Abstract
Adaptive modifications of heteromeric proteins may involve genetically based changes in single subunit polypeptides or parallel changes in multiple genes that encode distinct, interacting subunits. Here we investigate these possibilities by conducting a combined evolutionary and functional analysis of duplicated globin genes in natural populations of deer mice (Peromyscus maniculatus) that are adapted to different elevational zones. A multilocus analysis of nucleotide polymorphism and linkage disequilibrium revealed that high-altitude adaptation of deer mouse hemoglobin involves parallel functional differentiation at multiple unlinked gene duplicates: two α-globin paralogs on chromosome 8 and two β-globin paralogs on chromosome 1. Differences in O2-binding affinity of the alternative β-chain hemoglobin isoforms were entirely attributable to allelic differences in sensitivity to 2,3-diphosphoglycerate (DPG), an allosteric cofactor that stabilizes the low-affinity, deoxygenated conformation of the hemoglobin tetramer. The two-locus β-globin haplotype that predominates at high altitude is associated with suppressed DPG-sensitivity (and hence, increased hemoglobin-O2 affinity), which enhances pulmonary O2 loading under hypoxia. The discovery that allelic differences in DPG-sensitivity contribute to adaptive variation in hemoglobin–O2 affinity illustrates the value of integrating evolutionary analyses of sequence variation with mechanistic appraisals of protein function. Investigation into the functional significance of the deer mouse β-globin polymorphism was motivated by the results of population genetic analyses which revealed evidence for a history of divergent selection between elevational zones. The experimental measures of O2-binding properties corroborated the tests of selection by demonstrating a functional difference between the products of alternative alleles.
Keywords: gene duplication, hypoxia, molecular adaptation, Peromyscus, positive selection
Adaptive modifications of heteromeric proteins may involve genetically based changes in single subunit polypeptides or parallel changes in multiple genes that encode distinct, interacting subunits. The tetrameric hemoglobin (Hb) protein, which transports O2 from the respiratory surfaces to metabolizing tissues, is an ideal molecule for addressing questions about the functional evolution of allosteric, multimeric proteins. In jawed vertebrates, Hb is a heterotetramer, consisting of two α-chain subunits and two β-chain subunits that form two semirigid αβ dimers (α1β1 and α2β2). A mutual rotation of the two αβ dimers occurs during the oxygenation-linked transition in quaternary structure between the deoxy- and oxyHb conformations that is basic to cooperativity (1). Because modifications of Hb function are often implicated in adaptation to environmental hypoxia, and because much is known about structure–function relationships of vertebrate Hbs and their role in blood–O2 transport (2, 3), the study of Hb function in vertebrate species that are native to hypoxic environments provides an opportunity to elucidate detailed molecular mechanisms of physiological adaptation.
In high-altitude mammals, fine-tuned adjustments in Hb–O2 affinity are known to play a key role in adaptive modifications of the O2-transport system (3–7). Under conditions of extreme hypoxia when pulmonary O2 loading is at a premium, an increased Hb–O2 affinity can help maximize the level of tissue oxygenation for a given difference in O2 tension between the sites of O2-loading in the lungs and the sites of O2-unloading in the tissue capillaries (6, 7). A common mechanism for increasing Hb–O2 affinity involves decreasing the sensitivity of Hb to allosteric cofactors, particularly organic and inorganic anions such as 2,3-diphosphoglycerate (DPG) and Cl− that preferentially bind to positively charged sites in deoxyHb (8, 9). Because the binding of DPG and Cl− helps stabilize the low-affinity deoxyHb structure through the formation of salt bridges within and between subunits, diminished sensitivity to these RBC cofactors results in an increased O2 affinity by shifting the allosteric equilibrium in favor of the high-affinity oxyHb conformation. Evolutionary adjustments in the position of this allosteric equilibrium may require coordinated changes in both α- and β-chain subunits, thereby necessitating coordinated changes in two or more unlinked genes. Here we investigate this possibility by conducting a functional and evolutionary analysis of duplicated globin genes in natural populations of deer mice (Peromyscus maniculatus) that are adapted to different elevational zones.
