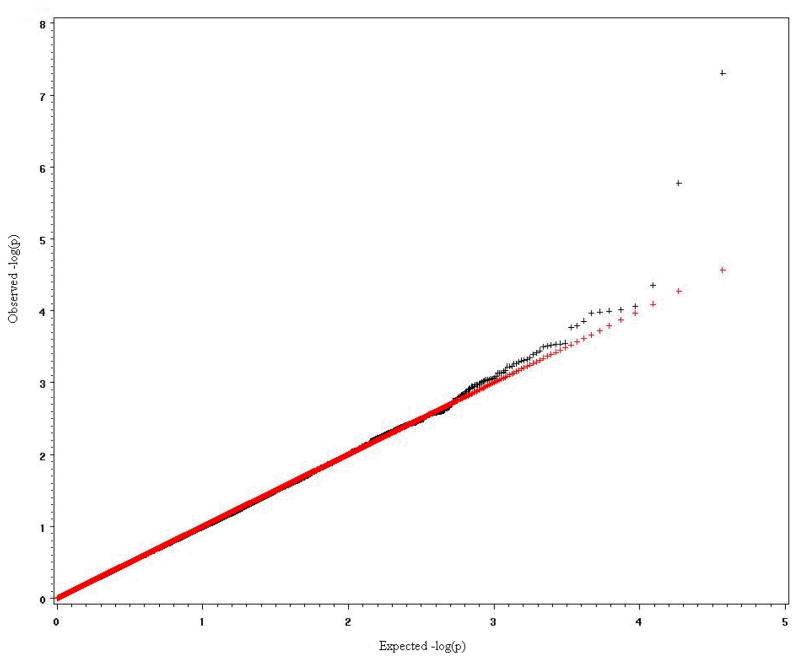
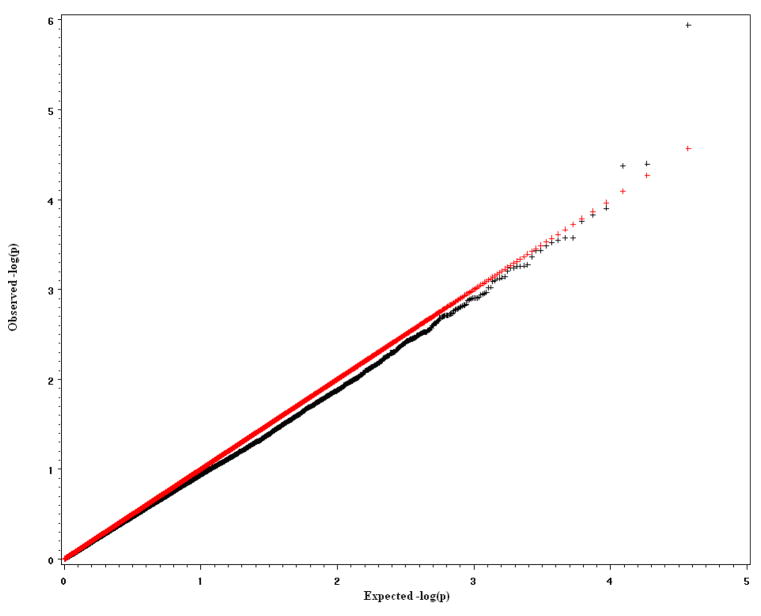
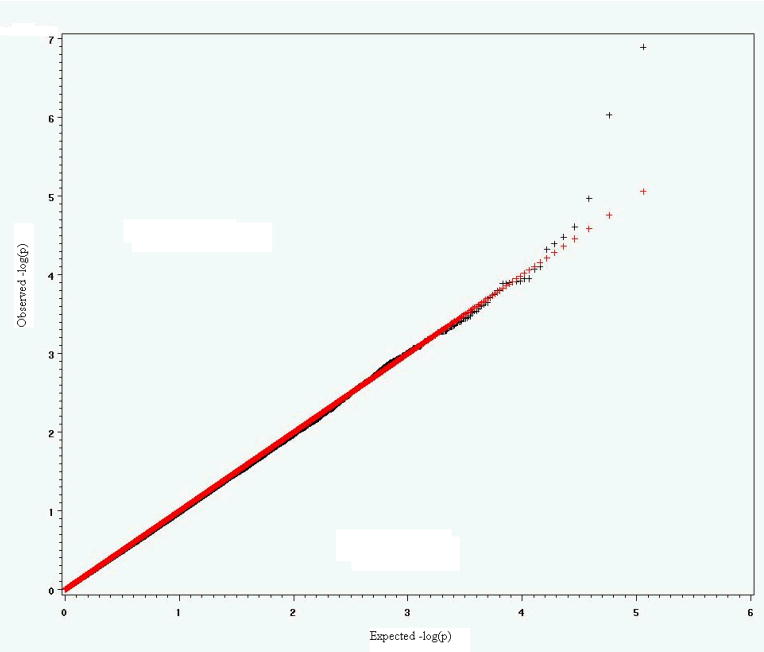
To verify the effectiveness of the MFMR empirically, we conducted several simulation studies to compare power and type I error for the proposed method, the Bonferroni procedure and the more powerful FDR method. The first and second simulation studies are based on data from genotyping that has been performed using Illumina bead arrays. There were 6922 Caucasian samples from Canada and North America, which included 2755 males and 4167 females. The data were extracted from an Illumina Infinium Human Hap300 SNP arrays [Plenge et al., 2007]. All SNPs in the extracted data set have p-value greater than 0.0001 for Hardy-Weinberg Equilibrium (HWE) test, minor allele frequency (MAF) greater than 0.01 and genotyping rate greater than 0.999. To generate 100 replicates, 500 subjects were randomly selected without replacement from the original 6922 samples to form a dataset until we obtained 100 datasets. For each dataset, a complex trait for each subject was simulated based on several artificial causal SNPs. The causal SNPs were chosen from different chromosomes and were uncorrelated (The maximum r2 among them is 4.8×10−4). In simulation study 1, we simulated a quantitative complex trait with three causal SNP effects. In simulation study 2, we simulated a complex dichotomous disease trait with three different causal SNPs. The minor allele frequencies of these causal SNPs varied from 9.5% to 27.7%. The genetic effects of the causal SNPs were additive, and contributed an average 1% to 5% of total trait variance in 100 replicates. The population is homogeneous in simulation studies 1 and 2. The inflation factors (the observed mean of test statistics divided by the expected mean of test statistics under H0 [Devlin et