Abstract
Emergence of antiestrogen-resistant cells in MCF-7 cells during suppression of estrogen signaling is a widely accepted model of acquired breast cancer resistance to endocrine therapy. To obtain insight into the genomic basis of endocrine therapy resistance, we characterized MCF-7 monoclonal sublines that survived 21-day exposure to tamoxifen (T-series sublines) or fulvestrant (F-series sublines) and sublines unselected by drugs (U-series). All T/F-sublines were resistant to the cytocidal effects of both tamoxifen and fulvestrant. However, their responses to the cytostatic effects of fulvestrant varied greatly, and their remarkably diversified morphology showed no correlation with drug resistance. mRNA expression profiles of the U-sublines differed significantly from those of the T/F-sublines, whose transcriptomal responsiveness to fulvestrant was largely lost. A set of genes strongly expressed in the U-sublines successfully predicted metastasis-free survival of breast cancer patients. Most T/F-sublines shared highly homogeneous genomic DNA aberration patterns that were distinct from those of the U-sublines. Genomic DNA of the U-sublines harbored many aberrations that were not found in the T/F-sublines. These results suggest that the T/F-sublines are derived from a common monoclonal progenitor that lost transcriptomal responsiveness to antiestrogens as a consequence of genetic abnormalities many population doublings ago, not from the antiestrogen-sensitive cells in the same culture during the exposure to antiestrogens. Thus, the apparent acquisition of antiestrogen resistance by MCF-7 cells reflects selection of preexisting drug-resistant subpopulations without involving changes in individual cells. Our results suggest the importance of clonal selection in endocrine therapy resistance of breast cancer.
Keywords: acquired resistance, clonal selection, DNA copy number, fulvestrant, tumor heterogeneity
Approximately 50% to 70% of breast cancers are estrogen receptor (ERα) positive without amplification of the ERBB2/HER2 gene (1). Such ER+/HER2− breast cancers typically respond well to endocrine therapy (ET), which suppresses estrogen signaling in cancer cells and halts their proliferation (cytostatic effect) and/or induces apoptosis (cytocidal effect). Although ET is proven effective, its clinical benefit is seriously limited by drug resistance. Approximately 50% of ER+ metastatic breast cancers do not respond to ET at all (de novo resistance); even when they initially respond to ET, most eventually become resistant during the course of therapy (acquired resistance). Nearly 40% of early-stage breast cancers treated with ET relapse with ET-resistant disease. Thus, ET resistance is an important clinical challenge in breast cancer treatment.
The MCF-7 cell line is an extensively studied cell culture model of ER+/HER2− breast cancer (2). Because proliferation and survival of MCF-7 cells in culture or as xenograft are strictly dependent on estrogen (3–5), this cell line has been widely used to study mechanisms of ET resistance. Reflecting the extensive intratumoral heterogeneity of malignant epithelial cells in human breast cancer (6), MCF-7 is a polyclonal cell line consisting of highly heterogeneous cancer cells that differ remarkably in their phenotypic and cytogenetic characteristics (7, 8). It also has been demonstrated by several studies that different MCF-7 cell stocks maintained in different laboratories are greatly diversified in their estrogen-induced proliferation and cytogenetic characteristics (7, 9). However, previous studies on ET resistance of MCF-7 cells have rarely considered the highly heterogeneous nature of this model system as a significant factor.
To obtain insight into the possible importance of intratumoral malignant cell heterogeneity in the occurrence of ET resistance, we developed multiple monoclonal sublines from polyclonal MCF-7 cells and examined features of their antiestrogen (AE) resistance and genomic characteristics. We provide evidence that the ET resistance of MCF-7 cells does not result from the acquisition of drug resistance by individual cancer cells during drug exposure. Rather, the emergence of ET resistance during exposure to AE can be explained by the expansion of a monoclonal progenitor-derived, AE-resistant subpopulation already existing in the culture before initiation of ET. Thus, clonal selection seems to be important in the development of breast cancer resistance to ET.
Results
Phenotypic Features of MCF-7 Monoclonal Sublines.
We picked well-isolated colonies from the polyclonal MCF-7 cell culture to establish sublines (supporting information (SI) Fig. S1). All sublines characterized in the present study were confirmed as monoclonal by their homogeneous morphology. The unselected AE-sensitive U-series sublines were derived from colonies that emerged in low-density MCF-7 cultures without drug pressure. The AE-resistant sublines were isolated by exposing high-density MCF-7 cultures to fulvestrant (Fv, 10–100 nM) or 4-hydroxytamoxifen (Tam, 1 μM) for 21 days. Approximately 1 in 105 to 2 × 106 AE-exposed cells survived this drug pressure. After a 7-day drug-free recovery period, colonies were picked to establish the Tam-selected T-series and the Fv-selected F-series sublines. All T/F-sublines were maintained in the absence of AEs and were characterized within 20 population doublings. Morphology of the U-sublines was relatively monotonous and was similar to that of the original MCF-7 cells, whereas the appearance of the T/F-sublines varied greatly (Fig. 1A).