Experimental studies of wild-derived strains of deer mice have demonstrated that adaptive variation in blood–O2 affinity and aerobic performance is strongly associated with allelic variation at two tandemly duplicated genes (HBA-T1 and HBA-T2) that encode the α-chain subunits of adult Hb (10–13). These experiments revealed that the two-locus α-globin genotype that confers high blood–O2 affinity is associated with superior aerobic performance under hypoxia at high altitude, but is associated with poor performance (relative to the low-affinity genotype) under normoxic conditions at low altitude. In both altitudinal extremes, double heterozygotes were intermediate with respect to aerobic performance. This rank order of phenotypic effects appears to be attributable to the fact that the possession of high-affinity Hb facilitates pulmonary O2 loading under hypoxia, but hinders O2 delivery to aerobically metabolizing tissues under normoxic conditions. These tradeoffs in O2-transport efficiency at different O2 partial pressures (PO2's) suggest that the high-affinity α-globin genotype may confer highest fitness in high-altitude environments, whereas the low-affinity genotype may confer highest fitness in low-altitude environments (10–14). Consistent with this hypothesis, survivorship studies of free-ranging deer mice demonstrated that aerobic performance is subject to strong directional selection at high-altitude (15) and electrophoretic surveys of α-globin polymorphism in deer mice from western North America revealed striking altitudinal patterns of allele frequency variation that are consistent with the predicted rank-order of genotypic fitnesses across environments: The high-affinity α-globin electromorph is present at high frequency in high-altitude populations (>2,750 m), whereas the low-affinity electromorph is either fixed or nearly fixed in low-altitude populations [<1,750 m; (16)]. Moreover, patterns of nucleotide diversity and linkage disequilibrium (LD) at the two HBA paralogs are indicative of spatially varying selection between high- and low-altitude populations (17, 18).
Although the original surveys of Hb polymorphism revealed that the β-globin electromorphs also exhibited shifts in frequency between high- and low-altitude populations, the β-chain subunits did not appear to make significant contributions to variation in blood–O2 affinity or aerobic performance (13). To gain insight into the possible adaptive significance of the deer mouse β-globin polymorphism, we surveyed DNA sequence variation at each of two tandemly duplicated β-globin paralogs (HBB-T1 and HBB-T2) in a sample of high- and low-altitude deer mice from eastern Colorado. After identifying the specific amino acid substitutions that distinguish previously characterized β-globin electromorphs, we used a simulation-based analysis of multilocus polymorphism data to evaluate the role of spatially varying selection in shaping the observed patterns of altitudinal variation at each of the HBB gene duplicates. We then evaluated the functional significance of the observed changes in Hb structure by measuring O2-binding properties of hemolysates from mice with known Hb isoform composition. Measurements of O2-binding properties were conducted in the absence of allosteric cofactors, in the presence of either DPG or Cl− in isolation, and in the presence of both cofactors together.
Results and Discussion
Patterns of β-Globin Polymorphism.
We surveyed β-globin polymorphism at the protein level by means of thin-layer isoelectric focusing (TL-IEF) in a sample of 75 deer mice (n = 37 and 38 for the high- and low-altitude samples, respectively). Having previously characterized the genomic structure of the β-globin gene cluster of P. maniculatus (19), we also screened nucleotide variation at each of the two underlying β-globin genes (HBB-T1 and HBB-T2). These two tandem gene duplicates are separated by 16.2 kb of noncoding DNA on Peromyscus chromosome 1.
The TL-IEF analysis revealed the presence of two highly distinct β-globin variants corresponding to the “d0” and “d1” electromorphs that were functionally characterized by previous workers (12, 20). Although the TL-IEF band profiles revealed a pattern of variation consistent with Mendelian segregation of two alleles at a single gene, our sequence data revealed that the d0 and d1 variants are actually the products of alternative two-locus β-globin haplotypes. One chromosome harbors the d0 allele at both HBB-T1 and HBB-T2, and the alternative chromosome harbors the d1 allele at both genes. This allele-sharing between the HBB-T1 and HBB-T2 genes is attributable to a history of interparalog gene conversion (17, 19, 21, 22). This same phenomenon has been documented for the two 5′ adult α-globin paralogs of deer mice, HBA-T1 and HBA-T2 (18). Thus, in both the α- and β-globin gene clusters of deer mouse (which are located on different chromosomes), linked pairs of tandem gene duplicates segregate the same pair of functionally distinct protein alleles.
We observed a perfectly nonrandom association between genotypes at the HBB-T1 and HBB-T2 genes (P < 0.0001), indicating that all sampled mice are either d0d0/d0d0 double homozygotes, d0d0/d1d1 double heterozygotes, or d1d1/d1d1 double homozygotes. Recombinant d0d1 chromosomes are either nonexistent or present at very low frequency. This perfect LD between the two HBB paralogs is especially remarkable given the 16.2-kb distance between them. By contrast, the HBA-T1 and HBA-T2 paralogs are characterized by a far less pronounced level of intergenic LD even though the two genes are separated by only ≈5 kb (21).
