Abstract
Maturity-onset diabetes of the young (MODY) is a subtype of diabetes defined by an autosomal pattern of inheritance and a young age at onset, often before age 25. MODY is genetically heterogeneous, with 8 distinct MODY genes identified to date and more believed to exist. We resequenced 732 kb of genomic sequence at 8p23 in 6 MODY families unlinked to known MODY genes that showed evidence of linkage at that location. Of the 410 sequence differences that we identified, 5 had a frequency <1% in the general population and segregated with diabetes in 3 of the families, including the 2 showing the strongest support for linkage at this location. The 5 mutations were all placed within 100 kb corresponding to the BLK gene. One resulted in an Ala71Thr substitution; the other 4 were noncoding and determined decreased in vitro promoter activity in reporter gene experiments. We found that BLK—a nonreceptor tyrosine-kinase of the src family of proto-oncogenes—is expressed in β-cells where it enhances insulin synthesis and secretion in response to glucose by up-regulating transcription factors Pdx1 and Nkx6.1. These actions are greatly attenuated by the Ala71Thr mutation. These findings point to BLK as a previously unrecognized modulator of β-cell function, the deficit of which may lead to the development of diabetes.
Keywords: beta cells, genetics, MODY, tyrosine kinase
Maturity-onset diabetes of the young or MODY (MIM 606391) is a form of diabetes characterized by an autosomal dominant pattern of inheritance and a relatively young onset (1). The availability of large families with multiple affected members has facilitated studies of this type of diabetes, leading to the identification of 8 distinct MODY genes: HNF4A, encoding hepatocyte nuclear factor 4α (2); GCK, encoding glucokinase (3); TCF1, encoding hepatocyte nuclear factor 1α (4); IPF1, encoding insulin promoter factor 1 (5); TCF2, encoding hepatocyte nuclear factor 1β (6); NEUROD1, encoding neurogenic differentiation 1 (7); KLF11, encoding for kruppel-like factor 11 (8); and CEL, encoding carboxyl-ester lipase (9). However, 15% or more of MODY cases are not accounted for by mutations in these genes, suggesting the existence of as yet undiscovered MODY genes in addition to those identified to date (10, 11). Here we report the identification of mutations at the B-lymphocyte kinase (BLK) locus that segregate with diabetes in MODY families unlinked to known MODY genes and have detrimental effects on BLK expression or activity in insulin secreting cells. We further show that BLK is a previously unrecognized modulator of insulin synthesis and secretion that enhances the expression of key β-cell transcription factors Pdx-1 and Nkx6.1.
Results
We previously described a 2.5 Mb region on chromosome 8p23 that segregated with diabetes in extended families with MODY not caused by mutations in known MODY genes (12). To identify causal mutations, we resequenced all transcripts described in this interval as of January 2008 (corresponding to 15 RefSeq genes and 20 EST-derived genes) in 2 diabetic members from each of 6 families supporting linkage at this location. All exons and exon-intron boundaries of each gene or EST, at least 2 Kb of the 5′ and 3′ flanking regions, all conserved intronic segments, and some intergenic regions were included in the screening. A total of 732 kb was resequenced (Table S1). Of the 410 sequence differences that were identified, 5 co-segregated with diabetes and had a frequency <1% in the general population. The 5 mutations—all placed within 100 kb of genomic sequence—were found in 3 families, including the 2 showing the strongest support for linkage at this location (Table 1). Three of the mutations occurred together as a haplotype in family F8; the other 2 mutations occurred uniquely in families F9 and F17. The unique mutations (families F9 and F17) were not detected among nondiabetic subjects, whereas the haplotype of family F8 was also found in 2 of 336 unrelated nondiabetic subjects (Table 1). Both nondiabetic haplotype carriers were lean (BMI = 20 and 24) and relatively young at examination (age 39 and 45). The segregation of the mutations in the 3 families is shown in Fig. S1. In agreement with the results of the linkage analysis (12), 21 out of 25 mutation carriers had diabetes or IGT (84% penetrance). One of the individuals carrying the risk mutation but not expressing abnormal glucose tolerance was a 10-year-old individual from family F9. The other 3 carriers belonged to family F8 and were older. Two of them had BMI <28. The LOD scores for segregation of the mutations with diabetes, estimated with the model specified in (12), were 1.16 for family F8, 1.63 for family F9, and 0.97 for family F17. The LOD score of family F8 went up to 1.90 if the reduced penetrance observed for the mutated haplotype among nonobese subjects (BMI <28) was factored into the model. The phenotype of affected mutation carriers resembled that of a previously described family with autosomal dominant diabetes linked to a NEUROD1 mutation (7), being characterized by overweight and a relative, rather than an absolute, insulin secretion deficit as observed instead in MODY3 (Table 2). Indeed, the serum insulin levels of mutation carriers who were not insulin-treated were similar to those of nondiabetic individuals, although such levels were not sufficient for the maintenance of normoglycemia (Table 2).