Fig. 1.
AE sensitivities of MCF-7 monoclonal sublines. (A) Phase-contrast images of the original MCF-7 cell culture and its monoclonal sublines isolated in the presence of Fv: 40 nM (F40–3, -6, -7) or 100 nM (F100–1, -3, -7, -16). (B and C) Drug resistance of Tam- or Fv-selected sublines. Cells were exposed to Fv (0.1 μM), 4-hydroxyTam (1 μM), paclitaxel (1 μM), or vehicle (0.1% ethanol) for 10 days, followed by visualization (B) and quantitation (C) of survived cells by sulforhodamine-B staining. Survived cell numbers relative to vehicle controls (mean ± SEM) are shown for individual sublines (open symbols) and each group (closed symbols). (D) AE-induced activation of caspase-7. Relative numbers of DEVDase-positive cells (mean ± SEM, n ≥ 3) after 48-hour exposure to 0.1 μM Fv (open symbols) or vehicle (closed symbols) are plotted. Small symbols represent individual sublines. Large symbols represent groups. (E) Cell cycle arrest by Fv. Unselected, Tam-selected, and Fv-selected sublines were exposed to Fv (0.1 μM) or vehicle (0.1% ethanol) for 48 h. Symbols indicate the relative S-phase cell population in cultures exposed to vehicle (open symbols) or to Fv (closed symbols). All data indicate the mean of at least 3 independent assays whose SEs were not greater than 10% of the means.
A 10-day exposure of the polyclonal MCF-7 cells and U-sublines to AEs (0.1 μM Fv or 1 μM Tam) caused massive cell death (Fig. 1 B and C). Two U-sublines (U-8 and U-13) showed significant resistance to Tam but still were sensitive to Fv, presumably reflecting the stronger cytocidal effects of Fv than Tam (10). Most T/F-sublines showed strong resistance to both Tam and Fv. However, a few F-sublines showed measurable degrees of sensitivity to Fv despite their initial selection by Fv, suggesting that AE resistance in the F-sublines is somewhat fluctuant. Supporting this interpretation, the F40–6 and F100–16 sublines, which initially showed strong Fv resistance, lost up to 50% of Fv resistance after a 30-day culture in the absence of AE (Fig. S2). However, extending the AE-free culture by an additional month did not significantly reduce the AE resistance of these sublines further, suggesting that the AE resistance of the F-sublines involves both irreversible and reversible mechanisms. All sublines retained strong sensitivity to paclitaxel (11), indicating that the resistance of the T/F-sublines was specific to AEs.
Because MCF-7 cells are null for the CASP3 gene, their apoptosis often involves sequential activation of caspases 9, 7, and 6 (12). The DEVDase assay demonstrated intact Fv-induced caspase-7 activation in all MCF-7 sublines (Fig. 1D). Therefore, the AE resistance of the T/F-sublines was not attributed directly to inhibition of the executioner caspases.
AEs have cytostatic and cytocidal effects. When the U-sublines were exposed to 0.1 μM Fv for 48 h, S-phase cells disappeared almost completely (Fig. 1E). In contrast, the cytostatic effects of Fv on the T/F-sublines varied remarkably (Figs. 1E and S3). Because the cytocidal Fv action requires a longer period of drug exposure (4, 5), cell viability was intact when cell cycle analysis was performed. The lack of correlation between the cytostatic and cytocidal effects of Fv suggests independent mechanisms for these 2 Fv effects.
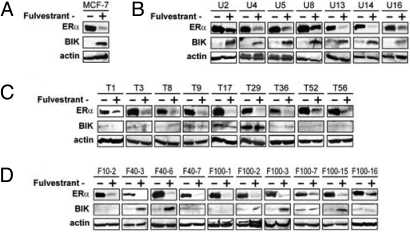
We previously demonstrated that the cytocidal effect of Fv in MCF-7 cells requires induced expression of the BIK proapoptotic protein (4, 5, 13, 14). In all AE-sensitive sublines and in the polyclonal MCF-7 cells, Fv strongly induced BIK expression and suppressed ERα expression (Fig. 2 A and B). Fv suppressed ERα expression in all AE-resistant sublines as strongly as in AE-sensitive cells, indicating that the AE resistance was not caused by enhanced drug metabolism or active exclusion from cells. In contrast, Fv failed to induce BIK in any of the T-sublines (Fig. 2C) and in 5 of 10 F-sublines examined (Fig. 2D). These results suggest that the AE resistance of the T/F-sublines may be related to the failed BIK induction.