In addition to this highly unusual haplotype structure, the two HBB genes also exhibited dramatic allele frequency differences between the high- and low-altitude population samples. Due to the perfect LD between the HBB-T1 and HBB-T2 genes, the altitudinal pattern of β-globin polymorphism can be described in terms of two-locus haplotype frequencies. Frequencies of the d1d1 haplotype were 0.973 (n = 74 chromosomes) and 0.263 (n = 76 chromosomes) in the high- and low-altitude samples, respectively. The observed level of genotypic differentiation between high- and low-altitude samples was highly significant (P < 0.0001). The level of altitudinal differentiation in two-locus HBB haplotype frequencies (FST = 0.691, n = 150 chromosomes) far exceeds levels of allele frequency differentiation at the HBA genes in the same sample of mice (FST = 0.255 and 0.180 for HBA-T1 and HBA-T2, respectively). Relative to the expected genotype frequencies at Hardy–Weinberg equilibrium, the altitudinal differences in two-locus HBB haplotype frequencies produced a highly significant excess of double homozygotes in the total sample (P < 0.0001). By contrast, genotype frequencies at the HBA genes exhibited no significant deviation from Hardy–Weinberg expectations (18).
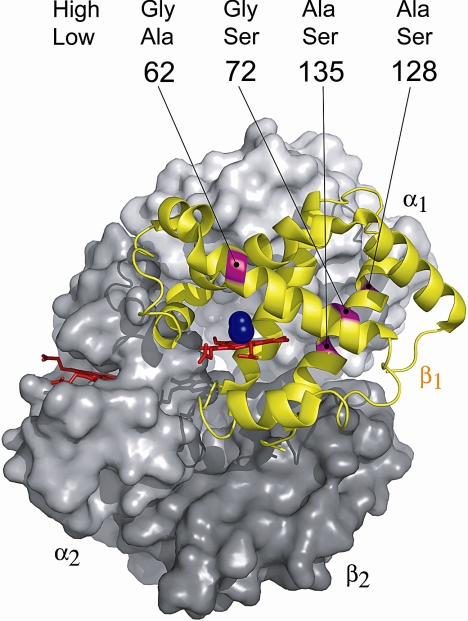
By cloning and sequencing the HBB-T1 and HBB-T2 genes of mice with known TL-IEF phenotypes, we discovered that the two main β-globin allele classes are distinguished by a total of four amino acid substitutions: 62(E6)Ala/Gly, 72(E16)Gly/Ser, 128(H6)Ser/Ala, and 135(H13)Ala/Ser (Fig. 1). In the notation used here, the ancestral residue is listed first, and the derived residue (relative to P. leucopus) is listed second. The d1 allele is defined by the four-site haplotype “62Gly/72Gly/128Ala/135Ala,” whereas the d0 allele is defined by the alternative four-site haplotype “62Ala/72Ser/128Ser/135Ser.” At both HBB genes, the alternative allele classes are distinguished from one another by extremely high levels of nucleotide divergence (Dxy with Jukes and Cantor correction = 0.0356 [SD = 0.0020] and 0.0341 [SD = 0.0020] for HBB-T1 and HBB-T2, respectively). The high sequence divergence between d1 and d0 allele classes is also clearly evident in the reconstructed haplotype network of HBB sequences (Fig. 2).
Fig. 1.
Homology-based structural model of deer mouse oxy-hemoglobin showing the location of four amino acid mutations located on the E- and H-helices of the β-chain subunit.
Fig. 2.
Haplotype network of HBB-T1 and HBB-T2 coding sequences in Colorado deer mice. The structure of the network reveals the high net sequence divergence between the d0 and d1 allele classes as well as the extensive allele-sharing between the two HBB paralogs due to gene conversion.
At the HBA-T1 and HBA-T2 genes, the two alternative allele classes are distinguished from one another by a total of five closely linked amino acid substitutions that span the highly conserved E-helix domain of the α-globin polypeptide (17, 18). Relative to the two alternative HBB allele classes, the HBA allele classes are much more heterogeneous in amino acid composition, as there are several intermediate-frequency amino acid polymorphisms that are not perfectly associated with TL-IEF electromorphs (17). The five main amino acid polymorphisms at the HBA genes and the four main amino acid polymorphisms at the HBB genes exhibited parallel shifts in allele frequency between high- and low-altitude populations (Fig. S1).
Tests of Spatially Varying Selection Between High- and Low-Altitude Populations.
To gain insight into the possible adaptive significance of the two-locus β-globin polymorphism, we used a simulation-based approach to test for evidence of spatially varying selection. Specifically, we conducted forward simulations under a neutral model of population structure to generate null distributions for between-sample measures of nucleotide divergence, δST, and LD, Zg (18). These summary statistics were computed for comparisons between population samples and between functionally defined allele classes. The simulation model was parameterized with polymorphism data from a panel of eight unlinked, autosomal reference loci (18).
