Abstract
Basonuclin 2 is a recently discovered zinc finger protein of unknown function. Its paralog, basonuclin 1, is associated with the ability of keratinocytes to multiply. The basonuclin zinc fingers are closely related to those of the Drosophila proteins disco and discorelated, but the relation between disco proteins and basonuclins has remained elusive because the function of the disco proteins in larval head development seems to have no relation to that of basonuclin 1 and because the amino acid sequence of disco, apart from the zinc fingers, also has no similarity to that of the basonuclins. We have generated mice lacking basonuclin 2. These mice die within 24 h of birth with a cleft palate and abnormalities of craniofacial bones and tongue. In the embryonic head, expression of the basonuclin 2 gene is restricted to mesenchymal cells in the palate, at the periphery of the tongue, and in the mesenchymal sheaths that surround the brain and the osteocartilagineous structures. In late embryos, the rate of multiplication of these mesenchymal cells is greatly diminished. Therefore, basonuclin 2 is essential for the multiplication of craniofacial mesenchymal cells during embryogenesis. Non-Drosophila insect databases available since 2008 reveal that the basonuclins and the disco proteins share much more extensive sequence and gene structure similarity than noted when only Drosophila sequences were examined. We conclude that basonuclin 2 is both structurally and functionally the vertebrate ortholog of the disco proteins. We also note the possibility that some human craniofacial abnormalities are due to a lack of basonuclin 2.
Keywords: cell multiplication, cleft palate
Basonuclin 1 and 2 (bnc1 and bnc2) are C2H2 zinc finger proteins found only in vertebrates. Bnc1 and bnc2 share the same general structure: they are about 1,000 residues in length and contain 3 separate pairs of zinc fingers and a nuclear localization signal (NLS). The deduced amino acid sequences of bnc1 and bnc2 are only slightly more than 40% identical, but the sequence identity is much higher in the N-terminal region, the zinc fingers, and the region of the NLS than in the rest of the molecule (1, 2). The basonuclin zinc fingers are related to those of the Drosophila proteins disconnected (3) and discorelated (the disco proteins), which have an essential function in larval head development (4).
Bnc1 is highly restricted in its tissue distribution; it is found mainly in basal keratinocytes of stratified squamous epithelium and in reproductive germ cells (5, 6). Although the evidence is by no means conclusive, it seems that the function of bnc1 is related to the potential for cell proliferation (7, 8). The only known function of bnc1 is that of a transcription factor in the synthesis of ribosomal RNA (9, 10), but it is possible that bnc1 possesses a nucleoplasmic function in the regulation of expression of genes transcribed by RNA polymerase II (11, 12). Knock-down of the mouse bnc1 gene in oocytes shows that bnc1 is required for oogenesis and possibly early embryogenesis (13).
The gene encoding bnc2 was discovered in 2004 (1, 2). The bnc2 mRNA is abundant in cell types that possess bnc1, but it is also found in tissues that lack bnc1, such as kidney, intestine, and uterus. The genes for bnc1 and bnc2 differ greatly in size and are located on different chromosomes, but it is clear that they have a common evolutionary origin. Bnc1 and bnc2 are thought to possess different functions, since bnc2 but not bnc1 localizes to nuclear speckles and therefore is likely to have a function in nuclear processing of mRNA (14).
The extreme evolutionary stability of the bnc2 sequence suggests that the protein possesses an important function (2). To elucidate this function, we have generated mice lacking bnc2. These mice die within 24 h of birth with a cleft palate and abnormalities of craniofacial bones and tongue. We show here that this phenotype results from a direct effect of bnc2 on the multiplication of craniofacial mesenchymal cells. From the study of GenBank data, we demonstrate that the basonuclins are the vertebrate orthologs of the insect disco proteins. We note the similarity between the function of bnc2 in the mouse and that of the disco proteins in Drosophila. We also note that some human craniofacial abnormalities such as cleft palate might be due to lack of bnc2.
Results
Ayu21–18, an ES Cell Line with a Gene-Trap Insertion in the bnc2 Gene.