Table 1.
Characteristics of mutations segregating with diabetes in the linked interval at 8p23
| Family | Origin | Chr | Position* | Substitution | Nondiabetic subjects |
|
|---|---|---|---|---|---|---|
| Whites (n = 672)† | AA (n = 1,154)† | |||||
| F17 | AA | 8 | 11,369,157 | G > A | 0 (0.00) | 0 (0.00) |
| F8 | W | 8 | 11,442,985 | G > A (A71T) | 2 (0.003)‡ | NT |
| F8 | W | 8 | 11,459,364 | T > G | 2 (0.003)‡ | NT |
| F9 | W | 8 | 11,459,531 | G > T | 0 (0.00) | NT |
| F8 | W | 8 | 11,468,050 | C > T | 2 (0.003)‡ | NT |
AA, African-American; W, White; NT, not tested.
*Position according to NCBI Build 36.1.
†Number of chromosomes that were tested.
‡These 3 mutations always occurred together as a haplotype.
Table 2.
Clinical characteristics of affected BLK mutation carriers as compared with carriers of NEUROD1 R111L and MODY3 mutations and nondiabetic subjects
| BLK mutations | NEUROD1 R111L* | HNF-1α (MODY3) mutations† | Nondiabetic subjects† | |
|---|---|---|---|---|
| Families (n) | 3 | 1 | 13 | 36 |
| Individuals (M/F) | 21 (9/12) | 5 (1/4) | 100 (44/56) | 223 (117/106) |
| Age at diagnosis, years | 31 ± 16 | 37 ± 5 | 21 ± 10 | - |
| Age at examination, years | 45 ± 20 | 46 ± 17 | 39 ± 17 | 44 ± 18 |
| BMI, kg/m2 | 28.7 ± 5 | 30.1 ± 2 | 24.6 ± 5 | 26.3 ± 4.8 |
| Treatment | ||||
| Diet only, % | 18.2 | 40.0 | 34.0 | - |
| Oral agents, % | 22.7 | 20.0 | 25.0 | - |
| Insulin, % | 59.1 | 40.0 | 41.0 | - |
| Hba1c, %‡ | 7.7 ± 1.6 | 6.1 ± 0.5 | 6.9 ± 1.5 | 5.4 ± 0.4 |
| Fasting glucose, mg/dl‡ | 166 ± 60 | 124 ± 22 | 126 ± 45 | 87 ± 14 |
| 2 hr glucose, mg/dl§ | 312 ± 79 | 202 ± 53 | 252 ± 95 | 90 ± 24 |
| Fasting Insulin, μu/ml‡ | 14.9 ± 7 | 17.0 ± 2 | 6.5 ± 2.4 | 11.7 ± 7.1 |
| 2 hr–insulin, μu/ml§ | 40.8 ± 24 | 59.7 ± 16 | 12.0 ± 7.5 | 39.8 ± 39.4 |
| Fasting C-peptide, ng/ml¶ | 0.87 ± 0.4 | 1.05 ± 0.2 | 0.71 ± 0.5 | ND |
Data are means ± SD.
*From reference 7.
†From reference 11. Non-diabetic subjects are non-diabetic members of MODY families.
‡Measured on non-insulin treated subjects (n = 7 for BLK, n = 3 for NEUROD1, n = 59 for MODY3, n = 156 for non-diabetic subjects).
§Measured on non-insulin treated subjects (n = 6 for BLK, n = 3 for NEUROD1, n = 59 for MODY3, n = 126 for non-diabetic subjects).
¶Measured on insulin-treated subjects (n = 6 for BLK, n = 2 for NEUROD1, and n = 34 for MODY3).