Fig. 2.
Expression of ERα and BIK proteins in MCF-7 sublines. Cells were exposed to Fv (0.1 μM) or vehicle (0.1% ethanol) for 48 h and subjected to Western blotting for detection of ER↑ and BIK. (A) Original MCF-7 cells, (B) unselected, AE-sensitive sublines, (C) Tam-selected, and (D) Fv-selected AE-resistant sublines.
AE-Resistant MCF-7 Sublines Lack Transcriptomal Responses to Fv Exposure.
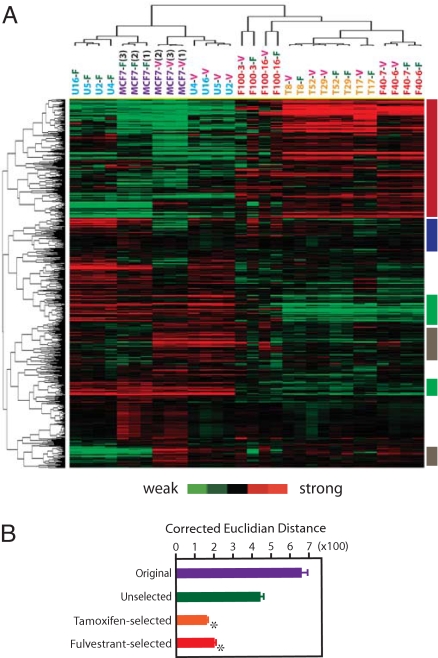
To determine transcriptomal responses of the MCF-7 sublines to Fv, cells were exposed to 0.1 μM Fv or vehicle for 48 h and subjected to microarray determination of mRNA expression. Hierarchical clustering analysis demonstrated clear transcriptomal separation between the AE-sensitive and -resistant cells (Fig. 3A). Supervised permutation tests (14) confirmed significant transcriptomal differences between the AE-sensitive and -resistant cells (P < 0.01). Transcriptomal profiles of the AE-sensitive cells differed remarkably between the Fv-exposed (-F suffix) and vehicle-exposed (-V suffix) conditions (P < 0.01). The strong transcriptomal similarity between the monoclonal U-sublines and the polyclonal MCF-7 cells suggested that the U-sublines represented a dominant population in the polyclonal MCF-7 cell culture. In contrast, the AE-resistant sublines showed remarkably diminished transcriptomal responses to Fv exposure, as indicated by the failed separation of Fv- and vehicle-exposed AE-resistant cells by hierarchical clustering (Fig. 3A). Transcriptomal distances calculated between Fv- and vehicle-exposed cells confirmed significant reduction in Fv responsiveness of the T/F-sublines (Fig. 3B, P < 0.001).
Fig. 3.
Transcriptomal profiles of MCF-7 monoclonal sublines. (A) Two-way hierarchical clustering of Tam-selected (T8, T17, T29, and T52), Fv-selected (F40–6, F40–7, F100–3, and F100–16), and unselected (U2, U4, U5, and U16) sublines and original MCF-7 culture (3 independent cultures) exposed to Fv (-F suffix; 0.1 μM) or vehicle (-V suffix; 0.1% ethanol) for 48 h. Columns represent cell culture samples; rows represent genes. Red and green vertical color bars indicate genes differentially expressed between AE-sensitive and -resistant cells; blue and gray vertical color bars represent genes that do not respond to Fv in AE-resistant cells. (B) Corrected Euclidian transcriptomal distances between the Fv- and vehicle-exposed cultures. Asterisks indicate significant difference (P < 0.001) from unselected sublines.
Genomic DNA Copy Number Analysis Reflects the Monoclonal Origin of the AE-Resistant MCF-7 Sublines.