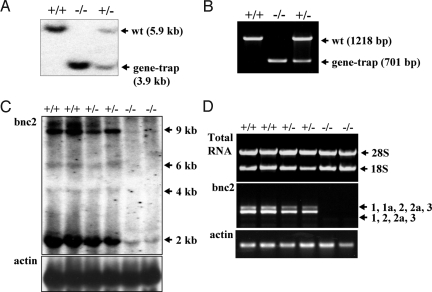
We searched gene-trap ES cell repositories for bnc2 insertions. A BLAST search using the mouse cDNA sequence identified line Ayu21–18 (15). The description of the generation of Ayu21–18 can be found at http://egtc.jp. Inverse genomic PCR (16) and nucleotide sequencing located the vector insertion site of Ayu21–18 in the intron that separates bnc2 exons 2a and 3. This allowed us to generate a probe and to design primers for genotyping by Southern analysis (Fig. 1A) and multiplex PCR (Fig. 1B). Probing of BamHI, PstI, ScaI and BglII digests of bnc2+/− genomic DNA with a fragment of the vector demonstrated that Ayu21–18 mice contained no other copy of the transgene inserted elsewhere in the genome.
Fig. 1.
Analysis of mice by Southern blotting, multiplex PCR, northern blotting, and RT-PCR. (A) Genotyping by Southern blot analysis, using BamHI. (B) Genotyping by multiplex PCR. (C) Northern analysis demonstrating the virtual absence of bnc2 mRNA in bnc2−/− embryos at E17.5. (D) RT-PCR with primers in exons 1 and 3 showing absence of bnc2 mRNA in bnc2−/− embryos.
To determine whether any bnc2 mRNA remained in bnc2−/− mice, we carried out northern analysis and RT-PCR on RNA prepared from embryonic day 17.5 (E17.5) embryos. The 4 mRNAs of 2, 4, 6, and 9 kb previously identified (1) were detected by northern analysis in wild-type (wt) and heterozygotes. The 2-kb RNA produced the most intense band; this RNA is too small to encode bnc2 and its significance is unclear. In bnc2−/− embryos, the RNAs of 4, 6, and 9 kb were not detected, but a small amount of the 2-kb RNA remained (Fig. 1C). No bnc2 mRNA was detected in a total of 8 bnc2−/− embryos examined either by RT-PCR with primers in exons 1 and 3 (Fig. 1D), or by quantitative RT-PCR using primers in exons 2a and 3.
Since the gene-trap vector was inserted in a large bnc2 intron, it could conceivably have disrupted a regulatory element belonging to a gene other than bnc2, and whose altered expression could then contribute to any observed phenotype. To eliminate such a possibility, we examined the expression of all genes located within 2 Mb of the vector insertion site in 3 bnc2−/− and 3 bnc2+/+ embryos by RT-PCR. No alteration of expression was detected in homozygous mutants for any of the 12 genes examined. Microarray analyses confirmed that among the genes located within 23 Mb of the gene-trap insertion site, only bnc2 showed altered expression in bnc2−/− mice.
Neonatal Lethality, Reduced Size of the Head, Abnormalities of the Tongue, and Cleft Palate in bnc2−/− Mice.
Bnc2+/− males crossed with bnc2+/− females produced litters of normal size, in which about 1/4 of the offspring were abnormally small and died within 24 h of birth with aerial distention of their digestive tract. All dead newborns were bnc2−/−, whereas surviving ones were either bnc2+/− or bnc2+/+. A cause of aerial distention of the digestive tract in mice is cleft palate. All bnc2−/− mice had a cleft of the secondary palate, which was complete in about half the newborns and posterior-only in the other half.
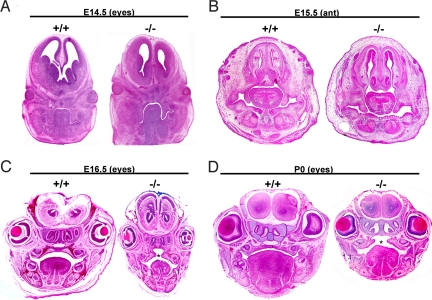
To determine whether lack of bnc2 impaired either elevation or growth of the palatal processes, heads of embryos and neonates were embedded in paraffin and sectioned in the coronal plane. No obvious difference was observed between bnc2−/− and wt mice either at E14.5, when palatal processes were still in a vertical position (Fig. 2A), or at E15.5, when the anterior part of the processes had begun to elevate (Fig. 2B). At E16.5, the palatal processes had elevated and fused in wt embryos, but in homozygous mutants, although the processes had elevated, they failed to establish contact because of insufficient growth after their elevation. There was also a reduction in the overall size of the head and tongue (Fig. 2C). At birth, a wide palatal cleft was obvious and the heads and tongues were clearly smaller than those of their wt littermates (Fig. 2D).