In comparison with the reference loci, the four adult globin genes were characterized by uniformly higher levels of nucleotide diversity at silent sites (Table S1). Although overall levels of nucleotide diversity were quite similar at all four adult globin genes, the HBA and HBB genes exhibited pronounced differences in the partitioning of this diversity within and between populations. In the comparison between the high- and low-altitude population samples, the HBB-T1 and HBB-T2 genes exhibited levels of nucleotide divergence that were several times higher than those of the HBA genes (Table 1). In contrast to the HBA genes, both of the HBB genes exceeded the simulation-based critical value for rejecting the neutral model of population structure. Levels of altitudinal divergence at the HBB genes also greatly exceeded the upper range of values for the panel of autosomal reference loci (Table 1). Although δST values at the HBA genes did not exceed neutral expectations when averaged across the entire 1.68 kb of sequence, sliding window plots of nucleotide divergence did successfully pinpoint specific gene regions that appear to be targets of divergent selection between high- and low-altitude populations (18).
Table 1.
Locus-specific measures of nucleotide divergence and LD between high- and low-altitude populations of deer mice, and between protein allele classes
| Locus | Between populations |
Between allele classes |
||||
|---|---|---|---|---|---|---|
| k* | δST (×10) | Zg | k† | δST (×10) | Zg | |
| α-globin genes‡ (chromosome 8) | ||||||
| HBA-T1 (1,680 bp) | 45.24 | 0.0102 | 0.0015 | 38.27 | 0.0737 | 0.0170§ |
| HBA-T2 (1,680 bp) | 43.77 | 0.0186 | 0.0031§ | 37.57 | 0.0653 | 0.0160§ |
| β-globin genes (chromosome 1) | ||||||
| HBB-T1 (1,246 bp) | 33.87 | 0.0724§ | 0.0223§ | 39.05 | 0.1766 | 0.0540§ |
| HBB-T2 (1,246 bp) | 33.11 | 0.0667§ | 0.0263§ | 37.59 | 0.1678 | 0.0560§ |
| Reference loci (n = 8) | ||||||
| Mean | 11.98 | 0.0053 | 0.0020 | - | - | - |
| Minimum | 3.67 | 0.0006 | 0.0007 | - | - | - |
| Maximum | 26.56 | 0.0075 | 0.0033 | - | - | - |
*Average number of nucleotide differences between populations.
†Average number of nucleotide differences between allele classes.
‡Data for the HBA-T1 and HBA-T2 genes were reported previously (18) and are presented here for the purpose of comparison with the HBB genes.
§P < 0.05.
With regard to the between-population component of LD, the two HBB genes were characterized by Zg values that were one order of magnitude higher than those of the HBA genes (Table 1).Zg values for the HBB genes also exceeded the upper range of values for the reference loci by roughly the same margin. Both HBB genes far exceeded the simulation-based critical value for rejecting neutrality, as did the HBA-T2 gene. With regard to the component of LD between allele classes, the two HBB genes were characterized by Zg values that were more than two times higher than those of the HBA genes. In the comparison between allele classes, Zg values for all four adult globin genes were statistically significant.
In summary, the observed patterns of nucleotide polymorphism and LD at each of the four adult globin genes are clearly indicative of local adaptation to different elevational zones (see ref. 18 for detailed analysis of the HBA genes). Moreover, the significant excess of LD between allele classes at all four genes suggests that the alternative protein alleles have been maintained as a balanced polymorphism by spatially varying selection between elevational zones. This history of spatially varying selection had already been established for the HBA genes, and is consistent with their known effects on fitness (5, 10–13, 16–18). Although previous studies of wild-derived strains of deer mice demonstrated that the HBA genes make the greatest contribution to adaptive variation in blood–O2 affinity and aerobic performance under hypoxia, the HBB genes actually exhibit a much greater level of altitudinal differentiation, which is suggestive of stronger divergent selection. The strong evidence for adaptive altitudinal divergence at HBB-T1 and HBB-T2 suggested that the products of these genes may mediate some adaptive modification of Hb function that was previously overlooked.
Functional Significance of β-Globin Polymorphism.