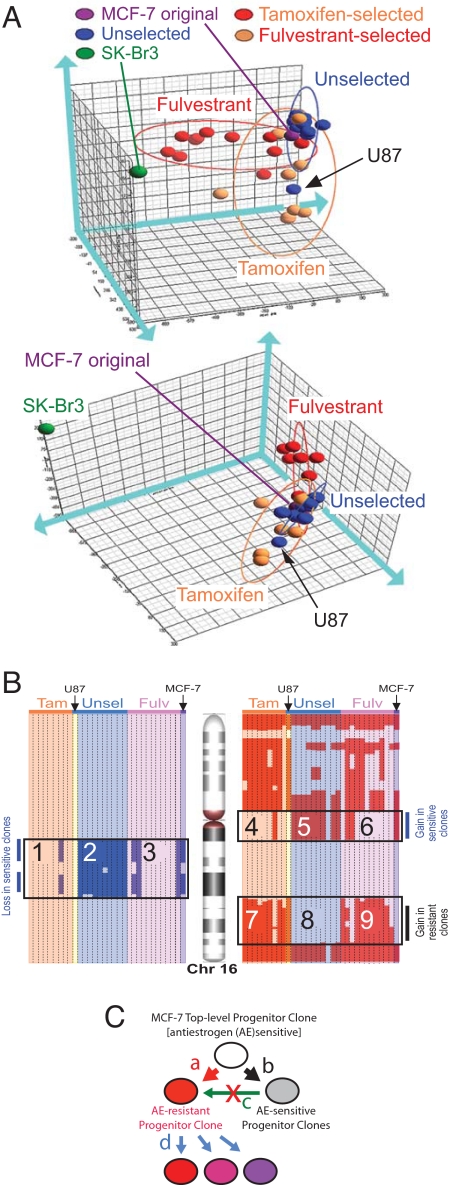
To obtain insight into the origin of the AE-resistant sublines, we performed genomic DNA (gDNA) copy number analysis using an Affymetrix 250K SNP microarray that had submegabase resolution power (15). The 3D plot of a principal component analysis (PCA) on distribution of the amplification/deletion events in the gDNA (Fig. 4A) showed clear separation of all MCF-7 sublines from another human breast cancer cell line (SKBr3). The overall similarity of all original and subline MCF-7 cells reflected 60 gDNA events shared by them (Table S1.1). These events included 14 out of 18 gDNA aberrations previously determined by Jones et al. for the MCF-7 BUS stock using comparative genomic hybridization (CGH) (9). In the PCA space, all U-sublines are located in the close vicinity of the original MCF-7 cells, indicating that the AE-sensitive MCF-7 cells share close gDNA aberration patterns. The T- and F-sublines formed independent ellipsoids, which partly overlapped with that of the U-sublines. The subline U87, which was a U-series clone but belonged to the T-subline ellipsoid in the PCA space, was an exception. Because subline U87 had weakest Fv sensitivity among all U-series sublines (Fig. S4), it might be appropriate to consider this subline a member of the T-series by genomic and AE-resistance criteria, although it originally was isolated without AE pressure.
Fig. 4.
gDNA copy number profiling of MCF-7 sublines. (A) Principal component analysis (PCA). Whole-genome copy number profiles of the MCF-7 sublines, the original MCF-7 culture, and SK-Br3 cells are subjected to dimensionality reduction and projected in a 3D space that is shown from 2 different angles of view. Blue, orange, and red ellipsoids represent the PCA spaces for the U-, T-, and F-series sublines, respectively. The arrow indicates the U87 subline, which belongs to the U-series but has its gDNA aberration profile and located within the T-series ellipsoid. (B) Copy number profiling of chromosome 16. Columns represent individual sublines. gDNA segments with increased and decreased copy numbers are indicated by red and blue, respectively. Fulv, fulvestrant selected; Tam, tamoxifen selected; Unsel, unselected series of monoclonal sublines. The original MCF-7 cells and the U-series sublines were sensitive to the cytocidal fulvestrant actions; the T- and F-series sublines and U87 subline (which belongs to the U-series) were resistant. (C) Evolution of the AE-resistant subpopulations in MCF-7 cells. From the top-level progenitor clone, an AE-resistant progenitor clone and an AE-sensitive progenitor clone were generated through accumulation of different sets of gDNA aberrations (arrows a and b, respectively). The AE-sensitive progenitor clone propagated to form the major subpopulation in MCF-7 cells (arrow d); the progenies of the AE-resistant progenitor clone remained as a minor subpopulation. The AE-resistant progenitor clone was not a progeny of the AE-sensitive progenitor clone (arrow c). Progenies of the AE-resistant clone evolved diversified phenotypic features through genetic and nongenetic changes.