al, 2001]) are close to 1 (0.996 in simulation study 1 and 0.934 in simulation study 2), which indicates no “overdispersion” was generated by population substructure. Supplementary figures 3 and 4 show the q-q plots of the expected negative log p-values of test statistics versus observed negative log p-values of test statistics. The third and fourth simulation studies are based on a 115K Affymetrix SNP data set that was collected by St. Jude Children’s Research Hospital from the germline DNA of 400 leukemia patients. There are 294 (73.5%) Caucasians, 69 (17.25%) Blacks, 13 (3.25%), Hispanics, and 24 (6.0%) others. Race was self-identified and consistent with genotype checking. 178 females and 222 males were distributed among the race groups and no significant gender difference was found in each race group. The data include 115,182 SNPs that were derived from analysis of an Affymetrix SNP array [Matsuzaki et al., 2004]. The BRLMM method was used to assign genotypes [Rabee and Speed, 2006]. The overall SNP call rate is more than 98%. There is a strong population stratification issue in simulation study 3 where the selected causal SNPs are highly associated with race. The inflation factor in this study is λ = 1.4. Supplementary Figure 5 shows the q-q plot of the expected negative log p-values of F statistics versus observed negative log p-values of F statistics. The purpose of simulation study 3 is to compare the influences of population stratification on the Bonferroni, FDR and MFMR tests. In simulation study 4, three selected causal SNPs are not correlated with race. The goal of the simulation study 4 is to check whether the inflation of false positives still exists when causal SNPs are not correlated with population substructure and whether MFMR still performs better than Bonferroni and FDR approaches in that situation.