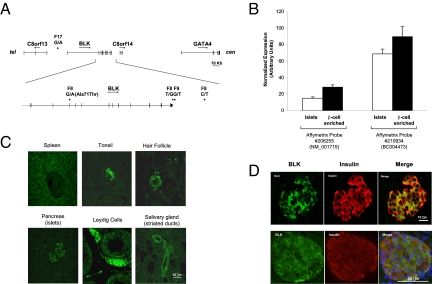
The 100-kb candidate region corresponds to the BLK gene, which codes for a nonreceptor tyrosine-kinase of the src family of proto-oncogenes involved in cell proliferation and differentiation (13). One of the mutations was placed 20 kb 5′ of the gene transcription start site, 1 in exon 4 where it determined an Ala to Thr substitution at position 71, 1 at the end of the 3′ UTR, 1 immediately 3′ of the polyadenylation signal, and 1 8 Kb from the gene on the 3′ side (Fig. 1A). Whereas BLK had been reported in the literature to be expressed only in B lymphocytes (13), analysis of existing expression data (14) revealed that this gene was also expressed in human pancreatic islets—a finding that was confirmed by RT-PCR (Fig. S2). Of note, both BLK probes in the array gave a stronger hybridization signal with RNA isolated from microdissected β cells rather than whole islets (Fig. 1B). Staining of a human tissue array with an anti-BLK antibody confirmed the microarray findings. In addition to lymphatic organs, BLK immunoreactivity was detected in pancreatic islets, striated ducts of salivary glands, hair follicles, and Leydig cells (Fig. 1C). In islets, BLK colocalized with insulin, indicating selective expression in this cell type as suggested by the microarray data (Fig. 1D).
Fig. 1.
BLK as a positional candidate gene for the 8p23 MODY locus. (A) Clustering of mutations at the BLK locus. Exons and introns are indicated by vertical and horizontal lines, respectively. The direction of transcription is indicated by an arrow for each gene. Mutations are designated with the name of the family in which they were found followed by the base substitution. Their position is indicated by a star. (B) BLK expression in islets and β cell-enriched tissue from laser capture microdissection of human pancreas sections. Data are from the experiments with the Affymetrix GeneChip Human Genome U133 described in ref. 14 and refer to 2 probes in the array corresponding to alternative transcripts of BLK. (C) Human tissues showing positive immunostaining for BLK. Images were obtained by staining a tissue array of normal human tissues with a rabbit antibody against human BLK. (D) Colocalization of BLK and insulin in human (Top) and murine (Bottom) islets. Pancreas sections were coimmunostained with anti-BLK (green) and anti-insulin (red) antibodies and images were taken with a confocal microscope. Murine sections were also stained with DAPI to highlight nuclei.
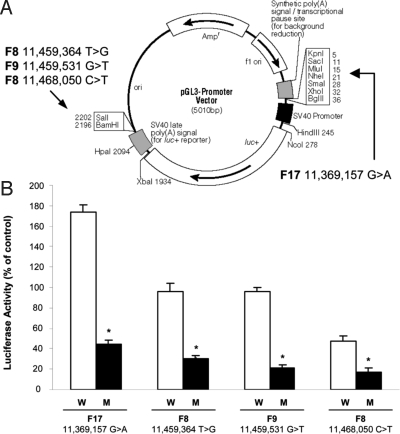
To evaluate the possible impact of mutations on BLK expression, reporter constructs were generated in which 500–900 bp surrounding each noncoding mutation were cloned upstream or downstream of the luciferase gene to mimic their position with respect to the BLK gene (Fig. 2A). Constructs were transfected into MIN6 β-cells—a highly differentiated β-cell line (15). The wild type DNA segment including position 11,369,157 enhanced luciferase expression, the one including positions 11,459,364 and 11,459,531 had no effect, and the one including position 11,468,050 decreased luciferase expression as compared with control constructs without any insert (Fig. 2B). Remarkably, all mutated forms were associated with a 60–80% decrease in luciferase expression with respect to both control and wild-type constructs, indicating functional significance of these sequence differences (Fig. 2B).
Fig. 2.
Effect of noncoding mutations at the BLK locus on promoter activity in vitro. (A) Schematic representation of the constructs that were used for the reporter gene assays. DNA fragments containing the mutated sites were cloned into a pGL3 promoter vector upstream or downstream of the luciferase gene according to their position in the genome with respect to BLK. (B) Luciferase activity from MIN6 β-cells transfected with luciferase reporter constructs. Data are expressed as normalized (firefly/Renilla luciferase) activities of wild type (W) and corresponding mutant (M) inserts relative to that of the control pGL3 promoter vector (100%). *, P < 0.01.