We identified 18 gDNA events relatively specific to the AE-sensitive cells (Fisher test, P < 0.05; Table S1.3). For example, in chr16q, copy number reduced selectively in the AE-sensitive cells in 1 region (Fig. 4B; compare and) but increased in another region (compare and). On the other hand, we identified 36 gDNA events relatively specific to the AE-resistant cells (Table S1.2) including a region amplified in chr16q (compare and). gDNA events distinguishing the AE-sensitive and -resistant cells were distributed widely in 19 chromosomes (Table S1.2 and S1.3). It is highly unlikely that such diverse gDNA events occurred in multiple independent clones during the 21-day AE selection period. There were 12 gDNA events relatively specific to the F-subline and 17 gDNA events relatively specific to the T-subline (Fig. S5 and Table S1.4 and S1.5), suggesting heterogeneity within the AE-resistant sublines. Interestingly, the gain at 9q31-q34 previously detected by CGH in the MCF-7 SOP stock but not in the BUS stock (9) was found in many U- and F-sublines but in only 1 T-subline (Fig. S5). A complete list of gDNA aberrations is provided as Fig. S5 and Table S1. These observations support the notion that most, if not all, AE-resistant sublines existing in the polyclonal MCF-7 cell line were derived from a common monoclonal progenitor and that most AE-sensitive sublines were derived from another monoclonal progenitor. The AE-resistant progenitor clone was not derived from the AE-sensitive cells that presently dominate the MCF-7 cell line because the AE-resistant sublines did not inherit many gDNA events characteristic to the AE-sensitive cells. Fig. 4C illustrates a model for evolution of MCF-7 subpopulations with discrete AE sensitivities.
Genes Selectively Expressed in the AE-Sensitive MCF-7 Sublines Predict Distant Metastasis-Free Prognosis of Breast Cancer Patients.
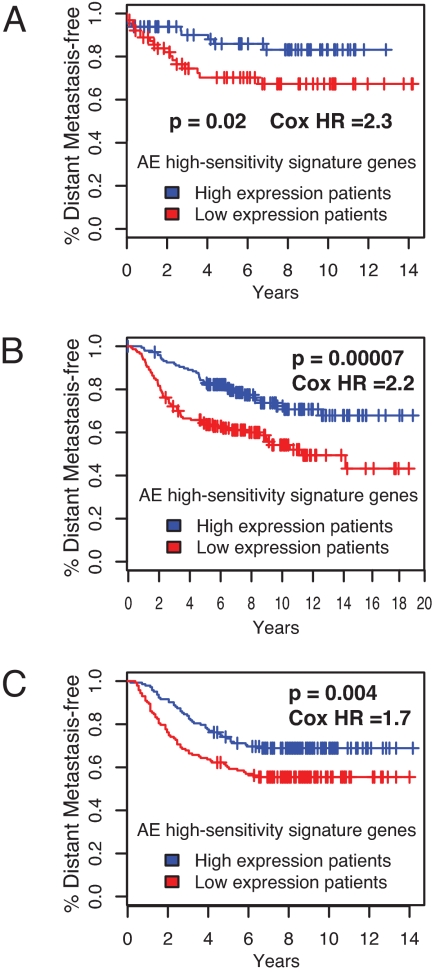
In an attempt to obtain clinically relevant information from the MCF-7 monoclonal subline model, we performed in silico prognosis prediction for breast cancer patients characterized in previously published studies using genes differentially expressed in the AE-sensitive and -resistant sublines. We identified 111 genes expressed more strongly in the AE-sensitive sublines than in the AE-resistant sublines (false discovery rate ≤ 0.01 and fold change > 5; Table S2). Thus, these 111 genes constituted a signature for the AE-sensitive phenotype. Reference data sets were obtained from 3 independent studies that determined transcriptomal profiles of breast cancer tissues and their relevance for the prognosis of patients, namely, the studies reported by Chin et al. (130 patients) (16), van de Vijver et al. (295 patients) (17), and Wang et al. (286 patients) (18). Patients were divided into 2 groups based on the strength of mRNA expression for the 111 AE-sensitivity signature genes. In all 3 studies, Kaplan-Meier curves drawn for these 2 groups of patients showed that periods of distant metastasis-free survival were significantly longer in the group more strongly expressing the AE-sensitivity signature genes than in the other group (Fig. 5), supporting the notion that information obtained from the MCF-7 subline profiling can be related to biological features of clinical breast cancer.
Fig. 5.
Prediction of a metastasis-free period of breast cancer patients by tumor expression of AE-sensitivity signature genes identified by MCF-7 monoclonal subline analysis. Patients were divided into 2 groups based on the expression of the AE-sensitivity signature genes. Kaplan-Meier curves for emergence of distant metastases in these 2 groups are based on large-scale clinical data described by (A) Chin et al. (16), (B) van de Vijver et al. (17), and (C) Wang et al. (18). One-sided p-values and Cox hazard ratios between the 2 groups are shown.
