Abstract
We consider detecting associations between a trait and multiple SNPs in linkage disequilibrium (LD). To maximize the use of information contained in multiple SNPs while minimizing the cost of large degrees of freedom (DF) in testing multiple parameters, we first theoretically explore the sum test derived under a working assumption of a common association strength between the trait and each SNP, testing on the corresponding parameter with only one DF. Under the scenarios that the association strengths between the trait and the SNPs are close to each other (and in the same direction), as considered by Wang and Elston (2007), we show with simulated data that the sum test was powerful as compared to several existing tests; otherwise, the sum test might have much reduced power. To overcome the limitation of the sum test, based on our theoretical analysis of the sum test, we propose five new tests that are closely related to each other and are shown to consistently perform similarly well across a wide range of scenarios. We point out the close connection of the proposed tests to the Goeman test. Furthermore, we derive the asymptotic distributions of the proposed tests so that p-values can be easily calculated, in contrast to the use of computationally demanding permutations or simulations for the Goeman test. A distinguishing feature of the five new tests is their use of a diagonal working covariance matrix, rather than a full covariance matrix as used in the usual Wald or score test. We recommend the routine use of two of the new tests, along with several other tests, to detect disease associations with multiple linked SNPs.
Keywords: genome-wide association study, logistic regression, multilocus analysis, permutation, single-locus analysis, SNP
INTRODUCTION
Due to the rapid development of large-scale genotyping technologies, it has become feasible to conduct genome-wide association studies (GWASs); in fact, quite a few GWASs have been completed while many others are underway. However, a remaining challenge is how to efficiently analyze data collected from GWASs. This is critical given that most complex diseases or traits are only weakly associated with causal genetic variants. The power of GWASs depends on linkage disequilibrium (LD) between DNA markers (e.g., genotyped SNPs) and causal loci that often are not genotyped: if there exists a causal locus in a DNA region, due to LD, it is likely that one or more SNPs nearby will be associated with the trait too. Hence, it is intuitively reasonable to consider multiple loci in LD in a region; this is the problem we consider here. Roughly speaking, most existing analysis methods fall into one of the two categories: single-locus versus multilocus approaches. The basic idea of the former is to analyze single SNPs one at a time, then combine such individual analyses (Roeder et al 2005 and references therein). For these methods, in addition to possibly failing to utilize information contained in high-order relationships among multiloci, another shortcoming is their reduced power due to the high cost of adjustments for multiple testing that are required to control family-wise error rates or false discovery rates at some nominal levels. On the other hand, to overcome the above two problems with single-locus approaches, multilocus methods jointly model the relationship between a trait and multiloci (e.g., Chapman et al 2003; Qin et al 2002; Schaid et al 2002; Stephens et al 2001; Wei et al 2008; Zhao et al 2003a, b, and references therein). However, there is also an inherent cost of large degrees of freedom (DF) when testing over multiple parameters involved in a joint model.
Wang and Elston (2007) fully recognized the two possibly conflicting goals of capturing information contained in multiloci in LD and reducing the cost of multiple testing or large DF, and thus proposed a weighted score test (WST) to achieve the two goals at the same time: although multiloci are involved, the DF of the WST is only 1. They used simulated data under various scenarios, including mimicking real LD patterns as drawn from HapMap data, to demonstrate improved power of the WST over other existing methods. Here we first study an alternative approach called the sum test in the same context: it is based on the familiar treatment of the trait as the response variable and modeling its mean (e.g. probability of having disease) in terms of multiple SNPs in LD in logistic regression or any generalized linear model (GLM). The basic idea is to test on only one parameter under a working assumption of a common association strength between each of multiple SNPs and the trait. Although in general the association strengths may vary with SNPs, the proposed methods can be more powerful than the WST and several other existing methods while controlling the test size at a specified nominal significance level; similar to the WST, the key is to utilize multiloci while controlling the DF at the minimum 1. Simply put, rather than testing on multiple parameters either individually (followed by a multiple test adjustment as in single-locus approaches) or jointly (with large DF as in most multilocus approaches), the sum test focuses on a scalar function of the multiple parameters with the resulting DF=1.
An original motivation of this work was to explore how the WST works. The numerical results of Chapman and Whittaker (2008) indicate that, if some of the coded SNPs are positively correlated while others are negatively correlated, the WST and the sum test may have low power. Here we exploit theoretical properties of the sum test. In particular, we provide a theoretical explanation for the possible failure of the sum test: if the association strengths between the outcome and the SNPs are not close, especially if some are in opposite directions, the sum test may have low power; this explanation differs from the heuristic argument given by Chapman and Whittaker (2008) that is based on the correlations among the coded SNPs. In addition, the theoretical result motivated the development of four new tests: two of them are asymptotically equivalent to an estimated most powerful test, and are weighted versions of the other two that are closely related to Goeman’s test (Goeman et al 2006), which was found to perform well across a wide range of scenarios by Chapman and Whittaker (2008). The proposed tests are all based on a sum of squared marginal regression coefficient estimates or of squared score statistics, possibly weighted inversely by the variances of the coefficient estimates or score statistics. In contrast to the use of computationally intensive permutations or simulations as for Goeman’s test, we derive asymptotic distributions of the proposed tests, easily yielding p-values. In spite of their simplicity, our numerical studies showed that the proposed tests performed similarly to Goeman’s test. A distinguishing feature of the new tests and Goeman’s test is their use of a diagonal working covariance matrix, rather than a full covariance matrix as used in the usual Wald or score test. We offer some intuitive explanations on the possibly improved power of the new tests over the usual Wald or score test, illustrating the well known fact that there is no uniformly most powerful test in the current context.