Fig. 2.
Small size of the head, cleft palate and abnormalities of the tongue in bnc2−/− neonates. Mouse heads were embedded in paraffin, sectioned and stained with hematoxylin/eosin. (A) At E14.5, the palatal processes are still in a vertical position. There is no obvious difference between bnc2−/− and wt embryos. (B) Anterior section at E15.5. Processes have begun to elevate in both bnc2−/− and wt. (C) E16.5 embryos. Palatal processes have elevated and fused in the wt. In the bnc2−/− embryo, processes have elevated but have failed to fuse because of insufficient growth toward the midline, thus causing cleft palate. (D) At P0, the wt mouse shows normal fused palate, whereas a cleft palate is evident in the mutant. Head and tongue of the homozygous mutant are smaller than those of the wt. Asterisks indicate cleft palate.
Developmental Abnormalities of Craniofacial Bones in bnc2−/− Mice.
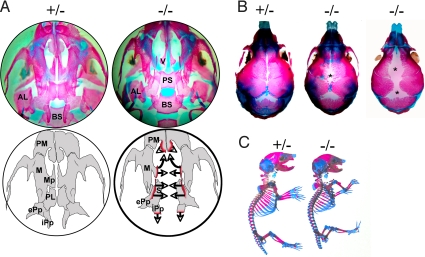
To determine the morphology of the craniofacial and skeletal bones of bnc2−/− newborns, we stained bone and cartilage with alizarin red and alcian blue. This showed that the maxillary processes had failed to grow toward the midline and to extend cranially. The palatine bone was virtually absent. The internal pterygoid processes had not extended caudally. The size of the alisphenoid was reduced (Fig. 3A). Diminished growth of the parietal and frontal bones with an excessively wide sagittal suture and an abnormally large posterior fontanel was clearly visible (Fig. 3B). The rest of the skeleton appeared normal (Fig. 3C). We conclude that lack of bnc2 specifically affects the growth of craniofacial bones.
Fig. 3.
Abnormalities of craniofacial bones in bnc2−/− newborn mice. Newborn mice were stained with alcian blue/alizarin red and cleared with KOH. (A) Inferior view of skull base after removal of the mandible. Anterior is to the top. The upper 2 circles are photographs. In the bnc2+/− mouse, the maxillary and palatine processes have converged toward the midline. In the bnc2−/− mouse, both processes are absent thus exposing the vomer and presphenoid. The internal pterygoid processes of the bnc2−/− mouse have failed to develop caudally. The lower 2 circles are schematic representations. Bone processes that have failed to grow in the bnc2−/− mouse are outlined in red. Arrows indicate direction of axial and rostral growths that did not occur. AL: alisphenoid, BS: basisphenoid, ePp: external pterygoid process, iPp: internal pterygoid process, M: maxillary, Mp: maxillary process, PL: palatine bone, PM: premaxillary, PS: presphenoid, S: sphenoid, V: vomer. (B) Superior view of skull showing that sutures and posterior fontanel are abnormally large in the homozygous mutants. (C) Shorter stature of mutant and absence of gross skeletal abnormalities. Tails have been cut for genotyping.
High Expression of the bnc2lacZ Gene Around the Palatal Cleft.
Although the cleft palate could result from endogenous abnormalities within the palate itself, it could also be secondary to other craniofacial bone defects (17). To investigate these alternatives, we analyzed the expression of the bnc2 gene in developing palatal processes. Both bnc2+/− and bnc2−/− mice express a fused bnc2-lacZ gene (bnc2lacz) whose transcription is under the control of the bnc2 promoter. Therefore, cell types expressing bnc2 could be identified by the presence of the β-galactosidase (β-gal) encoded by lacZ. RT-PCR in various tissues showed that the relative levels of the mRNAs for β-gal and bnc2 were correlated in heterozygotes, thus demonstrating that insertion of the vector did not affect the tissue-specific regulation of the bnc2 promoter. Whole-mount staining of bnc2 −/− newborns for β-gal activity using X-gal as a chromogenic substrate demonstrated that enzyme activity was strongest in the skull, particularly sutures and fontanels, and in the areas surrounding the palatal cleft (Fig. S1). Since the bnc2 gene is strongly expressed at the sites affected by lack of bnc2, the protein probably exerts a direct action on the growth of the palate and other affected bones.