Simulation study 1
In the first simulation study, we picked one SNP each from chromosomes 1, 5, and 13. The additive genetic effects −a, 0 and a are assigned to genotypes AA, AB and BB where AA is the common genotype. The parameter a is determined by the additive variance component σa2 = 2pqa2 where p and q are frequencies of alleles A and B [Hartl and Clark, 1997]. The quantitative trait is Y = 10 + 0.24X1 + 0.41X2 + 0.46X3 + ε, where X1, X2 and X3 are indicator variables for SNP1, SNP2 and SNP3 respectively, and each is coded -1, 0 and 1 for genotype AA, AB and BB. SNP1 contributes 5%, SNP2 contributes 3%, and SNP3 contributes 1% to the total trait variance. The normal residual e has mean 0 and contributes 91% of total trait variance. In 100 replicates, we use MFMR approach to obtain a multiple regression model for each replicate. The power calculated for detecting each causal SNP is the number of times detected in 100 replicates dividing by 100. If the causal SNP is not found but a SNP in the same LD region (with |r| > 0.8) of the causal SNP is detected, we count that SNP as a causal SNP (Table 1). The average number of false positives (ANFP) in each replicate is calculated by dividing the number of total false positive SNPs found in 100 replicates by 100. We also applied the Bonferroni and FDR procedures to the results of single SNP regression to estimate their power and the ANFP in each replicate.
Table 1.
Power and average number of false positives in each replicate.
| Method | Power Sample size = 500 |
ANFP | Total SNPs | ||
|---|---|---|---|---|---|
| SNP 1 | SNP 2 | SNP 3 | |||
| VC = 5% | VC = 3% | VC = 1% | |||
| Bonferroni (α=1.35×10−6) | 49% | 9% | 1% | 0.05 | 36,990 |
| FDR (q = 0.05) | 50% | 11% | 2% | 0.07 | 36,990 |
| FDR (q = 0.6) | 79% | 35% | 7% | 5.36 | 36,990 |
| MFMR (α = 10−4) | 81% | 41% | 5% | 5.41 | 36,990 |
| MFMR (α = 0.05) | 83% | 48% | 10% | 53.1 | 36,990 |
| MFMR (α = 1.35×10−6) | 50% | 10% | 1% | 0.06 | 36,990 |
Table 1 shows that Bonferroni and FDR (q = 0.05) approaches are too stringent to find weak genetic effects unless we reduce the significance criteria (such as increasing the q value in the FDR approach from 0.05 to 0.6 as shown in the Table 1) or increase the sample size. Although the Bonferroni and FDR (q = 0.05) approaches maintained FWER at 5% level (those 5 false positive SNPs were found from 5 different replicates) because of the relatively independent multiple testing in this simulation study, it is not necessary to have so stringent FWER in GWAS. MFMR has better power to detect weak genetic effects, and the number of false positives are acceptable when we take α = 10−4. Even if we take α = 0.05, the ANFP is still much less than that using separate SNP analysis, where about 1800 false positives are expected for α = 0.05. When we used the same significance criterion as Bonferroni procedure for MFMR, we obtained similar powers and false positive rates as found with the Bonferroni approach.
Simulation study 2
In the second simulation study, we picked three SNPs from chromosome 2, 4 and 8 separately. SNP1 is assigned additive effect −1, 0 and 1 to genotype AA, AB and BB. SNP2 and SNP3 are assigned additively −0.7, 0, 0.7 and −0.3, 0, 0.3 for genotype AA, AB and BB. Let the probability of complex disease for ith subject follow a logistic function pi = 1 − 1/[1 + exp(1 + X1i + 0.7X2i + 0.3X3i)], where Xki is the genotype coding (−1, 0 or 1 for genotype AA, AB or BB) for SNPk (k = 1, 2, 3) from ith subject. For each subject, a random variable u is generated from uniform distribution U(0, 1). If u < pi we treat the subject as a patient, otherwise we treat the subject as a healthy person. Because the probability of u < pi is pi, the ith subject has probability of pi to get disease. In simulation study 2, the average odds ratios of additive effects in 100 replicates are 2.6, 2.02 and 1.38 for SNP1, SNP2 and SNP3 respectively, and the average generalized r-squares in logistic regressions are 0.036, 0.025 and 0.01. The powers and ANFP from Bonferroni, FDR and MFMR approaches in 100 replicates are listed in Table 2.