METHODS
REGRESSION AND RELATED TESTS
Suppose that we have m independent observations (Yi, Xi), where subject i has trait (e.g. disease status) value Yi and genotype Xi = (Xi1, …, Xik). As in Wang and Elston (2007), we consider the dosage coding of Xij for an additive model: Xij = 0, 1 or 2, representing the copy number of one of the two alleles present in SNP j of subject i; other choices include a binary coding of Xij = 0 or 1 for a dominant or recessive genetic model, and a more general coding scheme can be implemented, albeit more complicated and will be skipped. To test any possible association between the trait and SNPs, we entertain a generalized linear model (GLM) (McCullagh and Nelder 1983)
where E(Yi) is the mean of Yi, h( ) is a link function and β = (β1, …, βk)′. Two commonly used models are the linear model with the identity link for continuous or quantitative traits, and the logistic regression model with the Logit link for a binary trait. In this paper, we focus on the logistic model with the dosage coding of Xij,
| (1) |
where Yi = 0 or 1 indicates whether subject i is a control (i.e. without disease) or a case (i.e. with disease). A global test of any possible association between the trait and SNPs can be formulated as jointly testing on the multiple parameters βj’s with the null hypothesis H0: β = 0 by one of the likelihood ratio test (LRT), Wald test and score test in the context of logistic regression (or more generally, of GLM); under H0, any of the three test statistics has an asymptotically chi-squared distribution with degrees of freedom DF=k. We call the LRT that was used in our numerical examples as the logistic-global (L-G) test. For a large k, the test can be low-powered because of the cost of the large DF.
In contrast to the global test, another extreme is to conduct individual SNP-by-SNP tests: rather than including all the k SNPs, we include only one SNP in
| (2) |
where we explicitly distinguish βM = (βM,1, …, βM,k)′ in marginal models (2) from β in joint model (1). Then we test H0: βM,j = 0 for each j = 1, …, k sequentially. Each test can be done with only one DF, but a multiple test adjustment has to be made, based on either permutation or Bonferroni adjustment. Because the Bonferroni adjustment is known to be conservative, permutation is more widely used, though it is computationally demanding; we call the univariate test based on permutation U-B. The multiple test adjustment may reduce the power of the test, as shown by Wang and Elston (2007).
Most of the existing methods fall into one of the above two extreme categories. For example, the generalized Hotelling’s T2 test (Fan and Knapp 2003; Xiong et al 2002) and haplotype analyses (e.g., Chapman et al 2003; Schaid et al 2002; Stephens et al 2001; Zhao et al 2003a, b, and references therein) belong to the first category for their joint testing on multiple parameters simultaneously, while other univariate approaches build on SNP-by-SNP tests (Roeder et al 2004 and references therein). The limitations of the two extremes have been well recognized and motivated the development of new methods. A general strategy is to capture full information in all βj’s while reducing the cost of large DF or adjustment for multiple testing, but the key is how to reconcile the two possibly conflicting goals. Wang and Elston (2007) proposed such an approach, called weighted score test (WST): the WST puts higher weights on more important components of Fourier-transformed genotypes and minimizes the DF at one. Below we first study a simple alternative to WST, the sum test; a theoretical analysis clearly indicates a possible limitation of the sum test, which further motivates the development of our new tests.
THE SUM TEST
We first formulate the sum test from a new perspective of striking a compromise between joint modeling and its resulting large DF: while all the SNPs in LD are to be used, we make a key and possibly incorrect working assumption that the SNPs are all equally associated with the trait; that is, rather than unnecessarily aiming to estimate separate βj’s in (1), we use a common βc in the logistic regression model:
| (3) |
To address the question of whether there is any association between the trait Yi and any SNP, we test H0: βc = 0, which can be easily done by the LRT, Wald or score test in fitting model (3); we used the LRT throughout. Note that fitting model (3) is equivalent to regress Y on a new covariate that is the sum of the genotypes of the multiple SNPs, and hence we call the resulting test the sum test.
Below we explore the advantages and limitations of the sum test. Intuitively, because it tests on only one parameter βc, there will be no power loss due to large DF (or multiple testing adjustment). Generally, the common association parameter βc in (3) is an “average” (or more precisely, a function) of the individual β1, …, βk; see Appendices A–C for the theory. It is most illustrative to consider the case with the linear model, in which it can be shown (in Appendix A) that
| (4) |
where β̂c is the MLE of βc in a linear model analogous to (3), and β̂M is the MLE of βM in the marginal linear models analogous to (2). Note that, because of the consistency of the MLE, with a large sample size in a typical genome-wide association study, equation (4) still approximately holds if β̂c and β̂M are replaced by their true values βc and βM respectively. Note that the weight in (4) is inversely proportional to the variance of β̂M,j.
Under the global H0: βM = 0, it is trivial to see that H0: βc = 0 also holds, guaranteeing that the sum test will have (asymptotically) correct test size. On the other hand, if the global H0 is false, then it is likely to have βc ≠ 0, leading to good power. For this latter purpose, because of the always non-negative weights for β̂M,j’s, it is best to have β̂M,j’s (or equivalently, true βM,j’s for large samples) with the same sign; this is also a requirement for the WST. Hence, to avoid the consequence of that positive and negative components of βM cancel out, a heuristic approach as discussed by Wang and Elston (2007) is the following: before applying the WST (and the sum test), one needs to adjust the coding of Xij’s so that there are as few negative correlations as possible among the SNPs; for example, if X.l of SNP l is negatively correlated with many other X.j’s, we will take new Xil as 2 − Xil for the dosage coding (or switching the values for the two categories in the binary coding). We developed an algorithm as the following: 1) calculate all pairwise correlations; 2) see which SNP s has the largest number, say ns, of negative correlations; 3) if ns > #SNPs/2, then flip the coding of SNP s, and repeat the above process; otherwise, stop. Nevertheless, at the end, it is not guaranteed that all SNP pairs will be non-negatively correlated; more importantly, even if they are, it may not help: equation (4) clearly shows that the key is on the signs of the components of β̂M; a positively correlated pair of X.j and X.l does not imply that sign(β̂M,i) = sign(β̂M,l). The theory clearly illustrates a possible limitation in inferring the performance of the sum test with the use of pairwise SNP correlations, a heuristic suggested by Chapman and Whittaker (2008). Below we propose a class of five new tests that overcome the above issue of the sum test.
TWO NEW TESTS BASED ON β̂M
The problem with the sum test is that, as shown in equation (4), because the estimated common effect β̂c is a linear combination of the components of β̂M with always positive coefficients, the test may have reduced power with a small β̂c when the components of β̂M have different signs. Hence, to eliminate the sign problem, we replace the components of β̂M in the linear combination by their squares. Specifically, we propose a test statistic
or, its weighted version with a weight assigned to each component of β̂M based on its variance:
where Diag(VM) = diag(vM,1, …, vM,k) is a diagonal matrix with the elements vM,j as the (estimated) variance of β̂M,,j from marginal model (2). Note that, albeit related to the Wald test, the above tests differ from the usual Wald test, , in that VM = Cov(β̂M) is replaced by a diagonal matrix, either an identity matrix I or Diag(VM).