To obtain insight into how these differentially expressed genes affect MCF-7 cell biology, we performed pathway-enrichment analyses using the Gene Ontology (GO) and the KEGG databases through the GeneSifter server. The GO analysis suggested strong suppression of cellular metabolisms—both anabolic and catabolic—in the AE- resistant cells (Fig. S6A). The KEGG analysis suggested enhanced cell adhesion and motility (Fig. S6 B–D; Axon guidance pathway represents cell migration guided by adhesion molecules and cytokines). These results suggest that the AE-resistant MCF-7 sublines are metabolically less active than the AE-sensitive sublines, but their cell surface activities (i.e., adhesion and migration) are augmented. These predicted features may be consistent with the notion that AE-resistant cells also have metastatic potency.
Discussion
Breast cancers often show pronounced intratumoral heterogeneity (6), and the MCF-7 cell line also consists of highly heterogeneous breast cancer cells with remarkable phenotypic and genetic variations (7–9). MCF-7 cells and their variants seem to “acquire” ET resistance after prolonged AE exposure or estrogen deprivation in cell culture (19) or as xenografts (3). This phenomenon is widely accepted as a model for clinically observed acquisition of ET resistance by breast cancers. However, prior studies often overlooked the highly heterogeneous nature of the MCF-7 model, leaving it unclear whether molecular-level alterations observed during the acquisition of ET resistance by tumors or cell cultures truly occur in each individual cell.
Our present study provides genetic evidence that the ET resistance observed with MCF-7 cells exposed to AEs for relatively short periods of time (21 days) is not acquired by individual cells. The gDNA aberration pattern is strongly conserved among the AE-resistant sublines, whereas the AE-sensitive sublines share a distinct pattern of gDNA aberrations. Many gDNA events shared by the AE-sensitive sublines are not found in the AE-resistant sublines (Fig. 4). These results challenge the prevailing but somewhat untested assumption that the AE-resistant MCF-7 variants are derived from the AE-sensitive cells concomitantly present in the same cell culture or xenograft. Our data support an alternative explanation, namely that the apparent “acquisition” of ET resistance by MCF-7 cells results primarily from the selection of AE-resistant cells already existing in the polyclonal MCF-7 cell culture before initiation of the drug challenge. It would be interesting to test the clonality of other ET-resistant MCF-7 cell variants that were isolated using protocols involving prolonged estrogen-deprived cell culture conditions [e.g., the Long-Term Estrogen Deprivation (LTED) cells, which were isolated through estrogen deprivation for 6 months (19)]. If their clonality profiles are indistinguishable from the ET-sensitive MCF-7 cells, the notion that these cells acquired ET resistance by changing their biological features during estrogen deprivation will be supported. If their clonality profiles differ significantly (as was the case in our present study), the dominant role of clonal selection in the process through which MCF-7 cell cultures gain ET resistance will be supported. It is possible, however, that certain MCF-7 subpopulations sharing a specific gDNA aberration profile are initially drug sensitive but are highly predisposed to acquire resistance during estrogen deprivation. It also is plausible that the well-known cytogenetic diversity observed among different MCF-7 polyclonal stocks (7, 9) might be caused by unintended selection resulting from the use of different cell culture conditions in different laboratories. The cytogenetic diversity of MCF-7 cells might be enhanced further by estrogen-induced genomic instability, which involves defective chromosomal segregation caused by interference with regulatory signaling kinases (20). Thus, future studies involving high-resolution gDNA copy number analyses of breast cancer cell cultures and tissues may provide important insight into the relative importance of the clonal selection and the gain of drug resistance in the individual cell in the clinically observed ET resistance.
The relatively homogeneous gDNA aberration pattern conserved among the AE-resistant sublines suggests that these cells were derived from a monoclonal progenitor that presumably lost responsiveness to induce transcriptomal changes upon exposure to AE (Fig. 3) and the AE-induced BIK protein expression (Fig. 2). Lack of the transcriptomal responsiveness to Fv exposure observed with our AE-resistant sublines (Fig. 3) agrees with a recent report by Iorns et al. that Tam-resistant MCF7 variants artificially produced by RNAi knockdown of CDK10 showed estrogen-independent cell proliferation and survival (21). This finding is in contrast to the estrogen hypersensitivity of AE-resistant MCF-7 variants produced by long-term estrogen deprivation as reported by another laboratory (19). Other biological characteristics of the AE-resistant MCF-7 sublines also may be relevant to the process of clinical breast cancer metastasis, as suggested by the segregation of the metastasis-free survival curves between 2 groups of breast cancer patients that were classified based on expression of a set of genes differentially expressed between the AE-sensitive and -resistant sublines (Fig. 5).