Table 2.
Power and average number of false positives to detect qualitative trait loci
| Method | Power Sample size = 500 |
ANFP | Total SNPs | ||
|---|---|---|---|---|---|
| SNP1 R2 = 0.036 | SNP2 R2 = 0.025 | SNP3 R2 = 0.01 | |||
| Bonferroni (α = 1.35×10−6) | 18% | 1% | 1% | 0.01 | 36,990 |
| FDR (q = 0.05) | 18% | 3% | 1% | 0.05 | 36,990 |
| FDR (q = 0.6) | 30% | 11% | 3% | 0.73 | 36,990 |
| MFMR (α = 10−4) | 46% | 13% | 3% | 2.32 | 36,990 |
| MFMR (α = 0.05) | 52% | 29% | 7% | 29.9 | 36,990 |
| MFMR (α = 1.35×10−6) | 19% | 1% | 1% | 0.00 | 36,990 |
Simulation study 2 for dichotomous traits shows consistent results with the simulation study 1 for continuous trait: if a genetic effect contributes less than 3% of total trait variation, Bonferroni and the commonly used FDR (q = 0.05) approaches have very little power to find the effect in a sample size of 500. We may use a less stringent criterion in FDR approach, such as q = 0.6, to obtain a close power as in MFMR, but which criterion should be adopted is very subjective and depends on how many positives are allowed. MFMR may use a less stringent criterion and has showed acceptable number of positives (even if α = 0.05), so it has more power to detect weak genetic effects.
Simulation study 3
In the third simulation study, three SNPs were selected from chromosomes 2, 15, and 19 separately. Each of them is significantly correlated with race to create a population stratification issue. The three causal SNPs are uncorrelated (the maximum r2 is 0.006). The quantitative trait Y is simulated by a model Y = 80 + SNP1*N(3, 4) + SNP2*N(1.4, 1) + SNP3*N(2, 1) + N(0, 4), where SNP1 (on chromosome 2 with MAF 0.10) contributes average 10.4% of total trait variation in 100 replicates and is coded additively 0, 0.5, and 1 for genotype AA, AB and BB separately; SNP2 (on chromosome 19 with MAF 0.31) contributes average 5.2% of total trait variation and is coded recessively 0, 0, and 1 for genotype AA, AB and BB; SNP3 (on chromosome 15 with MFA 0.21) contributes average 5.6% of total trait variation and is coded additively 0, 0.5, and 1 for genotype AA, AB and BB; N(3, 4) is a normal random variable with mean 3 and variance 4, et cetera. In simulation study 3, three causal SNPs are significantly correlated with 8,093 SNPs (|r| > 0.2, p < 0.0001) in the genome data. 8,078 of them are not in the same LD regions of the three causal SNPs. 6,008 of them are correlated with SNP1. The results from Bonferroni, FDR and MFMR procedures are listed in Table 3.
Table 3.
Power and ANFP under the condition that causal SNPs are correlated with thousands of SNPs in the data
| Method | Power Sample size = 400 |
ANFP | Total SNPs | ||
|---|---|---|---|---|---|
| SNP 1 R2=10.4% | SNP 2 R2=5.2% | SNP 3 R2=5.6% | |||
| Bonferroni(α = 4.3×10−7) | 92% | 21% | 21% | 31.74 | 115,182 |
| FDR (q=0.05) | 97% | 66% | 76% | 2075 | 115,182 |
| FDR (q = 0.001) | 75% | 20% | 21% | 57 | 115,182 |
| MFMR (α = 10−4) | 85% | 33% | 33% | 13.25 | 115,182 |
| MFMR (α = 0.05) | 88% | 37% | 34% | 48 | 115,182 |
Simulation study 3 shows that the ANFP from MFMR is not influenced by population stratification. Once the causal SNP that is correlated with population structure is selected into regression model, population stratification has been controlled through the causal SNP. Therefore, false positive SNPs caused by correlation with the causal SNP in univariate analysis will no longer be significant. Simulation study 3 also indicates that if there is population stratification in genome data and causal SNPs are correlated with population structure, separate SNP analyses such as Bonferroni and FDR approaches cannot control FWER at a theoretical level especially for FDR approach. In this case, the number of positives (both power and Type I error) will be inflated as shown in Table 3 and Figure 5. To deal with this problem, people do analysis stratified by population structure or controlling for population stratification [Price et al, 2006], but this may reduce power to detect causal SNPs. Table 4 lists the results after controlling for race in the same simulation study.