Below we describe the asymptotic distributions of the above two test statistics. Based on the theory of generalized estimating equations as shown in Appendix D, under H0: (βM = 0, β̂M asymptotically has a Normal distribution with mean 0, and its covariance matrix can be consistently estimated by a sandwich (or robust) estimator VM. Hence, asymptotically, each of the two test statistics has a quadratic form of Normal variates, , with W = I and W = Diag(VM) respectively. It is well known (e.g. Johnson and Kotz, 1970, p.150) that the distribution of Q is a weighted sum of k independent chi-squared variates with DF=1, , where cj’s are the eigen values of VMW−1. Furthermore, by the results of Zhang (2005), can be well approximated by with
To calculate a p-value, for example for the SumSqB test,
Note that in the above two new tests, we used β̂M from marginal models (2), not β̂ from joint model (1). Intuitively, because of collinearity, the components of β̂ have much larger variances than their counter parts of β̂M. Our numerical results (not shown) confirmed the much worse performance of the resulting SumSqB or SumSqBw if β̂M was substituted by β̂. In contrast, the Wald test based on β̂M and that based on β̂ are asymptotically equivalent because their corresponding score tests are the same, as to be shown later.
TWO NEW TESTS BASED ON MARGINAL SCORE STATISTICS
By the asymptotic connection between the MLE and the score statistic (Cox and Hinkley 1974, p.315), mimicking SumSqB and SumSqBw, we can construct a modified score test based on the marginal model (2). It is easy to verify that the marginal score statistic for βM,j under H0: βM,j = 0 in (2) is
where and 1 is a column vector with elements all 1’s. Denote UM = (UM,1, …, UM,k)′. We propose a new test based on the sum of the squares of the marginal score statistics,
where Y and X are the response vector and the design matrix respectively. A weighted form is
where If = Cov(UM) = Ȳ(1 − Ȳ)(X − X̄)′(X − X̄) is the expected Fisher information matrix, and X̄ is the sample mean of each SNP.
As for SumSqB and SumSqBw, the null distributions of the two test statistics have quadratic forms and can be approximated by . Furthermore, as for SumSqB and SumSqBw, the above two test statistics differ from the usual score test statistic with If = Cov(UM) replaced by a diagonal matrix. Because of the asymptotic equivalence between the Wald test and score test (see equation (5) below for a rigorous proof of their asymptotic equivalence), we expect SumSqB and SumSqU (or, SumSqBw and SumSqUw) to perform similarly, as to be verified numerically next.
Note that the SumSqU test itself can be motivated as a modification to the sum test. It is easy to derive that the score statistic of the sum test for H0: βc = 0 in model (3) is , which may be sensitive to varying signs of the components of UM (and thus of β̂M), leading to reduced power; as the SumSqB test, is invariant to varying signs of UM,j’s.
A NEW TEST AS AN ESTIMATED MOST POWERFUL TEST: A UNIFICATION
Below we propose a new test by estimating the most powerful test, which is to be proved to be asymptotically equivalent to the SumSqBw and SumSqUw tests, serving to unify the five new tests proposed here.
It is well known that there is no uniformly most powerful unbiased (UMPU) test on multiple parameters (Cox and Hinkley 1974). On the other hand, under local alternatives with b as a fixed vector and δ a scalar, the most powerful test is based on the test statistic TMP = b′U (Cox and Hinkley 1974, p.319). Because the true β, and hence b are unknown, we aim to estimate TMP by replacing b by an estimate of β:
By a standard Taylor expansion of the marginal score equation UM,j(β̂M,j) = 0, it is easy to verify that
| (5) |
where IM,d is a diagonal matrix with −∂UM,j(βM,j)/∂βM,j|βM,j=0 as the jth diagonal element, and UM,j(βM,j) is the score function of marginal model (2). Hence, we have
As before we approximate the asymptotic distribution of TEMP using a scaled and shifted chi-squared distribution.
By the relationship between an observed and an expected information matrix, we have IM,d ≈ Diag(If), and thus
which suggests that the SumSqUw is asymptotically equivalent to the estimated most powerful (EMP) test. Similarly, using (5), it is easy to verify that SumSqBw, SumSqUw and EMP are all asymptotically equivalent.
Equation (5) also directly suggests an estimator for Cov(β̂M) as used in deriving the asymptotic distributions of SumSqB and SumSqBw:
which is the sandwich (also called robust or empirical) covariance estimator used in the GEE for β̂M (Appendix D).
Interestingly, because the score statistic of the sum test for H0: βc = 0 in model (3) is , the sum test can be viewed as an estimated most powerful test by estimating β as c1, where c is a constant scalar and 1 is a vector with elements all 1’s. This is in agreement with the intuition that the sum test is most powerful when the components of the true β are all equal (and thus with the same sign).
Finally, we note that the usual joint score test, and thus its asymptotically equivalent LRT and Wald test, can be also regarded as estimated most powerful tests. If we use β̂, instead of β̂M of TEMP, to estimate TMP, we have , where the last two expressions correspond to the score test statistics for joint model (1) and marginal models (2) respectively; the proof is based on two simple facts that, similar to (5), we have , and UM = U as shown next.
GOEMAN’S TEST AND ITS CONNECTION TO THE SUMSQU TEST
Goeman et al (2006) proposed a general empirical Bayes method to test on a large number of parameters, as applicable to the βj’s in the joint logistic regression model (1). A key idea is to treat βj’s as random, rather than as fixed as considered in joint logistic regression. Specifically, β = (β1, …, βk)′ is assumed a priori from a distribution with mean E(β) = 0 and covariance Cov(β) = τ2I. Thus, to test on the original H0: β = 0, one can instead test on a new H0: τ2 = 0; with the logistic regression model (1), the test statistic turns out to be (Chapman and Whittaker 2008)
where U is the score statistic for β under H0: β = 0 in joint model (1), and If is the corresponding Fisher information matrix: U = X′(Y − Ȳ) = UM, and If = Cov(U) = Ȳ(1 − Ȳ) (X − X̄)′ (X − X̄).
The null distribution of TGO is unknown and has to be estimated by permutation or simulation, which is computationally intensive.
As noted by Chapman and Whittaker (2008), given Y (as under permutation), the second term of TGO is non-random while the first term has a quadratic form of asymptotic Normal variates. Noting that U = UM, the Goeman test is equivalent to the SumSqU test if permutations are used in the former; more generally, the two tests are different.