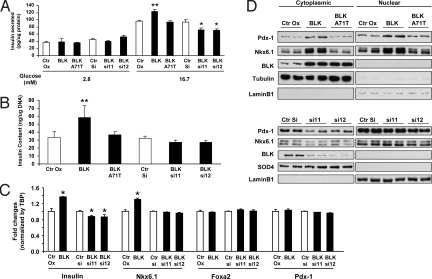
To examine the effects of BLK on insulin secretion and synthesis, BLK was either overexpressed or knocked-down by means of retrovirus or lentivirus shRNAs transiently in the parental MIN6 β-cells and then in stable MIN6 β-cell lines. These cells and the corresponding controls were exposed to low (2.8 mM) or high (16.7 mM) glucose; similar findings were seen in both the transient and stable infections. In low glucose, neither BLK overexpression nor its downregulation had significant effects on insulin secretion. However, at high glucose concentrations, BLK overexpression significantly enhanced insulin secretion, whereas the opposite effect was noted in cells in which BLK had been downregulated (Fig. 3A). The enhancement of insulin secretion induced by BLK overexpression was accompanied by a 70% increase in insulin content as compared with control cells (Fig. 3B). Conversely, the BLK knockdown was associated with a tendency to lower insulin content, although this effect did not reach statistical significance (Fig. 3B). The enhancing effect of BLK on insulin content and secretion was largely attenuated, to the point of being undetectable, when the alanine at position 71 was mutated to a threonine, as observed in family F8 (Fig. 3 A and B). Thus, BLK may enhance insulin response to glucose at least in part by increasing the amount of insulin available for secretion. This effect is blunted by the Ala71Thr mutation.
Fig. 3.
Modulation of β-cell function by BLK. All experiments were conducted on MIN6 β-cell lines in which the wild-type BLK was overexpressed (BLK), knocked-down (BLK si11 and BLKsi12), or overexpressed in a mutated form (BLK A71T), along with the corresponding controls (Ctr OX = pBabe-puro empty vector, Ctr Si = scrambled shRNA). (A) Effect of BLK on glucose-stimulated insulin secretion (GSIS). Cell lines were stimulated with either 2.8 mM (basal) or 16.7 mM (stimulatory) glucose and cell media were sampled at 60 min for determination of insulin levels. Data are expressed as picograms of secreted insulin per micrograms of total cellular proteins. *, P < 0.05 vs. control; **, P < 0.01 vs. control. (B) Effect of BLK on total intracellular insulin content. Data are expressed as nanograms of intracellular insulin to micrograms of total DNA content. **, P < 0.01 vs. control. (C) Effect of BLK on β-cell mRNA levels for insulin and selected modulators of insulin biosynthesis as determined by RT-PCR analysis. Data are normalized to TBP expression and expressed as fold-change relative to control cells. *, P < 0.05 vs. control. (D) Effect of BLK on β-cell expression of Pdx1 and Nkx6.1. Western blot analysis was performed on cytosolic and nuclear fractions prepared from MIN6 β-cells using antibodies specific for each factor. Lamin B1 and tubulin or SOD4 were used as markers of the nuclear and cytosolic fractions, respectively, to control for purity and gel loading.
In view of these findings, we examined the protein and gene expression of various transcription factors implicated in insulin biosynthesis. In agreement with the insulin secretion and content data, we found a 40% increase in insulin transcript abundance in MIN6 β-cells overexpressing BLK and a 15% decrease in cells in which BLK had been knocked-down (Fig. 3C). These findings suggested that the changes in insulin content modulated by BLK occurred at the level of transcription. Next, we observed a significant increase in the expression of transcription factor Nkx6.1 mRNA (Fig. 3C) and protein in both cytoplasmic and nuclear fractions (Fig. 3D). We also detected an increase in Pdx-1 protein in both cytoplasmic and nuclear fractions (Fig. 3D), although no changes were observed in mRNA levels (Fig. 3C). The changes in Nkx6.1 and Pdx-1 seemed to be specific because other transcription factors previously reported to modulate insulin transcription (Foxa2, HNF1α, and HNF4α) were unchanged (Fig. S3). As observed with insulin content and secretion, the inducing effect of BLK on Pdx-1 and Nkx6.1 expression was abolished by the Ala71Thr mutation (Fig. 3D). We did not find significant effects of BLK overexpression or downregulation on pathways involved in glucose sensing, glucose metabolism, channel coupling, or insulin exocytosis (Figs. S4 and S5).