Figure 5.
q-q plot of expected negative log p-value vs observed negative log p-value in simulation study 3.
Table 4.
Power and ANFP under the condition of controlling for race
| Method | Power Sample size = 400 |
ANFP | Total SNPs | ||
|---|---|---|---|---|---|
| SNP 1 R2=10.4% | SNP 2 R2=5.2% | SNP 3 R2=5.6% | |||
| Bonferroni (α = 4.3×10−7) | 45% | 5% | 7% | 0.94 | 115,182 |
| FDR (q = 0.05) | 50% | 10% | 9% | 7.3 | 115,182 |
| FDR (q = 0.15) | 69% | 19% | 13% | 20.49 | 115,182 |
| MFMR (α = 10−4) | 77% | 31% | 26% | 18.2 | 115,182 |
| MFMR (α = 0.05) | 78% | 32% | 28% | 51.2 | 115,182 |
| MFMR (α = 4.3×10−7) | 50% | 6% | 8% | 0.13 | 115,182 |
After controlling for race, the inflation factor is close to 1 (1.02). Although the ANFP from Bonferroni and FDR are well controlled, the power from Bonferroni and FDR approaches are significantly reduced. The power from MFMR is influenced less and it is higher than that from Bonferroni and FDR approaches. Comparing the results of MFMR before and after controlling for race, the power is reduced but the ANFP is not. This result suggests no need to control population stratification for MFMR approach. The ANFP from MFMR is greater than that from Bonferroni and FDR methods because MFMR uses a less stringent α level. If one uses Bonferroni criterion for MFMR or uses less stringent q value (0.15) for FDR correction, the ANFP from MFMR is slightly lower than that obtained using other approaches in this simulation study.
Simulation study 4
In simulation study 4, the quantitative trait Y was simulated by using the same model as in simulation study 3, but the three causal SNPs were not associated with race (p > 0.41) and not significantly correlated with each other (the maximum r2 is 0.01). SNP1 (coded additively) is selected from chromosome 8 with MAF 0.10 and contributes average 6.8% of total trait variation in 100 replicates; SNP2 (coded recessively) is selected from chromosome 1 with MAF 0.17 and contributes average 5.3% of total trait variation; SNP3 (coded additively) is selected from chromosome 4 with MAF 0.37 and contributes 5.2% of total trait variation. Because the three causal SNPs are not associated with population substructure, they are only significantly correlated (|r| > 0.2, p < 0.0001) with 41 SNPs in the genome. 33 of them (0.2 < |r| < 0.6) are not in the same LD regions of the causal SNPs. The results from genome-wide testing for 100 replicates by using Bonferroni, FDR and MFMR approaches are shown in Table 5.
Table 5.