The Goeman test was originally proposed for a large number of parameters with a relatively small sample size, which is not true in the current context: we have a sample size of hundreds to thousands while the number of parameters is less than 100, e.g. only around 20 in our examples. Hence, the good performance of the Goeman test in the current context as obtained by Chapman and Whittaker (2008) and confirmed here later is somewhat puzzling. Here we offer an explanation. By the specified prior distribution of βj’s as iid with mean 0 and variance τ2, we have as an empirical estimate of τ2, thus SumSqB can be naturally used to test H0: τ2 = 0; on the other hand, the asymptotic equivalence between SumSqB and SumSqU, and their close connection to the EMP test, provide a partial justification for SumSqU, and thus for the Goeman test.
RESULTS
SIMULATION SET-UPS
We performed simulation studies by largely following the set-ups given in Wang and Elston (2007) with k = 4, 10 and 20 marker SNPs, and sample size n = 100, 200 and 500. The disease-causing SNP was assumed to be in the center of the marker SNPs, but was removed from the data. First, we generated a latent vector from a multivariate normal distribution with one of three covariance structures: a compound symmetry (CS) with an equal pairwise correlation ρ = 0.4, an AR-1 with the correlation ρij = 0.8|i − j| between components i and j, and a correlation matrix with elements ρij randomly between 0.3 and 0.7. Second, the latent vector was dichotomized to yield a haplotype with allele frequencies randomly between 0.2 and 0.8 while the minor allele frequency (MAF) for the disease-causing SNP was fixed at 0.2, 0.3 or 0.4. Third, we combined two haplotypes and obtained marker genotype data Xi = (Xi1, …, Xik)′ (and X0i for disease-causing SNP) for subject i. Fourth, the disease status Yi of subject i was generated from a logistic regression model:
| (6) |
where we chose β0 = −log 4 to give a background (i.e. not caused by the SNP) disease probability of 0.2, and the odds ratio (OR) ranged from 1 (i.e. no association) to 2. Finally, following the case-control design, we sampled n cases (with Yi = 1) and n controls (with Yi = 0). We excluded the disease-causing SNP, supplying {(Yi, Xi): i = 1,2, …, 2n} as a dataset to various statistical tests. For each set-up, we simulated 1000 datasets, from which we obtained an empirical size or power for each test as its proportion of correctly or incorrectly rejecting its H0; in particular, the Monte Carlo standard error of an empirical size/power p̂ is .
In addition to the sum test and the weighted score test, we also considered a few other ones: the first (denoted as L-G) was a global LRT on H0: β = 0 based on the joint model (1); the second was the generalized Hotelling’s T2 test; third, we included the univariate test and Goeman’s test based on permutations, U-P and Go-P respectively; finally, we had our five asymptotic tests based on the sum of (weighted) squares of the components of β̂M or of the score statistics.
SIMULATION RESULTS
We considered k = 4, 10 and 20 marker SNPs, sample size n = 100, 200 and 500, MAF=0.2, 0.3 and 0.4 for the disease-causing SNP, and used nominal significance levels α = 0.05 and α = 0.01; because the conclusions were similar, to save space, we only gave results for k = 10, n = 500, MAF=0.2 and α = 0.05.
First, with the CS correlation structure suggesting all the marker SNPs contained the same amount of information about the disease-causing SNP, as shown in Table 1, it is obvious that the sum test and the WST were the winners, closely followed by the Goeman test and our proposed five tests. Second, for the AR-1 correlation structure (Table 2), the sum test, the Goeman test and our proposed five tests performed similarly and better than other methods. Third, with random pairwise SNP correlations (Table 3), the conclusion was similar to that for the CS: the sum test and WST performed best, having a slight edge over the Goeman test and our proposed five tests. Fourth, across all the scenarios, our proposed five tests performed similarly to each other and to the Goeman test, which however depends on the use of computationally intensive permutations or simulations. Note that for the AR-1 or the random correlation structure, pairwise correlations ρij were not constant, representing a situation in which the working assumption of “a common association strength” underlying the sum test is violated; nevertheless, the key for the good performance of the sum test (and WST) is that the associations between the response and SNPs were (mostly) in the same direction.
Table 2.
Empirical sizes and powers of various tests with nominal significance level α = 0.05 for simulated data based on the real LD pattern of gene CHI3L2 with MAF> 0.2 (#SNP=16).
| SumSq
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | Sum | WST | L-G | T2 | U-P | Go-P | Bw | B | Uw | U | EMP |
| 200 | 1.0 | .050 | .041 | .094 | .036 | .053 | .052 | .051 | .049 | .052 | .053 | .055 |
| 200 | 1.2 | .181 | .160 | .142 | .058 | .169 | .182 | .177 | .181 | .177 | .179 | .180 |
| 200 | 1.4 | .521 | .480 | .292 | .173 | .483 | .516 | .512 | .513 | .512 | .513 | .518 |
| 200 | 1.6 | .803 | .774 | .521 | .375 | .764 | .818 | .814 | .816 | .813 | .811 | .818 |
|
| ||||||||||||
| 500 | 1.0 | .051 | .043 | .074 | .032 | .054 | .057 | .056 | .056 | .056 | .054 | .057 |
| 500 | 1.2 | .387 | .356 | .188 | .113 | .333 | .381 | .370 | .376 | .370 | .370 | .371 |
| 500 | 1.4 | .886 | .867 | .606 | .483 | .886 | .899 | .901 | .901 | .901 | .896 | .901 |
| 500 | 1.6 | .994 | .992 | .927 | .879 | .997 | .995 | .995 | .997 | .995 | .994 | .995 |
Table 3.