Discussion
Our findings point to BLK as a MODY gene encoding a previously unrecognized modulator of β-cell function, which acts as a stimulator of insulin synthesis and secretion in response to glucose. These effects of BLK appear to be mediated by an up-regulation of Pdx-1—one of the key modulators of β-cell function and itself a MODY gene (5, 16, 17). Another contributing mechanism is the up-regulation of the transcription factor Nkx6.1, which is involved in the control of glucose-stimulated insulin secretion in pancreatic β cells (18). It is possible that the BLK-induced increase in protein levels of Pdx-1 directly promotes the expression of Nkx6.1 (19) and the 2 transcription factors together enhance β-cell function and mass (20).
Basedon our functional data, we envision a scenario in which the mutations identified in our families decrease BLK activity and/or expression, which in turn reduces insulin content and makes β cell less responsive to glucose, resulting into a relative insulin secretion deficit and diabetes. The reason why a small proportion of the mutation carriers, especially of the F8 haplotype, remain normoglycemic is unclear, but variable mutation penetrance and expressivity have been described for monogenic disorders, including MODY, and are thought to result from environmental as well as genetic modifiers (21, 22). An important role in this case might be played by body weight. The penetrance of the F8 haplotype was 0.33 (2 affected out of 6) among carriers with a BMI <28 as compared with 0.89 (8 affected out of 9) among carriers with BMI greater than or equal to 28, and 4 out of 5 nonpenetrants had a BMI below this value. Thus, the diabetogenic environment conferred by an increased body weight (perhaps in the form of insulin resistance and increased insulin demands) might be necessary for translation of the β-cell abnormalities caused by the F8 haplotype into diabetes.
Three of the families that were found linked to 8p23 in the original report, albeit with a relatively low LOD score, were negative for mutations in the BLK gene. The possibility that these families harbor large deletions of the BLK gene that went undetected on sequencing seems unlikely because heterozygosis at polymorphic sites was found in most exons in these families (Fig. S6). These families might carry mutations placed in distant regulatory elements that were not screened or could have been false positives in the linkage analysis.
Mice homozygous for targeted disruption of the BLK gene have been generated and studied for 8 weeks with a focus on investigating the role of BLK in B-lymphocyte physiology (23). However, no phenotypes relevant to diabetes have been described for these mutants, and no phenotypic data are available with regard to responses to exposure to a diabetogenic environment such as a high-fat diet, or cross breeding with an insulin-resistant strain. In light of our findings, further detailed studies are warranted to explore the phenotypes of global KO mice and/or β cell-specific knockouts, in the context of glucose homeostasis.
Whether variability at the BLK locus also contributes to common forms of type 2 diabetes remains to be determined. In the DIAGRAM meta-analysis of genome-wide association data concerning type 2 diabetes, none of the SNPs in the BLK gene and flanking regions exceeded the threshold for genome-wide significance (P = 0.0088 for rs13248109) (24). Those data, however, refer to common polymorphisms and do not exclude the existence of rare variants at this locus contributing to common forms of type 2 diabetes, similar to what has been shown for other metabolic traits (25).
Noncoding SNPs at the BLK locus have been recently found to be associated with increased susceptibility to systemic lupus erythematosus (SLE) and with reduced BLK mRNA levels in B-lymphocyte cells lines (26). When asked about the presence of other health problems in addition to diabetes, none of the carriers of BLK mutations in our MODY families reported a history of SLE or other autoimmune disorders. However, because we did not specifically ask about SLE, we cannot categorically exclude that mutation carriers are also at increased risk for this disease. On the other hand, genetic variants, especially noncoding or nonsynonymous ones affecting phosphorylation sites, may have tissue-specific effects. It is therefore possible that mutations decreasing BLK activity and/or expression in β cells do not have such effects in cells relevant to the etiology of SLE and vice versa.
In summary, the finding of mutations segregating with autosomal dominant diabetes at 8p23 has led us to the discovery of BLK as an unsuspected player in the regulation of insulin synthesis and its secretion in response to glucose. These findings illustrate the continuing value of investigating Mendelian forms of diabetes to gain insights into the molecular mechanisms of glucose homeostasis and identify potential targets for the development of new therapeutic agents. They also indicate that BLK should be added to the list of genes that should be screened in autosomal dominant diabetes, especially those forms characterized by overweight and apparently preserved β-cell function.
Materials and Methods
Resequencing of the 8p23-Linked Interval.