Power and ANFP under the condition that causal SNPs are not associated with race
| Method | Power Sample size = 400 |
ANFP | Total SNPs | ||
|---|---|---|---|---|---|
| SNP 1 R2=6.8% | SNP 2 R2=5.3% | SNP 3 R2=5.2% | |||
| Bonferroni (4.3×10−7) | 40% | 19% | 19% | 0.20 | 115,182 |
| FDR (q = 0.05) | 44% | 27% | 25% | 0.51 | 115,182 |
| FDR (q = 0.50) | 70% | 60% | 59% | 122 | 115,182 |
| MFMR (α = 0.05) | 84% | 62% | 72% | 49.6 | 115,182 |
| MFMR (α = 10−4) | 82% | 61% | 67% | 15.25 | 115,182 |
| MFMR (α = 4.3×10−7) | 52% | 27% | 34% | 0.06 | 115,182 |
In this simulation study, because population substructure is not associated with causal SNPs, it is also not associated with the quantitative trait generated by the causal SNPs. The average p-value is 0.52 for the association tests between race and quantitative trait in 100 replicates. We did not find inflation of false positives in this simulation study. The inflation factor is λ = 0.985. The q-q plot of the expected negative log p-values versus observed negative log p-values is shown in Supplementary Figure 6. This study indicates that the population stratification due to effects from a few large strata may not be an issue if population substructure is not associated with disease trait. From Table 5, the power from MFMR is significantly higher than that from Bonferroni and FDR procedures, and the ANFP from MFMR is still acceptable although a less stringent a level is used. If the Bonferroni significance criterion is adopted by MFMR, the MFMR shows higher power and lower Type I error rate than the other two procedures.
DISCUSSION
Compared to the Bonferroni and FDR procedures, MFMR shows higher power to detect weak and moderate genetic effects and an acceptable number of false positives. According to Figures 1 and 2, and the formula to calculate the power of the maximum order statistic, when a is reduced from 0.05 to 0.0001, the Type I error rate of MFMR will reduce greatly while the power from MFMR will not reduce similarly if the specified power from a single SNP test is not too low, especially when there are multiple true positive effects. This expectation was confirmed in simulation studies 1 to 4. Table 1 through Table 5 show when α is reduced from 0.05 to 10−4, the power from MFMR changes little if the power from a single SNP test is not very low, while the Type I error rate from MFMR reduces greatly. Therefore, using α = 10−4 for MFMR may be a better option.
For simulation studies 1 and 2, we extracted the dataset that includes only SNPs that are not, or modestly correlated with each other. If higher correlated SNPs are studied, the power of the Bonferroni test can be expected to deteriorate further because the procedure will be even more conservative, given that it assumes the tests are independent. The power for the MFMR test is not as influenced by correlations because it is not adjusting the critical value for the total number of independent tests.
When causal SNPs are correlated with many SNPs out of LD regions due to population stratification or other factors, single SNP analysis without adjusting for population stratification will inflate the false positives rate, while MFMR still maintains better power and well-controls the Type I error rate. When causal SNPs are not correlated with population substructure and population substructure is not associated with disease trait as shown in simulation study 4, there is no inflation of false positives. In that case, the performances of all three approaches are similar to their performances in simulation studies 1 and 2 where there is no population stratification. If causal SNPs are not correlated with population substructure but there is an unknown environmental risk factor that is correlated with population substructure, the inflation of false positives may still be an issue. In that case, investigators may adjust population substructure to control the inflation of false positives in separate SNP analyses. MFMR is very likely to select a SNP, which is highly associated with the unknown environmental risk factor, into regression model to effectively control population stratification. Because there are hundreds of thousands of SNPs in genome, we assume such a SNP always can be found. Since in any case MFMR has higher power to detect weak genetic effects and has limited number of false positives, it may be a good choice for a first-stage analysis in a two-stage GWAS design.
There is another advantage in MFMR. In practice, if a trait is caused by several independent factors, then the tests for each factor in multiple regressions are usually more significant than in univariate regression analysis. That is, when multiple predictors influencing a trait are not included in the model, their effects are subsumed by the residual error. Therefore, the standard error for each variable in univariate regression will be larger than that in multiple regressions. That’s why MFMR has better power than the Bonferroni approach when using the same significant criterion in simulation studies 1 to 4.