Despite their relatively homogeneous genomic DNA aberration profiles, the AE-resistant sublines showed significantly heterogeneous phenotypic characteristics in morphology (Fig. 1A), relative strength of AE resistance (Fig. 1C), sensitivity to the cytostatic action of AE (Figs. 1E and S3), and AE-induced BIK expression (Fig. 2). Such phenotypic heterogeneity might indicate the existence of other mechanisms, in addition to the gDNA amplifications and deletions, that modify the AE sensitivity of MCF-7 cells, and such mechanisms could be relevant to the partially reversible nature of the AE resistance of some MCF-7 sublines (Fig. S2). Epigenetic changes such as DNA methylation at the promoter CpG sequences or alterations in chromatic structures are possible candidates of such mechanisms.
In summary, our study provides genetic evidence that all AE-resistant MCF-7 sublines obtained through AE selection of the polyclonal MCF-7 cells were derived from a common monoclonal ancestor. The commonly observed acquisition of AE resistance by MCF-7 cells is not explained by changes in the individual cancer cell during the AE challenge; rather, it seems to reflect selection of minor, preexisting, AE-resistant subpopulations. To obtain a better understanding of the basis of the ET resistance, future studies are needed to determine the intratumoral malignant cell clonality during ET of human breast cancer tissues.
Materials and Methods
Cell Culture.
Maintenance of MCF-7 cells (BUS stock) in a xenoestrogen-controlled environment and the determination of cytocidal AE effects and caspase-7 activity were described in our previous studies (13, 14). The cell-cycle profile was determined by propidium iodide staining and FACS (Becton Dickinson). Fv was provided from AstraZeneca. 4-hydroxyTam and paclitaxel were purchased from Sigma.
Gene Expression Profiling.
ERα and BIK protein expression was determined by Western blotting (4, 5). Determination of MCF-7 transcriptome using Affymetrix U133-plus 2.0 microarray and 2D hierarchical clustering were performed as described in our previous studies (13, 14). To evaluate transcriptomal distances, the Euclidian distance of log-transformed Affymetrix microarray signal intensities was calculated for each gene and then divided by the average signal intensities of the gene. Summation of these corrected distances was used as the indicator of transcriptomal changes induced by Fv exposure.
gDNA Copy Number Analysis.
Copy numbers of gDNA segments were determined using Affymetrix Human Mapping 250K/StyI SNP microarray and Partek Genomics Suite software, which implemented the BRLMM algorithm to detect copy number aberrations against the HapMap normal human genome standards (22).
Statistics and Bioinformatics.
PCA was performed using Partek Genomics Suite. Specificity of gDNA aberrations to the U-, T-, and F-series sublines was determined by Fisher's exact probability test (α < 0.05). Kaplan-Meier survival studies and Cox proportional hazard risk analyses for evaluation of signature gene sets were performed using an approach similar to that we described previously (23, 24). Briefly, differential mRNA expression between the Fv-sensitive and -resistant MCF-7 sublines was characterized by a t test with the Benjamin-Hochberg adjustment for multiple hypothesis testing. We selected 111 genes (Table S2) whose mRNA expression was at least 5-fold greater in the Fv-sensitive sublines than in Fv-resistant sublines (adjusted p value ≤ 0.01). These genes were mapped to the appropriate platform for each survival data set. For each sample, the expression values of the genes were averaged to form a sensitivity signature score. The samples were divided into those with sensitivity signature score above and below the median.
Acknowledgments.
We thank Ana Soto and Carlos Sonnenschein (Tufts University) for MCF-7 BUS cell culture, Ai Muto (Kyoto University) for bioinformatics, and Haley Ellis for technical support. We also thank Steven Isakoff, Paul Goss, and Jeff Settleman for useful discussions. This study was supported by Grants FAS0703860 and KG090515 from Susan G. Komen for Cure, from the AVON Breast Cancer Crusade Fund (AVON Foundation), and from AstraZeneca Development Compound Research Fund (to T.S). S.R. and B.S.W. were supported by National Cancer Institute Breast Cancer Specialized Program of Research Excellence Award 2P50 CA89393–07 to the Dana-Farber/Harvard Cancer Center and a Department of Defense Center of Excellence in Breast Cancer Award BC05-COE W81XH-06-2-0033.
Footnotes
The authors declare no conflict of interest.
Data deposition footnote: The microarray data presented in this paper have been deposited in the Gene Expression Omnibus (GEO) (accession no. GSE14986).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907560106/DCSupplemental.