Expression of the bnc2lacZ Gene in Embryonic Head Is Specific to Mesenchymal Cells in the Developing Palatal Processes, the Tongue, and around Bone and Cartilage.
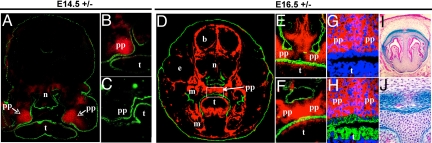
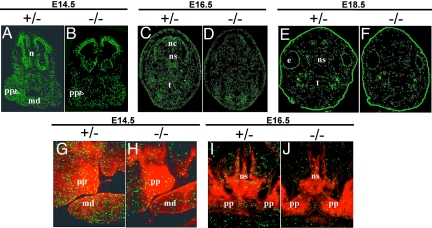
To investigate whether mesenchymal or epithelial cells of the palate expressed bnc2lacZ, we double-stained coronal sections of bnc2+/− embryonic heads with antibodies to β-gal and keratins (Fig. 4). At E14.5, a moderate β-gal staining was virtually confined to the anterior part of the palatal processes; little staining was detected in the rest of the head (Fig. 4 A–C). At E16.5, a much stronger β-gal staining was present along the entire anteroposterior axis of the palate and in sheaths of cells around the brain, the nasal cartilage, and the molar tooth buds, as well as at the base and the periphery of the tongue (Fig. 4 D–F). Examination of the double-stained sections at high magnification clearly established that the β-gal protein was found in mesenchymal cells and was absent from epithelial cells (Fig. 4 G and H).
Fig. 4.
Detection of β-gal and keratins in coronal sections of embryonic heads. (A–H) Frozen sections of bnc2+/− embryonic heads fixed in methanol/acetone and double-stained with anti-β-gal (red) and anti-pankeratin (green) antibodies. (A–C) Embryo at E14.5. (A) View of the whole section showing β-gal staining virtually limited to palatal processes. Staining is present in the anterior region of the palatal processes (B), but not in the posterior part (C). (D–H) Embryo at E16.5. (D) shows strong β-gal staining in the palate and in sheaths of cells around the brain, the molar teeth, and the nasal cartilages as well as at the periphery of the tongue. (E) illustrates the anterior and (F) the posterior part of the palatal processes, which have elevated into a horizontal position and fused at the midline; β-gal staining is present in both parts of the processes. (G and H) Enlargement of the palatal processes boxed in D, counterstained with DAPI (blue) for DNA. β-gal is found in mesenchymal cells and is absent from the epithelia lining the palate and the tongue (G), stained green by the anti-keratin antibody (H). (I and J) Paraffin-embedded sections of nasal region at E16.5, after whole-mount staining with X-gal and counterstaining with eosin. Regions stained with X-gal appear blue. β-gal is specific to the mesenchymal cells surrounding cartilage. b: brain, e: eye, m: molar tooth, n: nasal region, pp: palatal processes, t: tongue.
To confirm the specific expression of the bnc2 gene in mesenchymal cells, heads of bnc2+/− embryos were fixed at E16.5, skinned, and incubated in the presence of X-gal. Heads were then embedded in paraffin and sectioned. β-gal activity was found to be restricted to the mesenchymal cells in the palate, and to the sheaths of mesenchymal cells surrounding cartilage, bone (Fig. 4 I and J) and brain. No β-gal activity was detected in other mesenchymal cells, in the differentiated progeny of mesenchymal cells, such as cartilage and bone, or in non-mesenchymal cell types such as epithelial and brain cells.
Bnc2 Controls the Multiplication of Mesenchymal Cells in the Late Embryo.
The diminished size of mesenchyme-derived structures in the bnc2−/− mice suggests that bnc2 might control the multiplication of mesenchymal cells. The proportion of cells in mitosis can be estimated by counting the cells containing phosphohistone H3 since phosphorylation of histone H3 is strictly associated with the chromosomal condensation that occurs during mitosis (18, 19). Staining of embryonic heads for histone H3 phosphorylated on serine 10 revealed that bnc2−/− embryos possessed 30–50% fewer phosphohistone-containing cells (Fig. 5 A–F). Double staining with the anti-β-gal antibody showed that the reduction in the number of cells possessing phosphohistone H3 affected predominantly areas rich in β-gal-containing mesenchymal cells, particularly the palatal processes at E14.5 (Fig. 5 G–J). These results were confirmed by measuring the mitotic index in paraffin-embedded sections stained with hematoxylin/eosin. The proportion of cells in metaphase, anaphase or telophase in the palatal processes was decreased by nearly 50% at E14.5 and E16.5 in homozygous mutants.