Empirical sizes and powers of various tests with nominal significance level α = 0.05 for simulated data based on the real LD pattern of gene CHI3L2 with MAF> 0.05 (#SNP=22).
| SumSq
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | Sum | WST | L-G | T2 | U-P | Go-P | Bw | B | Uw | U | EMP |
| 200 | 1.0 | .047 | .040 | .103 | .034 | .048 | .058 | .047 | .055 | .049 | .053 | .052 |
| 200 | 1.2 | .174 | .151 | .148 | .068 | .148 | .177 | .162 | .161 | .168 | .174 | .173 |
| 200 | 1.4 | .481 | .438 | .302 | .164 | .424 | .520 | .498 | .472 | .507 | .512 | .512 |
| 200 | 1.6 | .743 | .719 | .500 | .353 | .735 | .808 | .789 | .766 | .791 | .801 | .796 |
|
| ||||||||||||
| 500 | 1.0 | .051 | .043 | .072 | .034 | .050 | .056 | .049 | .049 | .049 | .050 | .050 |
| 500 | 1.2 | .350 | .315 | .173 | .113 | .290 | .372 | .357 | .342 | .358 | .363 | .360 |
| 500 | 1.4 | .849 | .819 | .527 | .413 | .863 | .894 | .894 | .877 | .894 | .891 | .895 |
| 500 | 1.6 | .991 | .985 | .894 | .836 | .991 | .995 | .996 | .992 | .996 | .995 | .996 |
HAPMAP DATA FOR GENE CHI3L2
As in Wang and Elston, we conducted a simulation study based on real LD patterns within the CHI3L2 gene as observed in HapMap data. We downloaded the SNPs of the CHI3L2 gene for the 90 CEU (Utah residents with ancestry from northern and western Europe) individuals from the HapMap site in June 2008. As in Wang and Elston (2007), first, we excluded SNPs with MAF ≤ 0.2, leaving 23 SNPs. Second, we did a single imputation for each of the missing genotypes by randomly drawing an observed genotype of the same SNP. Third, we used the dosage coding for the SNPs and tried to minimize the number of negative correlations among them. Fourth, we deleted redundant SNPs that were perfectly correlated with other SNPs. There was still substantial LD among the remaining 17 SNPs as indicated by the distribution of their pairwise Pearson’s correlation coefficients, which ranged from −0.388 to 0.989 with the three quartiles Q1, Q2 and Q3 as 0.364, 0.544 and 0.738. Fifth, we repeatedly sampled (with replacement) subjects from the 90 CEU individuals. Finally, as Wang and Elston, we chose the SNP rs2182114 as disease-causing and generated disease indicators from the logistic regression model (6) with the same β0 and four possible values of OR. The results for two sample sizes are shown in Table 2. It is clear that the sum test, Goeman’s test and our proposed five tests performed similarly and were the winners; in particular, they could have an extra 3% power over that of WTS or the univariate test. The global LRT and the Hotelling’s T2 test had low power, and even possibly inflated test sizes.
For the CHI3L2 gene, we also considered including all the SNPs with MAF> 0.05, rather than MAF> 0.2, and followed exactly the same steps, including choosing SNP rs2182114 as disease-causing. In this way, we ended up with 23 SNPs. Among the 253 SNP pairs, we had 42 pairs with negative correlation coefficients. The distribution of the correlation coefficients for all the pairs was summarized below: min= −0.388, the first quartile Q1 = 0.142, Q2 = 0.375, Q3 = 0.602, and max= 0.989. Hence, compared to that with only 17 SNPs, now some SNPs had weaker correlations, possibly leading to the power loss for the sum test (Table 3). The Goeman test and our proposed tests seemed to be the winners, slightly more powerful than the univariate test, then closely followed by the WST. The other two tests again had much lower power. It seems that the weighted version SumSqBw had a slight edge over SumSqB, albeit not necessarily significant; in contrast, the power difference between SumSqUw and SumSqU was hardly visible.
HAPMAP DATA FOR GENE IL21R
As in Chapman and Whittaker (2008), we also considered the region of gene IL21R, in which LD was low. We followed exactly the same steps as for gene CHI3L2 except that as in Chapman and Whittaker (2008), the disease-causing SNP was selected randomly and then excluded from the data in each simulation run. At the end, we had 28 SNPs. There were 54 SNP pairs with negative correlation coefficients. A summary of the correlation coefficients among all the SNP pairs was the following: min= −0.564, Q1 = 0.097, Q2 = 0.212, Q3 = 0.459, and max= 0.991. Hence, the LD pattern in this region was more extreme with some negative or low pairwise correlations, which might explain why the power of the sum test (and WST) was much lower than Goeman’s test, our proposed five tests and the univariate test (Table 4). It is interesting to note that the univariate test appeared to be the winner, more powerful than Goeman’s test and our proposed five tests in this case.
Table 4.
Empirical sizes and powers of various tests with nominal significance level α = 0.05 for simulated data based on the real LD pattern of gene IL21R with MAF>0.2 (#SNP=27).
| SumSq
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | OR | Sum | WST | L-G | T2 | U-P | Go-P | Bw | B | Uw | U | EMP |
| 200 | 1.0 | .046 | .050 | .098 | .063 | .057 | .052 | .046 | .047 | .047 | .047 | .048 |
| 200 | 1.2 | .078 | .078 | .107 | .078 | .087 | .087 | .078 | .078 | .079 | .084 | .082 |
| 200 | 1.4 | .204 | .215 | .200 | .148 | .256 | .265 | .260 | .264 | .265 | .261 | .267 |
| 200 | 1.6 | .351 | .366 | .344 | .275 | .500 | .474 | .451 | .457 | .457 | .464 | .470 |
|
| ||||||||||||
| 500 | 1.0 | .050 | .049 | .054 | .031 | .055 | .047 | .042 | .045 | .044 | .042 | .045 |
| 500 | 1.2 | .165 | .174 | .142 | .107 | .183 | .204 | .207 | .202 | .208 | .202 | .211 |
| 500 | 1.4 | .432 | .444 | .408 | .333 | .652 | .600 | .587 | .582 | .589 | .594 | .594 |
| 500 | 1.6 | .607 | .611 | .717 | .667 | .908 | .831 | .833 | .836 | .836 | .828 | .839 |
POWER ANALYSIS UNDER A SIMPLE SCENARIO
We have two immediate observations based on the above numerical studies. First, there is no uniform winner, though often Goeman’s test and our proposed five tests have high power and sometimes, the sum test is most powerful. In fact, when testing on multiple parameters, it is well known that no (asymptotically) uniformly most powerful unbiased (UMPU) test exists; if and only if the true value of the parameter vector is known, then an asymptotically UMPU test can be constructed (Cox and Hinkley 1974, p.319). In practice, of course, we do not know the true values of the parameters and hence have no UMPU test, suggesting that, without much prior knowledge, we perhaps should try more than one test. Second, it may be surprising that our proposed five tests can be more powerful than the traditional Wald, score or likelihood ratio test. To be concrete, we contrast the SumSqB test and the standard Wald test; the former uses an identity matrix, rather than a covariance estimate as used in the latter. Intuitively it seems that using a good covariance matrix as in the Wald test should be always more productive, but our numerical results suggest otherwise. We first thought that it might be due to bad covariance estimates, which however was not likely given that we had a large sample size and relatively a much smaller number of parameters. Below we offer an explanation under a simplified scenario, which in addition vividly illustrates the above point that the power of a test depends on the true parameter values.