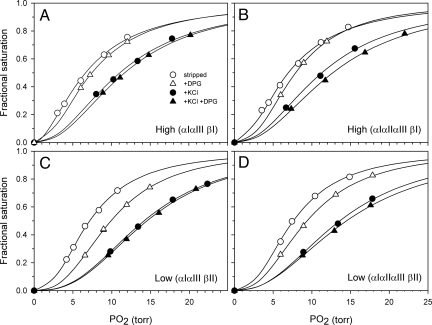
To assess the functional significance of the two-locus HBB polymorphism, we compared O2-binding properties of hemolysates from high- and low-altitude deer mice that possessed Hbs with the same α-chains but different β-chains. This permitted an assessment of functional differences between the β-chain products of d0d0 and d1d1 haplotypes against uniform HBA backgrounds. In each of two comparisons involving matched pairs of high- and low-altitude mice with identical α-chain Hb isoforms, hemolysates of high-altitude mice containing the βI subunit isoform (product of the d1d1/d1d1 genotype) were characterized by markedly suppressed DPG-sensitivies compared with those of low-altitude mice containing the βII subunit isoform (product of the d0d0/d0d0 genotype) (Fig. 3 and Table S2). In the case of the low-altitude d0d0/d0d0 mice, the addition of DPG produced a pronounced rightward shift in the O2 equilibrium curve (log-P50, (DPG) − log-P50, (str) = 0.09–0.14, where P50 is the PO2 at which Hb is 50% saturated). By contrast, in the case of the high-altitude d1d1/d1d1 mice, the addition of DPG produced a scarcely detectable rightward shift (log-P50, (DPG) − log-P50, (str) = 0.04–0.06; see Fig. 3). A less pronounced difference in Hb–O2 affinity was seen in the presence of Cl− ions (log-P50, (DPG) − log-P50, (0.1M Cl) = 0.21 vs. 0.25–0.30 for the Hbs of high- and low-altitude mice, respectively), indicating that Cl− has similar effects on Hb–O2 affinity in mice from both elevational zones. The suppressed DPG-sensitivity of mice with the d1d1/d1d1 genotype helps to maintain an elevated Hb–O2 affinity (reduced P50) in the presence of RBC cofactors, which enhances pulmonary O2-loading under hypoxia. It thus appears that allelic differences in DPG-sensitivity make a significant contribution to the well-documented differences in blood–O2 affinity between high- and low-altitude deer mice (10, 11, 14). Interestingly, the effect of Cl− on Hb–O2 affinity is much greater than that of DPG in both high- and low-altitude mice (Fig. 3), which is uncommon in vertebrates.
Fig. 3.
O2 equilibrium curves of stripped deer mouse Hbs at pH 7.4 and 37 °C in the presence and absence of allosteric cofactors ([Cl−], 0.1 M; [NaHEPES], 0.1 M; DPG/Hb tetramer ratio, 2.0; [Heme], 0.16 mM). Curves for high-altitude mice that express the βI isoform (product of the d1d1/d1d1 genotype) are shown in A and B, and curves for low-altitude mice that express the βII isoform (product of the d0d0/d0d0 genotype) are shown in C and D. Comparisons of A vs. C and B vs. D reveal differences in O2 equilibrium curves for matched pairs of high- and low-altitude mice that possessed Hbs with the same α-chains but different β-chains. The three α-chain Hb isoforms are defined by the following five-site amino acid combinations (sites 50, 57, 60, 64, 71): αI = PGAGS, αII = HGAGS, αIII = HAGDG. The two β-chain Hb isoforms are defined by the following four-site amino acid combinations (sites 62, 72, 128, 135): βI = GGAA, βII = ASSS.
Suppressed DPG-sensitivity is also responsible for the increased O2 affinities of fetal Hb in humans (8) and the adult Hbs of Andean camelids such as the llama and vicuña (23). In the case of fetal Hb, the suppressed DPG-sensitivity is attributable to an amino acid substitution in the β-chain product of the prenatally expressed γA- and γG-globin genes, β143(H21)His → γSer, which eliminates a DPG-binding site near the C terminus of the β-type subunit. However, additional substitutions remote from the DPG-binding site also exert significant second-order effects on DPG-binding affinity (24). In the case of Andean camelids, the suppressed DPG-sensitivity is primarily attributable to a substitution, β2(NA2)His → Asn, that eliminates a DPG-binding site at the N terminus of the β-chain. Although none of the four HBB substitutions that distinguish the alternative deer mouse Hb isoforms directly affect the set of charge–charge interactions responsible for DPG binding, the pair of Ser → Ala substitutions at sites β128(H6) and β135(H13) alter the orientation of the E- and H-helices, and may therefore indirectly affect DPG-binding through stereochemical alterations of the binding cleft between the β-chain subunits.