The MODY families investigated in this study have been previously described (12). The study protocol and informed consent procedures were approved by the Joslin Committee on Human Studies. DNA fragments from the critical interval (Table S1) were amplified from 2 affected individuals per family by PCR and sequenced with an ABI Prism 3100 Avant using dye terminator cycle sequencing chemistry (Applied Biosystems). All sequences were analyzed by 2 observers using Sequencing Analysis 3.3 (Applied Biosystems) and then aligned using Sequencher version 4.1.2 (Gene Codes Corp.). Heterozygous sites were validated by a second round of sequencing and by following their segregation in families. Mutation frequencies in the general population were determined by Taqman (Applied Biosystems) or iPlex (Sequenom) assays including a mutation carrier as a positive control.
Luciferase Reporter Studies.
Three DNA fragments spanning positions 11,369,157 (960 bp), 11,459,364 and 11,459,531 (527 bp), and 11,468,050 (707 bp) were amplified by means of PCR from a nondiabetic human control (without mutations) using the primers described in the SI Text. Fragments were subcloned into a pCR-TOPO Vector (Invitrogen), subjected to targeted in vitro mutagenesis (QuikChange II XL, Stratagene), and sequenced to exclude artifacts. Wild-type and mutated inserts were then subcloned into a pGL3 promoter vector (Promega). The fragment spanning position 11,369,157 was subcloned between the KpnI-XhoI sites upstream of the luciferase gene, whereas the 2 other fragments were subcloned between the BamHI-SalI sites downstream of the luciferase gene to reproduce the position of the mutated sites with respect to BLK in the human genome. After confirming correct cloning and resequencing of critical regions, each construct was cotransfected with a Renilla luciferase reporter into MIN6 β-cells in duplicate using Lipofactamine (Invitrogen). A Dual-Luciferase Reporter Assay (Promega) was performed according to manufacturer's suggested protocol using Monolight, 3010 luminometer. pGL3 promoter vector without any insert (control) was very active in MIN6 β-cells (>200,000RLU). The normalized activity of firefly to Renilla luciferase for the control was set at 100, and the results are presented as the relative activity of plasmids with wild type and mutant insert compared with the control pGL3 promoter plasmid. Experiments were repeated 3 times, each time in duplicate, and the mean luciferase/Renilla ratios were compared between wild-type and mutated constructs by t test.
Immunofluorescence Staining.
Immunostaining for paraffin-embedded human tissue used microwaving as antigen retrieval, then rabbit anti-human BLK (alias 3262) (Origene Technologies) in a 1:50 dilution overnight at 4°C, biotinylated donkey anti-rabbit IgG and then Alexafluor 488 conjugated streptavidin. Insulin immunostaining used guinea pig anti-human insulin (1:200, Linco) for 2 h RT incubation followed by Texas Red conjugated donkey anti-guinea pig IgG. Imgenex tissue array of normal human tissues were similarly stained. Mouse pancreas was fixed in Z-fix containing 4% paraformaldehyde, and 5 μm-thick paraffin sections were coimmunostained with anti-BLK antibody (R&D Systems), anti-insulin (Linco) and DAPI (Sigma) at 4°C overnight. Secondary antibodies included donkey anti-goat-cy2 and donkey anti-guinea pig (Jackson ImmunoResearch) for 1 h at RT incubation. Images were taken confocally on a Zeiss LSM 410 microscope. To rule out nonspecificity, sections were immunostained with a primary antibody that had been absorbed with poly-L-lysine (Sigma) (27).
Stable BLK Overexpression and Knockdown Cell Lines.
Full-length cDNA of human BLK was cloned into pBabe-puro retroviral vector. Site directed mutagenesis was performed with PCR using oligos from Integrated DNA Technologies and constructs were sequenced to confirm the mutagenesis and to exclude additional mutations in the BLK ORF. An empty pBabe-puro vector was used as control. shRNA constructs (TRCN0000023411 [hairpin sequence: CCGGCCAGTAGAGACTCTGGAAGTACTCGAGTACTTCCAGAGTCTCTACTGGTTTTT] and TRCN0000023412 [hairpin sequence: CCGGGACAGTGAATACACTGCCCAACTCGAGTTGGGCAGTGTATTCACTGTCTTTTT]) in pLKO.1 were obtained from the RNAi Consortium through Open Biosystems. A scrambled shRNA construct was used as a control. Both retroviral and lentiviral constructs were first used for transient overexpression or knockdown experiments for pilot studies in mixed population of MIN6 β-cells and subsequently for stable cell lines production by infecting mouse insulinoma-derived MIN6 β-cells used between passages 26 and 40 and grown in high-glucose DMEM containing 15% (vol/vol) heat-inactivated FBS, 50 U/mL penicillin, and 10 μg/mL streptomycin, followed by puromycin selection. The stable cell lines used in the experiments were mixed clones from at least 3 independent viral production, infections, and selection. Similar data were obtained with the parental population of MIN6 β-cells.