We consider k = 2 parameters, β = (β1, β2)′, and its estimate β̂ ~ N(β, V) with V known: Var(β̂1) = Var(β̂2) = 1/500 and corr(β̂1, β̂2) = ρ. To test H0: β = 0, we can compare the following four tests: the Wald test with a test statistic β̂′V−lβ̂, the SumSqB test with SumSqB = β̂′β̂, a univariate test with Max = max(|β̂1|, |β̂2|), and a simplified sum test with Sum = β̂1 + β̂2. The Max test is similar to the U-P test except that the multiple testing on the components of β is analytically adjusted, rather than by permutation. The previous sum test (4) is simplified to the current special case if each covariate is standardized to have the same sample variance 1. For any of the above four test with test statistic T(β), the rejection region of the test is simply R(β) = {β: |T(β)| > c}. The constant c can be obtained either analytically or numerically (Conneely and Boehnke 2007) to guarantee that ∫R(β)f0(β)dβ1dβ2 = 1−α=0.95, where f0(β) is the density function of β̂ under H0, i.e., that of a bivariate N(0, V).
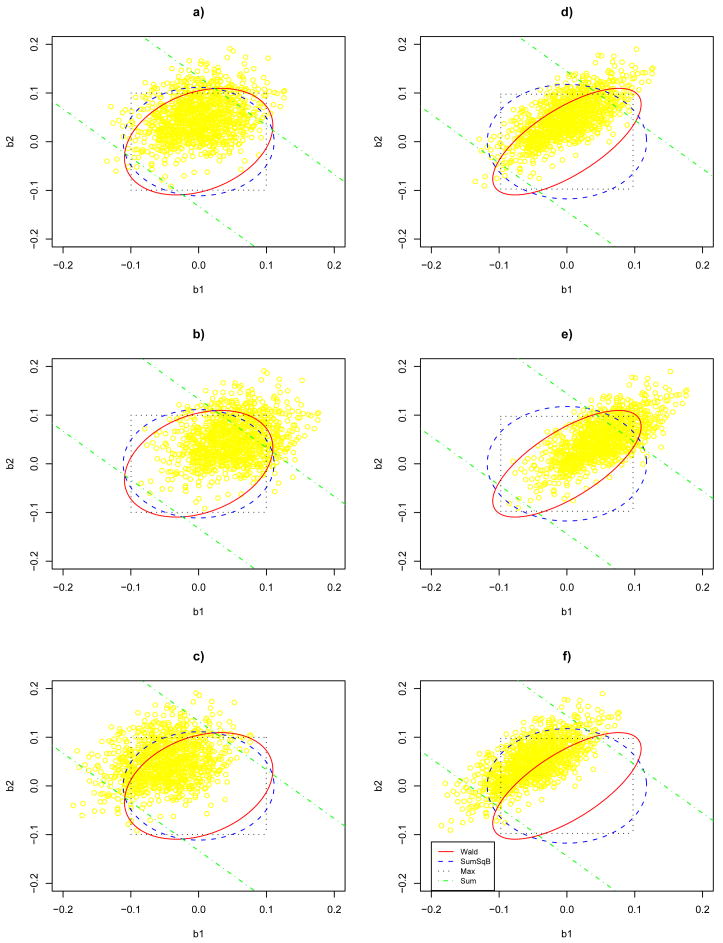
Figure 1 plots the rejection regions of the four tests with two different values of ρ = 0.3 and ρ = 0.7. In addition, 1000 random variates drawn from N(β, V) are also plotted on each panel. Three non-zero β values were considered: β = (0, 0.05)′, (0.05, 0.05) or (−0.05, 0.05). Table 5 gives the empirical power of each test under each set-up. It is most illustrative to notice the varying rejection regions of the tests, which determine the power of the tests. For example, along the identity (i.e. 45-degree) line, the rejection region of the Wald test covers that of the other tests while the sum test gives the smallest rejection region, suggesting that among the four tests, the Wald and sum tests are respectively the least and most powerful when β1 ≈ β2 ≠ 0; on the other hand, if β1 ≈ − β2 ≠ 0, the roles of the Wald and sum tests are reversed, as confirmed by Table 5. In particular, it is clear that the use of the (true!) covariance matrix of β̂ in the Wald test may lose power as compared to the SumSqB test (and sum test) if the components of the true β are indeed close to each other; furthermore, the power difference increases with the correlation between the components.
Figure 1.
Rejection regions of the four tests at significance level α = 0.05 for bivariate Normal data, along with 1000 points generated from the true distribution for each set-up; see Table 5 for more details on the set-ups.
Table 5.
Empirical powers of the four tests with nominal significance level α = 0.05 for simulated bivariate Normal data.
| Set-up | ρ | β | Wald | SumSqB | Max | Sum |
|---|---|---|---|---|---|---|
| a | 0.3 | (0, 0.05)′ | 0.164 | 0.143 | 0.158 | 0.121 |
| b | 0.3 | (0.05, 0.05)′ | 0.226 | 0.258 | 0.242 | 0.312 |
| c | 0.3 | (−0.05, 0.05)′ | 0.373 | 0.239 | 0.274 | 0.059 |
| d | 0.7 | (0, 0.05)′ | 0.263 | 0.102 | 0.158 | 0.133 |
| e | 0.7 | (0.05, 0.05)′ | 0.180 | 0.224 | 0.222 | 0.296 |
| f | 0.7 | (−0.05, 0.05)′ | 0.725 | 0.171 | 0.292 | 0.082 |
DISCUSSION
We have studied association mapping with multiple SNPs, possibly in LD. These SNPs may form an LD block, or more generally fall within a sliding window in a GWA scan. All the tests can be equally applied when other covariates are included in a regression model. We have mainly studied two ways of boosting statistical power: one is through dimension reduction as done in the sum test, while the other is to ignore possible correlations among the parameter estimates or score statistics as in the class of the SumSq tests. Interestingly, both the sum test and the class of the SumSq tests, along with the standard joint score test (and its asymptotically equivalent Wald test and LRT), can be all regarded as estimated most powerful tests with various estimates of the true association parameters.