In summary, results of this study indicate that the d1d1 haplotype that predominates at high altitudes confers an increased Hb–O2 affinity via suppressed DPG sensitivity, whereas the d0d0 haplotype that predominates at low altitudes is associated with a relatively low Hb–O2 affinity. These functional differences mirror the pattern of altitudinal differentiation that has been documented for the HBA genes (16) and indicates that the α- and β-chain subunits both contribute to the divergent fine-tuning of Hb–O2 affinity in deer mice that are native to different elevational zones.
Conclusions
Patterns of nucleotide polymorphism and LD at each of the four adult globin genes of deer mice (HBA-T1, HBA-T2, HBB-T1, and HBB-T2) are clearly indicative of local adaptation to different elevational zones. Thus, high-altitude adaptation of deer mouse Hb appears to involve parallel functional differentiation at multiple unlinked gene duplicates: The two HBA paralogs on chromosome 8 and the two HBB paralogs on chromosome 1. In deer mice that are native to different elevational zones, the divergent fine-tuning of Hb–O2 affinity appears to be attributable to the independent or joint effects of 5 amino acid mutations in the α-chain subunits and 4 amino acid mutations in the β-chain subunits. Although altitudinal variation in blood–O2 affinity has been well documented in deer mice (10, 11, 14), the functional (and adaptive) significance of the HBB polymorphism was previously unappreciated. The discovery that allelic differences in DPG-sensitivity contribute to adaptive variation in Hb–O2 affinity illustrates the value of integrating evolutionary analyses of sequence variation with mechanistic appraisals of protein function (25). The population genetic analysis revealed evidence that the observed patterns of HBB polymorphism have been shaped by a history of divergent selection between elevational zones, and this result motivated our experimental investigation into the functional significance of the allelic variation. The experimental measures of O2-binding properties corroborated the tests of selection by demonstrating a functional difference between the products of alternative HBB alleles.
Materials and Methods
Samples.
We sampled a total of 75 P. maniculatus from a pair of high- and low-altitude localities in eastern Colorado. The high-altitude sample (n = 37 mice) was collected from the summit of Mt. Evans, Clear Creek County, 4,347 m above sea level, and the low-altitude sample (n = 38 mice) was collected from a prairie grassland site in Yuma County, 1,158 m above sea level (17). The population genetic analysis combined new sequence data from the two duplicated β-globin genes, HBB-T1 and HBB-T2, with previously published sequence data from the two HBA paralogs and eight unlinked autosomal genes (18).
PCR, Cloning, and Sequencing.
Genomic DNA was extracted from frozen liver of each mouse by using DNeasy kits (QIAGEN). Paralog-specific primers were designed by aligning flanking sequences of HBB-T1 and HBB-T2 orthologs in P. maniculatus and P. leucopus (19). For each of the 75 mice that were subject to the TL-IEF analysis, we used an allele-specific PCR assay to confirm the identity of d0/d0, d0/d1, and d1/d1 genotypes at both HBB paralogs (see SI Text for detailed protocol).
For both HBB-T1 and HBB-T2, we cloned and sequenced both alleles from a subset of mice from each population (n = 13 from Mt. Evans and n = 12 from Yuma County). Diploid PCR products were cloned by using the pCR4-TOPO vector (Invitrogen) and 8–10 clones per gene were sequenced by using T3/T7 and M13F/M13R vector primers. Thus, in the case of both HBB-T1 and HBB-T2, the haplotype phase of all heterozygous sites was determined experimentally. For each mouse in our sample, we were able to confirm that we had cloned both alleles from each HBB paralog by matching the sequence data with results of the TL-IEF and allele-specific PCR assays.
Genetic Data Analysis.
DNA sequences were aligned and contigs assembled by using Clustal X (26). Summary statistics of nucleotide polymorphism and LD were computed with custom programs written in the C programming language (available on request). For comparisons between the high- and low-altitude samples, we used variation at silent sites to calculate δST as a measure of the between-population component of nucleotide diversity (27) and Zg as a measure of the between-population component of LD (18). In the case of the HBB genes, we tested for differentiation between high- and low-altitude samples by using an exact test of genotype frequencies based on the log-likelihood G-statistic (28). The same Markov-chain contingency table approach was used to test for genotypic LD between the HBB-T1 and HBB-T2 paralogs. We also tested for deviations from Hardy–Weinberg genotype frequencies by performing the exact test of Guo and Thompson (29).
Simulation-Based Test of Spatially Varying Selection.