References
- 1.Jordan VC, Brodie AM. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids. 2007;72:7–25. doi: 10.1016/j.steroids.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: An update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 3.Brodie A, et al. Therapeutic observations in MCF-7 aromatase xenografts. Clin Cancer Res. 2005;11:884s–888s. [PubMed] [Google Scholar]
- 4.Hur J, et al. Regulation of expression of BIK proapoptotic protein in human breast cancer cells: p53-dependent induction of BIK mRNA by fulvestrant and proteasomal degradation of BIK protein. Cancer Res. 2006;66:10153–10161. doi: 10.1158/0008-5472.CAN-05-3696. [DOI] [PubMed] [Google Scholar]
- 5.Hur J, et al. The Bik BH3-only protein is induced in estrogen-starved and antiestrogen-exposed breast cancer cells and provokes apoptosis. Proc Natl Acad Sci USA. 2004;101:2351–2356. doi: 10.1073/pnas.0307337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polyak K. Breast cancer: Origins and evolution. J Clin Invest. 2007;117:3155–3163. doi: 10.1172/JCI33295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nugoli M, et al. Genetic variability in MCF-7 sublines: Evidence of rapid genomic and RNA expression profile modifications. BMC Cancer. 2003;3:13. doi: 10.1186/1471-2407-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Resnicoff M, et al. Subpopulations of MCF7 cells separated by Percoll gradient centrifugation: A model to analyze the heterogeneity of human breast cancer. Proc Natl Acad Sci USA. 1987;84:7295–7299. doi: 10.1073/pnas.84.20.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones C, et al. Comparative genomic hybridization reveals extensive variation among different MCF-7 cell stocks. Cancer Genet Cytogenet. 2000;117:153–158. doi: 10.1016/s0165-4608(99)00158-2. [DOI] [PubMed] [Google Scholar]
- 10.Diel P, Smolnikar K, Michna H. The pure antiestrogen ICI 182780 is more effective in the induction of apoptosis and down regulation of BCL-2 than tamoxifen in MCF-7 cells. Breast Cancer Res Treat. 1999;58:87–97. doi: 10.1023/a:1006338123126. [DOI] [PubMed] [Google Scholar]
- 11.Howell SJ, Anderson E, Hunter T, Farnie G, Clarke RB. Prolactin receptor antagonism reduces the clonogenic capacity of breast cancer cells and potentiates doxorubicin and paclitaxel cytotoxicity. Breast Cancer Research: BCR. 2008;10:R68. doi: 10.1186/bcr2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang Y, Yan C, Schor NF. Apoptosis in the absence of caspase 3. Oncogene. 2001;20:6570–6578. doi: 10.1038/sj.onc.1204815. [DOI] [PubMed] [Google Scholar]
- 13.Coser KR, et al. Global analysis of ligand sensitivity of estrogen inducible and suppressible genes in MCF7/BUS breast cancer cells by DNA microarray. Proc Natl Acad Sci USA. 2003;100:13994–13999. doi: 10.1073/pnas.2235866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shioda T, et al. Importance of dosage standardization for interpreting transcriptomal signature profiles: Evidence from studies of xenoestrogens. Proc Natl Acad Sci USA. 2006;103:12033–12038. doi: 10.1073/pnas.0605341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardina PJ, Lo KC, Lee W, Cowell JK, Turpaz Y. Ploidy status and copy number aberrations in primary glioblastomas defined by integrated analysis of allelic ratios, signal ratios and loss of heterozygosity using 500K SNP Mapping Arrays. BMC Genomics. 2008;9:489. doi: 10.1186/1471-2164-9-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chin K, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 17.van de Vijver MJ, et al. A gene-expression signature as a predictor of survival in breast cancer. New England Journal of Medicine. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 19.Santen RJ, Song RX, Zhang Z, Yue W, Kumar R. Adaptive hypersensitivity to estrogen: Mechanism for sequential responses to hormonal therapy in breast cancer. Clin Cancer Res. 2004;10:337S–345S. doi: 10.1158/1078-0432.ccr-031207. [DOI] [PubMed] [Google Scholar]
- 20.Kabil A, Silva E, Kortenkamp A. Estrogens and genomic instability in human breast cancer cells—involvement of Src/Raf/Erk signaling in micronucleus formation by estrogenic chemicals. Carcinogenesis. 2008;29:1862–1868. doi: 10.1093/carcin/bgn138. [DOI] [PubMed] [Google Scholar]
- 21.Iorns E, et al. Identification of CDK10 as an important determinant of resistance to endocrine therapy for breast cancer. Cancer Cell. 2008;13:91–104. doi: 10.1016/j.ccr.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, et al. Assessing batch effects of genotype calling algorithm BRLMM for the Affymetrix GeneChip Human Mapping 500 K array set using 270 HapMap samples. BMC Bioinformatics. 2008;9(Suppl 9):S17. doi: 10.1186/1471-2105-9-S9-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wittner BS, et al. Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res. 2008;14:2988–2993. doi: 10.1158/1078-0432.CCR-07-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]