When multiple SNPs are highly correlated with a positive SNP, each of the SNPs will tend to be identified when univariate analyses are conducted. In contrast, only the most strongly associated SNP will be identified in forward multiple regressions. Additional SNPs that are correlated with this SNP and without independent contributions to response variable will fail to explain a significant proportion of the residual variance conditional on the most associated SNP. Thus, these additional SNPs will fail to be incorporated in multiple regression models and false positives due to correlation with the true positive SNP can be excluded. In some situations, this characteristic of MFMR may not be beneficial. If two SNPs are highly correlated with each other and the SNP with weaker effect has a true effect, MFMR will pick the SNP with the stronger effect and have less power to detect the weaker effect. For example, in Table 3 we see MFMR has less power than the Bonferroni approach to detect SNP1’s effect. That was because in some replicates, some non-causal SNPs that were highly correlated with SNP1 had a stronger effect than SNP1 due to random fluctuation of samples. Those SNPs were first selected by MFMR, which reduced the power to detect SNP1. The same thing could happen to SNP2 and SNP3 when they are highly correlated with many other SNPs. If a causal SNP is not highly correlated with other SNPs in different genomic regions, as expected for most GWAS where homogeneous populations were used, MFMR has higher power than other methods as shown in simulation studies 1, 2 and 4.
One disadvantage of MFMR is that it is sensitive to missing genotype. Because MFMR jointly analyzes n SNPs after n steps, even if each SNP has only several missing genotypes, dozens of SNPs could have several hundred observations with missing genotypes. In that case multiple regressions will lead to removing any subject having missing genotype from the regression model, consequently influencing SNP selection. We suggest imputing missing genotypes with a method such as Mach [Scott et al., 2007] or Bimbam [Servin and Stephens, 2008] before using MFMR.
Figure 3.
q-q plot of expected negative log p-value vs observed negative log p-value in simulation study 1.
Figure 4.
q-q plot of expected negative log p-value vs observed negative log p-value in simulation study 2.
Figure 6.
q-q plot of expected negative log p-value vs observed negative log p-value in simulation study 4.
Acknowledgments
This work was supported by NCI CA 21765 and the NIH/NIGMS Pharmacogenetics Research Network and Database (U01 GM61393, U01GM61374 http://pharmgkb.org/) from the National Institutes of Health; and by American Lebanese Syrian Associated Charities (ALSAC). Special thanks for the help from Dr. Wenjian Yang, who collected and provided the 115K SNP data set.
References
- Amos CI, Wu X, Borderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai Y, Shete S, Spitz M, Houlston RS. A genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical Population Biology. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- Lin DY. An efficient Monte Carlo approach to assessing statistical significance in genomic studies. Bioinformatics. 2005;21:781–787. doi: 10.1093/bioinformatics/bti053. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Principles of Population Genetics. 3. Sinauer Associates; Sunderland, MA: 1997. [Google Scholar]
- Hocking RR. Methods and Applications of Linear Models. 2. Wiley- Interscience; Hoboken, NJ: 2003. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65– 70. [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima S, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzan merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Nagelkerke NJD. A note on a general definition of the coefficient of determination. Biometrika. 1991;78(3):691–692. [Google Scholar]
- Plenge RM, Seielstad M, Padyukov L, Lee AT, Remmers EF, Ding B, Liew A, Khalili H, Chandrasekaran A, Davies LR, Li W, Tan AK, Bonnard C, Ong RT, Thalamuthu A, Pettersson S, Liu C, Tian C, Chen WV, Carulli JP, Beckman EM, Altshuler D, Alfredsson L, Criswell LA, Amos CI, Seldin MF, Kastner DL, Klareskog L, Gregersen PK. TRAF1-C5 as a risk locus for rheumatoid arthritis--a genomewide study. N Engl J Med. 2007;357(12):1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Rabbee N, Speed TP. A genotype calling algorithm for affymetrix SNP arrays. Bioinformatics. 2006;22(1):7–12. doi: 10.1093/bioinformatics/bti741. [DOI] [PubMed] [Google Scholar]
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- Satagopan JM, Elston RC. Optimal two-stage genotyping in population-based association studies. Genet Epidemiol. 2003;25:149–157. doi: 10.1002/gepi.10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate genes and quantitative traits. PLoS Genetics. 2007 Jul;3(7):e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–5. doi: 10.1126/science.1142382. Epub 2007 Apr 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DC, Haile RW, Duggan D. Recent developments in genomewide association scans: A workshop summary and review. Am J Hum Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Young SS. Resampling-based multiple testing: Examples and methods for p-value adjustment. Wiley; NY: 1993. [Google Scholar]