To test for locus-specific evidence of spatially varying selection, we conducted forward simulations under a neutral model of population structure to generate null distributions for between-sample measures of δST and Zg (18). The measures of nucleotide divergence and LD were computed in comparisons between population samples and in comparisons between alternative classes of protein alleles. Simulations were parameterized with empirical data from eight autosomal reference loci that were cloned and sequenced in the same panel of mice. Two key nuisance parameters in the simulation model are the scaled mutation rate (4Nu) and the scaled recombination rate (4Nr), where N is the effective size of the total diploid population, u is the neutral mutation rate, and r is the rate of crossing-over. We conducted simulations by using a range of locus-specific estimates for these two parameters: 4Nu = 0.02–0.03 and 4Nr = 0.01–0.04 (Table S1). Simulation-based critical values for each test statistic were calculated according to the methods described in Storz and Kelly (18).
Determination of Hb Isoform Composition.
Hemolysates of wild-caught mice were prepared according to standard methods and were “stripped” of allosteric cofactors by ion exchange chromatography. Allelic variation at the HBB genes was resolved by using immobilized gradient TL-IEF as described previously (17). The Hb isoform composition of each hemolysate was determined by using TL-IEF native gels (PhastSystem, GE Healthcare Bio-Sciences), and the individual globin chain subunits were separated by means of 2D gel electrophoresis. The 2D gel analysis involved separating native Hb isoforms by TL-IEF in the first dimension, followed by the separation of dissociated α- and β-globin monomers by means of SDS/PAGE in the second dimension. Globin chains excised from each 2D gel were then identified by means of electrospray ionization mass spectrometry. The peptide mass fingerprints derived from the mass spectrometry analysis were used to query a reference database of HBA and HBB DNA sequences from the same sample of mice.
Measurement of Hb–O2 Affinity and Cofactor Sensitivities.
O2 equilibrium curves of hemolysates were measured by using a modified diffusion chamber, where changes in the absorbance of thin-layer Hb solutions were recorded after stepwise changes in the O2 tension of gas mixtures in the chamber (30–33). The resultant measurements provide estimates of P50 (pH 7.4, 37 °C) and n50 (the cooperativity coefficient at 50% O2 saturation) in the presence and absence of added cofactors ([Cl], 0.10 M; [NaHEPES], 0.1 M; DPG/Hb tetramer ratio, 2.0; [Heme], 0.16 mM).
Supplementary Material
Acknowledgments.
We thank A. Bang, M.-B. Hemmingsen, B. Monteiro, and M. Yamanishi for valuable laboratory assistance and two anonymous reviewers for helpful suggestions. This work was funded by National Science Foundation Grant DEB-0614342 and National Institutes of Health/National Heart, Lung, and Blood Institute Grant R01 HL087216.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DNA sequence data reported in this article have been deposited in the GenBank database (accession numbers GQ139365–GQ139470).
This article contains supporting information online at www.pnas.org/cgi/content/full/0905224106/DCSupplemental.
References
- 1.Perutz MF. Mechanisms of cooperativity and allosteric regulation in proteins. Q RevBiophys. 1989;22:139–236. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 2.Perutz MF. Species adaptation in a protein molecule. Mol Biol Evol. 1983;1:1–28. doi: 10.1093/oxfordjournals.molbev.a040299. [DOI] [PubMed] [Google Scholar]
- 3.Weber RE, Fago A. Functional adaptation and its molecular basis in vertebrate hemoglobins, neuroglobins and cytoglobins. Respir Physiol Neurobiol. 2004;144:141–159. doi: 10.1016/j.resp.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 4.Monge C, León-Velarde F. Physiological adaptation to high altitude: Oxygen transport in mammals and birds. Physiol Rev. 1991;71:1135–1172. doi: 10.1152/physrev.1991.71.4.1135. [DOI] [PubMed] [Google Scholar]
- 5.Storz JF. Hemoglobin function and physiological adaptation to hypoxia in high-altitude mammals. J Mammal. 2007;88:24–31. [Google Scholar]
- 6.Weber RE. High-altitude adaptations in vertebrate hemoglobins. Respir Physiol Neurobiol. 2007;158:132–142. doi: 10.1016/j.resp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Storz JF, Moriyama H. Mechanisms of hemoglobin adaptation to high-altitude hypoxia. High Alt Med Biol. 2008;9:148–157. doi: 10.1089/ham.2007.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagel RL, Steinberg MH. Hemoglobins of the embryo and fetus and minor hemoglobins of adults. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of hemoglobin: genetics, pathophysiology, and clinical management. New York: Cambridge Univ Press; 2001. pp. 197–230. [Google Scholar]