Insulin Secretion and Insulin Content.
Stable MIN6 β-cell lines were seeded in 12-well plates and incubated for 24 h. Before glucose stimulated-insulin secretion (GSIS) studies, cells were incubated for 14 h with 2.8 mM glucose in DMEM with 0.1% BSA, followed by 2.8 mM glucose in KRB buffer [KRB, 125 mM NaCl, 4.74 mM KCl, 1 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5 mM NaHCO3, 25 mM HEPES (pH 7.4), and 1% BSA] for 1 h. Cells were then “stimulated” with further incubation in either the same KRB buffer containing 2.8 mM glucose (basal concentration) or KRB buffer containing 16.7 mM glucose (stimulatory concentration). Aliquots of the media were collected at 15, 30, or 60 min after stimulation. Insulin concentration in supernatant was measured by ELISA using rat insulin as a standard. Secretion data were normalized to total cellular protein levels. For the measurement of insulin content, stable MIN6 β-cell lines were seeded in 12-well plates and incubated for 24 h. After incubating them for 14 h with 2.8 mM glucose, the medium was removed and cells were washed twice with PBS. Cells were extracted with acid ethanol (18% 1 N HCl, 75% ethanol, and 7% H2O) solution for 16 h at 4°C. Insulin concentration was measured by ELISA, normalizing insulin content to total DNA content.
Western Blotting.
The antibodies that were used are described in the SI Text. For whole cell lysates, stable MIN6 β-cell lines cultured in 6-well plates were washed twice with ice-cold PBS and lysed on ice with 200 μL ice-cold RIPA lysis buffer. For nuclear and cytoplasmic fractions, cells culture on 6-cm dishes were collected by scraping into PBS and fractionated using the NE-PER isolation kit from Pierce according to the manufacturer's recommendation. Protein concentrations were determined using BCA (Pierce). Cell lysates (20–50 μg) were subjected to SDS/PAGE, followed by immunoblotting using specific antisera and detection with chemiluminescence (Roche). Multiple exposures were used to ensure signal linearity.
Quantitative RT-PCR Analysis.
Total RNA was isolated from cultured cells using the RNeasy kit (Qiagen). cDNA was prepared from 1 μg total RNA using the SuperScript III RT-PCR kit (Invitrogen) with random hexamer primers, according to the manufacturer's instructions. The resulting cDNA was diluted 10-fold, and a 2-μL aliquot was used in a 10 μL PCR (SYBR Green, PE Biosystems) containing primers at a concentration of 300 nM each. PCR reactions were run in triplicate and quantitated using the ABI Prism 7900 Sequence Detection System (ABI). Results were normalized to TATA box binding protein (TBP) expression and expressed as arbitrary units. Sequences of primers used in this study are available upon request.
Statistical Analyses.
All data from cellular studies are presented as mean ± SEM. and analyzed by 2-tailed Student's t test assuming unequal variances; a P value smaller than 0.05 was considered as evidence of a statistically significant difference between groups.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants DK55523 (to A.D.), DK 67536 (to R.N.K.), and DK36836 (Genetics Core, Specialized Assay Core, and Advanced Microscopy Core of the Diabetes and Endocrinology Research Center at the Joslin Diabetes Center). M.B. and S.H.K. were supported by a Mentor-based Postdoctoral Fellowship from the American Diabetes Association. C.L.W. was the recipient of an Iacocca Fellowship Award.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906474106/DCSupplemental.