A surprising result of this study is the confirmed good performance of the sum test in detecting possible associations between a trait and multiple SNPs in LD under some situations. The basic idea underlying the sum test is to adopt a working assumption on the existence of a common association strength between the trait and each of multiple SNPs. Intuitively, if any of the SNPs is associated with the trait, due to LD, other correlated SNPs should also be associated with the trait; even if their association strengths vary, their “average” (or more generally, some function of them) is likely to differ from 0, thus motivating the sum test with DF=1 on this “average” effect. Under situations with both positive and negative SNP associations with the trait, resulting in that the targeted “average” effect is much weaker than some individual effects, the sum test, as WST, may not be powerful. Largely for this reason, Chapman and Whittaker (2008) did not recommend the use of the sum test. However, because the sum test, as any SumSq test, is so easy and general to use with applicability to either continuous or discrete traits with or without other covariates, and to other study designs as long as a GLM can be employed, we do not dismiss its use. The good performance of the sum test is not limited to simulated data: Zhong and Pan (2008) applied it to a GAW16 Rheumatoid Arthritis (RA) dataset (Plenge et al 2007); for the four LD blocks containing well known RA-associated loci, among the sum test, Goeman’s test and univariate test, each test won in one case while all three performed similarly well in the fourth case (with empirical power close 1 due to too strong signals). Nevertheless, as shown here and in Zhong and Pan (2008), the performance of the sum test depends on the coding of SNPs in an unknown way; further studies are needed to identify optimal SNP codings for the sum test for any given dataset.
Our proposed five tests all have a quadratic form as a sum of squares of (weighted) marginal coefficient estimates or score statistics. It is perhaps surprising that, though the covariance matrix of the marginal coefficient estimates or score statistics can be consistently estimated, the new tests use only an identity matrix or only the diagonal elements of the corresponding covariance matrix, deviating from the standard Wald test and score test. We tried with the use of the full covariance matrices, and the results (not shown) were not good; in fact, the resulting Wald or score test was asymptotically equivalent to the LRT (L-G) for the joint model (1), which did not perform well as shown in our numerical examples. We emphasize that the possibly improved (or degraded) performance of our proposed tests over the standard LRT, Wald or score test should generally hold for any regression models, even with the presence of other covariates. Rather than using a sum of squares, we also tried a sum of absolute values of (weighted) marginal coefficient estimates; it performed no better than the sum of squares (results not shown). Importantly, all of the proposed five tests can be conducted by recoursing to their asymptotic null distributions, avoiding the use of time-consuming permutations or simulations as for Goeman’s test. We also compared the use of the asymptotic null distributions of the proposed tests to that of their permutational distributions, leading to similar results (not shown). Because of the consistently good and similar performance of Goeman’s test and our proposed tests, and computational advantages of our proposed tests, especially with the sum of the squared score statistics, over Goeman’s test, we recommend the routine use of SumSqU and SumSqUw, along with the sum test, the global LRT (or Wald or score test) and the univariate test (U-P) for the lack of any uniformly most powerful test.
Table 1.
Empirical sizes and powers of various tests with nominal significance level α = 0.05 for simulated data with three correlation structures (Corr).
| SumSq
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Corr | OR | Sum | WST | L-G | T2 | U-P | Go-P | Bw | B | Uw | U | EMP |
| CS | 1.0 | .051 | .053 | .047 | .049 | .046 | .047 | .044 | .046 | .044 | .043 | .045 |
| 1.2 | .098 | .096 | .059 | .062 | .072 | .084 | .076 | .076 | .077 | .080 | .080 | |
| 1.4 | .235 | .226 | .089 | .093 | .153 | .206 | .198 | .199 | .199 | .193 | .199 | |
| 1.6 | .395 | .399 | .145 | .153 | .239 | .366 | .357 | .363 | .358 | .356 | .360 | |
| 1.8 | .578 | .578 | .255 | .262 | .379 | .530 | .518 | .506 | .518 | .519 | .520 | |
| 2.0 | .711 | .713 | .357 | .366 | .480 | .670 | .661 | .657 | .661 | .662 | .666 | |
|
| ||||||||||||
| AR-1 | 1.0 | .055 | .048 | .053 | .054 | .037 | .049 | .047 | .047 | .048 | .048 | .049 |
| 1.2 | .132 | .115 | .078 | .080 | .107 | .131 | .123 | .123 | .124 | .125 | .127 | |
| 1.4 | .350 | .315 | .192 | .194 | .289 | .354 | .354 | .353 | .354 | .352 | .357 | |
| 1.6 | .599 | .549 | .361 | .370 | .504 | .583 | .584 | .583 | .585 | .577 | .589 | |
| 1.8 | .798 | .743 | .549 | .560 | .704 | .796 | .782 | .779 | .783 | .785 | .785 | |
| 2.0 | .895 | .868 | .726 | .727 | .845 | .907 | .897 | .891 | .896 | .901 | .898 | |
|
| ||||||||||||
| Rand | 1.0 | .044 | .043 | .048 | .051 | .050 | .048 | .044 | .046 | .044 | .046 | .046 |
| 1.2 | .134 | .130 | .078 | .079 | .087 | .121 | .116 | .113 | .116 | .114 | .117 | |
| 1.4 | .320 | .318 | .148 | .153 | .200 | .290 | .279 | .280 | .281 | .284 | .281 | |
| 1.6 | .546 | .550 | .243 | .246 | .360 | .523 | .505 | .510 | .505 | .500 | .506 | |
| 1.8 | .753 | .748 | .383 | .391 | .537 | .729 | .716 | .717 | .718 | .721 | .720 | |
| 2.0 | .863 | .864 | .530 | .540 | .688 | .848 | .837 | .835 | .837 | .836 | .840 | |
Acknowledgments
This research was partially supported by NIH grants GM081535 and HL65462. The author is grateful to Drs Tao Wang and Rob Elston for sharing their data and computer program, to Fang Han for help with Figure 1, and to Dr Melanie Wall for helpful discussions that motivated the technical development in Appendices A–C. The author thanks the reviewers for helpful comments.