- 9.Weber RE, et al. Novel mechanism for high-altitude adaptation in hemoglobin of the Andean frog Telmatobius peruvianus. Am J Physiol. 2002;283:R1052–R1060. doi: 10.1152/ajpregu.00292.2002. [DOI] [PubMed] [Google Scholar]
- 10.Snyder LRG. Deer mouse hemoglobins: Is there genetic adaptation to high altitude? Bioscience. 1981;31:299–304. [Google Scholar]
- 11.Snyder LRG, Born S, Lechner AJ. Blood oxygen affinity in high- and low-altitude populations of the deer mouse. Respir Physiol. 1982;48:89–105. doi: 10.1016/0034-5687(82)90052-4. [DOI] [PubMed] [Google Scholar]
- 12.Chappell MA, Snyder LRG. Biochemical and physiological correlates of deer mouse α chain hemoglobin polymorphisms. Proc Natl Acad Sci USA. 1984;81:5484–5488. doi: 10.1073/pnas.81.17.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chappell MA, Hayes JP, Snyder LRG. Hemoglobin polymorphisms in deer mice (Peromyscus maniculatus): Physiology of beta-globin variants and alpha-globin recombinants. Evolution. 1988;42:681–688. doi: 10.1111/j.1558-5646.1988.tb02486.x. [DOI] [PubMed] [Google Scholar]
- 14.Snyder LRG. Low P50 in deer mice native to high altitude. J Appl Physiol. 1985;58:193–199. doi: 10.1152/jappl.1985.58.1.193. [DOI] [PubMed] [Google Scholar]
- 15.Hayes JP, O'Connor CS. Natural selection on thermogenic capacity of high-altitude deer mice. Evolution. 1999;53:1280–1287. doi: 10.1111/j.1558-5646.1999.tb04540.x. [DOI] [PubMed] [Google Scholar]
- 16.Snyder LRG, Chappell MA, Hayes JP. α-chain hemoglobin polymorphisms are correlated with altitude in the deer mouse, Peromyscus maniculatus. Evolution. 1988;42:689–697. doi: 10.1111/j.1558-5646.1988.tb02487.x. [DOI] [PubMed] [Google Scholar]
- 17.Storz JF, et al. The molecular basis of high-altitude adaptation in deer mice. Plos Genetics. 2007;3(e45):448–459. doi: 10.1371/journal.pgen.0030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Storz JF, Kelly JK. Effects of spatially varying selection on nucleotide diversity and linkage disequilibrium: Insights from deer mouse globin genes. Genetics. 2008;180:367–379. doi: 10.1534/genetics.108.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol Biol Evol. 2008;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder LRG. Genetics of hemoglobin in the deer mouse, Peromyscus maniculatus. I. Multiple α- and β-globin structural loci. Genetics. 1978;89:511–530. doi: 10.1093/genetics/89.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storz JF, Hoffmann FG, Opazo JC, Moriyama H. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics. 2008;178:1623–1638. doi: 10.1534/genetics.107.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol Biol Evol. 2008;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- 23.Bauer C, Rollema HS, Till HW, Braunitzer G. Phosphate binding by llama and camel hemoglobin. J Comp Physiol. 1980;136:67–70. [Google Scholar]
- 24.Dumoulin A, Manning LR, Jenkins WT, Winslow RM, Manning JM. Exchange of subunit interfaces between recombinant adult and fetal hemoglobins. Evidence for a functional inter-relationship among regions of the tetramer. J Biol Chem. 1997;272:31326–31332. doi: 10.1074/jbc.272.50.31326. [DOI] [PubMed] [Google Scholar]
- 25.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: The functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
- 28.Goudet J, Raymond M, de Meeus T, Rousset F. Testing differentiation in diploid populations. Genetics. 1996;144:1933–1940. doi: 10.1093/genetics/144.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo SW, Thompson EA. Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 30.Weber RE. Cationic control of oxygen affinity in lugworm erythocruorin. Nature. 1981;292:386–387. [Google Scholar]
- 31.Weber RE. Use of ionic and zwitterionic (Tris/BisTris and Hepes) buffers in studies on hemoglobin function. J Appl Physiology. 1992;72:1611–1615. doi: 10.1152/jappl.1992.72.4.1611. [DOI] [PubMed] [Google Scholar]
- 32.Fago A, et al. Allosteric regulation and temperature dependence of oxygen binding in human neuroglobin and cytoglobin—Molecular mechanisms and physiological significance. J Biol Chem. 2004;279:44417–44426. doi: 10.1074/jbc.M407126200. [DOI] [PubMed] [Google Scholar]
- 33.Weber RE, et al. Modulation of red cell glycolysis: Interactions between vertebrate hemoglobins and cytoplasmic domains of band 3 red cell membrane proteins. Am J Physiol. 2004;287:R454–R464. doi: 10.1152/ajpregu.00060.2004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.