References
- 1.Tattersall RB. Mild familial diabetes with dominant inheritance. Q J Med. 1974;43:339–357. [PubMed] [Google Scholar]
- 2.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1) Nature. 1996;384:458–460. doi: 10.1038/384458a0. [DOI] [PubMed] [Google Scholar]
- 3.Froguel P, et al. Familial hyperglycemia due to mutations in glucokinase. Definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702. doi: 10.1056/NEJM199303113281005. [DOI] [PubMed] [Google Scholar]
- 4.Yamagata K, et al. Mutations in the hepatocyte nuclear factor-1alpha gene in maturity-onset diabetes of the young (MODY3) Nature. 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 5.Stoffers DA, Ferrer J, Clarke WL, Habener JF. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17:138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 6.Horikawa Y, et al. Mutation in hepatocyte nuclear factor-1 beta gene (TCF2) associated with MODY. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 7.Malecki MT, et al. Mutations in NEUROD1 are associated with the development of type 2 diabetes mellitus. Nat Genet. 1999;23:323–328. doi: 10.1038/15500. [DOI] [PubMed] [Google Scholar]
- 8.Neve B, et al. Role of transcription factor KLF11 and its diabetes-associated gene variants in pancreatic beta cell function. Proc Natl Acad Sci USA. 2005;102:4807–4812. doi: 10.1073/pnas.0409177102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raeder H, et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet. 2006;38:54–62. doi: 10.1038/ng1708. [DOI] [PubMed] [Google Scholar]
- 10.Frayling TM, et al. beta-cell genes and diabetes: molecular and clinical characterization of mutations in transcription factors. Diabetes 50 Suppl. 2001;1:S94–100. doi: 10.2337/diabetes.50.2007.s94. [DOI] [PubMed] [Google Scholar]
- 11.Klupa T, et al. Determinants of the development of diabetes (maturity-onset diabetes of the young-3) in carriers of HNF-1alpha mutations: Evidence for parent-of-origin effect. Diabetes Care. 2002;25:2292–2301. doi: 10.2337/diacare.25.12.2292. [DOI] [PubMed] [Google Scholar]
- 12.Kim SH, et al. Identification of a locus for maturity-onset diabetes of the young on chromosome 8p23. Diabetes. 2004;53:1375–1384. doi: 10.2337/diabetes.53.5.1375. [DOI] [PubMed] [Google Scholar]
- 13.Dymecki SM, Niederhuber JE, Desiderio SV. Specific expression of a tyrosine kinase gene, blk, in B lymphoid cells. Science. 1990;247:332–336. doi: 10.1126/science.2404338. [DOI] [PubMed] [Google Scholar]
- 14.Marselli L, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93:1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki J, et al. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: Special reference to expression of glucose transporter isoforms. Endocrinology. 1990;127:126–132. doi: 10.1210/endo-127-1-126. [DOI] [PubMed] [Google Scholar]
- 16.Dutta S, Bonner-Weir S, Montminy M, Wright C. Regulatory factor linked to late-onset diabetes? Nature. 1998;392:560. doi: 10.1038/33311. [DOI] [PubMed] [Google Scholar]
- 17.Edlund H. Factors controlling pancreatic cell differentiation and function. Diabetologia. 2001;44:1071–1079. doi: 10.1007/s001250100623. [DOI] [PubMed] [Google Scholar]
- 18.Schisler JC, et al. The Nkx6.1 homeodomain transcription factor suppresses glucagon expression and regulates glucose-stimulated insulin secretion in islet beta cells. Proc Natl Acad Sci USA. 2005;102:7297–7302. doi: 10.1073/pnas.0502168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedersen JK, et al. Endodermal expression of Nkx6 genes depends differentially on Pdx1. Dev Biol. 2005;288:487–501. doi: 10.1016/j.ydbio.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Schisler JC, et al. Stimulation of human and rat islet beta-cell proliferation with retention of function by the homeodomain transcription factor Nkx6.1. Mol Cell Biol. 2008;28:3465–3476. doi: 10.1128/MCB.01791-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dipple KM, McCabe ER. Phenotypes of patients with “simple” Mendelian disorders are complex traits: Thresholds, modifiers, and systems dynamics. Am J Hum Genet. 2000;66:1729–1735. doi: 10.1086/302938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SH, et al. Genetic modifiers of the age at diagnosis of diabetes (MODY3) in carriers of hepatocyte nuclear factor-1alpha mutations map to chromosomes 5p15, 9q22, and 14q24. Diabetes. 2003;52:2182–2186. doi: 10.2337/diabetes.52.8.2182. [DOI] [PubMed] [Google Scholar]
- 23.Texido G, et al. The B-cell-specific Src-family kinase Blk is dispensable for B-cell development and activation. Mol Cell Biol. 2000;20:1227–1233. doi: 10.1128/mcb.20.4.1227-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen JC, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci USA. 2006;103:1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 27.Scopsi L, Wang BL, Larsson LI. Nonspecific immunocytochemical reactions with certain neurohormonal peptides and basic peptide sequences. J Histochem Cytochem. 1986;34:1469–1475. doi: 10.1177/34.11.2877024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.