APPENDIX A
RELATIONSHIPS BETWEEN β̂c, β̂ and β̂M IN LINEAR REGRESSION
Denote Y = (Y1, …, Ym)′, X = (X.1, …, X.k), and β = (β1, …, βk)′. Note that Xc = X1 with 1 = (1, …, 1)′. Without loss of generality, assuming that Y and each predictor X.j have been centered at 0 such that the intercept term is 0. Because under the normality assumption, the maximum likelihood estimate (MLE) (and equivalently, the least squares estimate, LSE) of β is β̂ = (X′X)−1X′Y, we have X′Y = (X′X)β̂. On the other hand, the MLE of βc is
where is a row vector with the sum of its elements being
Thus β̂c is a linear combination of β̂1, …, β̂k. Because the sum of the coefficients equals to 1, it can be interpreted that β̂c is a weighted average of β̂1, …, β̂k. Furthermore, the weights equal to the column sums of X′X divided by , hence, the signs of the weights may not be positive; however, if all the X.j’s are positively correlated, then the weights are all positive.
With collinearity, it is well known that the components of β̂ may have large variances, which will not be able to explain the good performance of β̂c. In fact, by Cov(β̂) = σ2(X′X)−1 with Var(Yi) = σ2, we have
which is not directly related to possibly nearly singular (X′X)−1. Furthermore, because the MLE from the marginal models (2) is , where (X′X)d = Diag(X′X), it is easy to see that
APPENDIX B
RELATIONSHIPS BETWEEN β̂c, β̂ and β̂M IN GLM
For simplicity, we assume that there is no intercept term; otherwise, it can be handled similarly (but with more complicated notations). In general, there is no closed form solution for MLEs β̂ and β̂c, it is difficult to characterize the relationship between the two. However, with the canonical link function h( ), e.g., the logit link in logistic regression, the score equations for GLMs analogous to models (1)–(3) satisfy respectively (McCullagh and Nelder 1983)
where h−1( ) is the inverse of the link function and its operation on a vector or matrix is element-wise. Hence,
which implicitly determines a relationship between β̂c and β̂, and β̂c and β̂M.
APPENDIX C
CONVERGENCE RESULTS
Now we consider the following question: given that the joint model (1) is the true model, and thus (3) is possibly a misspecified model, what does β̂c converges to as the sample size goes to infinity? First, obviously, by the consistency of MLE under a correct model, β̂ converges to its true value β̂*. Second, based on the property of MLE under a mis-specified model (Huber 1967; White 1982), β̂c converges to , which is the value of β̂c that minimizes the Kullback-Leibler distance between the true distribution f(β*) of (Yi, Xi)’s and the distribution g(βc) implied by the misspecified model (3).
Because of the relationship between β̂ and β̂c, there is also an implied relation between β* and , say ; for example, r( ) is a linear function in linear regression. Therefore, testing H0: in model (3) is equivalent to testing H0: r(β*) = 0 on the individual parameters in model (1).
APPENDIX D
DISTRIBUTIONAL THEORY OF βM
We formulate fitting separate marginal models as joint modeling in the context of generalized estimating equations (GEE) to derive the asymptotic distribution of β̂M. In GEE, for “cluster” i, denote yi, = (yi1, …, yik)′ = Yi1k with 1k as a k-dimensional vector containing all ones, and zij = j. Fitting the k marginal models (2) is equivalent to fitting a GEE model:
with a working independence correlation structure. By the GEE theory (Liang and Zeger 1986), β̂M has an asymptotic Normal distribution with mean βM and a covariance matrix that can be consistently estimated by a sandwich (or robust) estimator, which is available from commonly used statistical packages such as R and SAS.
References
- Chapman JM, Whittaker J. Analysis of multiple SNPs in a candidate gene or region. Genetic Epidemiology. 2008;32:560–566. doi: 10.1002/gepi.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–428. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of p values for multiple correlated tests. Am J Hum Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR, Hinkley DV. Theoretical Statistics. Chapman and Hall; London: 1974. [Google Scholar]
- Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- Fan R, Knapp M. Genome association studies of complex diseases by case-control designs. Am J Hum Genet. 2003;72:850–868. doi: 10.1086/373966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer S, van Houwelingen HC. Testing against a high dimensional alternative. J Royal Stat Soc B. 2006;68:477–493. [Google Scholar]
- Huber PJ. Proceedings if the Fifth Berkeley Symposium in Mathematical Statistics and Probability. Berkeley: University of California Press; 1967. The behavior of maximum likelihood estimates under nonstandard conditions. [Google Scholar]
- Johnson NL, Kotz S. Distributions in Statistics, Continuous Univariate Distributions. 2. Boston: Houghton-Mifflin; 1970. [Google Scholar]
- Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- McCullagh P, Nelder JA. Generalized linear models. Chapman and Hall; London: 1983. [Google Scholar]
- Plenge RM, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin ZS, Niu T, Liu JS. Partition-ligation EM algorithm for haplotype inference with single nucleotide polymorphisms. Am J Hum Genet. 2002;71:1242–1247. doi: 10.1086/344207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder K, Bacanu SA, Sonpar V, Zhang X, Devlin B. Analysis of single-locus tests to detect gene/disease associations. Genet Epidemiol. 2005;28:207–219. doi: 10.1002/gepi.20050. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Elston RC. Improved power by use of a weighted score test for linkage disequilibrium mapping. Am J Hum Genet. 2007;80:353–360. doi: 10.1086/511312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Li M, Rebbeck T, Li H. U-statistics-based tests for multiple genes in genetic association studies. Annals of Human Genetics. 2008;72:821–833. doi: 10.1111/j.1469-1809.2008.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White H. Maximum likelihood estimation of misspecified models. Econometrica. 1982;50:1–25. [Google Scholar]
- Xiong M, Zhao J, Boerwinkle E. Generalized T2 test for genome association studies. Am J Hum Genet. 2002;70:1257–1268. doi: 10.1086/340392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-T. Approximate and asymptotic distributions of Chi-squared-type mixtures with applications. Journal of the American Statistical Association. 2005;100:273–285. [Google Scholar]
- Zhao H, Pfiffer R, Gail MH. Haplotype analysis in population genetics and association studies. Pharmacogenomics. 2003a;4:171–178. doi: 10.1517/phgs.4.2.171.22636. [DOI] [PubMed] [Google Scholar]
- Zhao LP, Li S, Khalid N. Assessing haplotype-based association with multiple SNPs in case-control studies. Am J Hum Genet. 2003b;72:1231–1250. doi: 10.1086/375140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Pan W. Power comparison of statistical tests of association with multiple SNPs with the GAW16 rheumatoid arthritis data. Research Report 2008–015, Division of Biostatistics, University of Minnesota. 2008 Available at www.biostat.umn.edu/rrs.php.