Fig. 5.
Staining of embryonic heads for phosphohistone H3 and β-gal. (A–F) Coronal sections stained for phosphohistone H3 (green), which is specific for mitotic cells. At E14.5 (A and B), E16.5 (C and D), and E18.5 (E and F), the number of cells containing the phosphorylated histone is decreased by 30–50% in bnc2−/− embryos. This diminution is particularly evident in the palatal processes at E14.5 (arrowheads). Notice in F the diminished size of the head of the homozygous mutant at E18.5. (G–J) Double-staining of palatal region for the phosphohistone and the β-gal (red). At E14.5 in the heterozygote, (G) the frequency of cells containing the phosphohistone is similar in areas expressing and not expressing bnc2lacZ. In the bnc2−/− embryo (H), the frequency of phosphohistone-containing cells is predominantly reduced in areas with high expression of the fusion gene. At E16.5 (I and J), the scarcity of mitoses in regions expressing the fusion gene is also observed, particularly in the nasal septum. m: mouth, md: mandible, n: nasal region, nc: nasal cavity, ns: nasal septum, pp: palatal processes, t: tongue.
To determine whether lack of bnc2 increased cell death, DNA strand breaks characteristic of apoptotic cells were labeled with biotin-dUTP (see SI Text). We found no evidence of increased apoptosis in bnc2−/− mice examined at E16.5 and E18.5. We conclude that lack of bnc2 does not decrease the number of mitotic cells by causing cell death.
Bnc2 Is Orthologous to the Disco Proteins.
The similarity between the zinc fingers of basonuclin and those of the Drosophila proteins disco and discorelated has long been recognized (1, 2, 5, 12), but the significance of this similarity has remained unclear because the sequence of the Drosophila disco proteins has no similarity to that of basonuclin, apart from the zinc fingers and the function of the disco proteins in larval head development also seems to have no relation to that of bnc1. In view of the similarity between the function of bnc2 in mouse head development and that of the Drosophila disco proteins in larval head development (4), we reexamined the relatedness of the basonuclins to the disco proteins. The recent sequencing of a number of insect genomes has shown that Drosophila and other dipterans have undergone accelerated evolution and have diverged considerably more from vertebrates than other arthropods (20). We reasoned that comparison of the basonuclin sequences with the nondipteran disco sequences could shed light on the relatedness of the 2 groups of proteins.
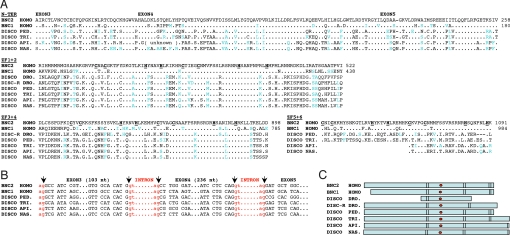
BLASTP and TBLASTN similarity searches of the nondipteran arthropod GenBank databases using the human bnc2 sequence revealed that, apart from the zinc fingers, the predicted disco proteins of Apis, Nasonia, Pediculus, and Tribolium possessed a sequence of about 150 residues with over 65% identity to the N-terminal region of bnc2 and bnc1 (Fig. 6A). In bnc1, bnc2, and disco genes, this region was encoded by 3 exons, all of which were interrupted by introns at exactly the same position, and 2 of which were of the same size in insects and vertebrates (Fig. 6B). The positions of the zinc finger pairs, their number, and the position of the NLS were also similar in the nondipteran disco proteins and in the basonuclins (Fig. 6C). Therefore the disco and the basonuclin genes must be derived from a common ancestor. All of the nondipteran arthropods examined possessed a single disco gene that resembled Drosophila discorelated more than disco. We conclude that discorelated is the ancestral arthropod gene, and disco its dipteran-specific paralog.
Fig. 6.
Relatedness of basonuclins and disco proteins. (A) Alignment of the N-terminal sequence and of the 3 pairs of zinc fingers of human bnc2 (GenBank NP_060107.3) with those of human bnc1 (NP_001708.3), of the Drosophila disco (NP_523362.2) and discorelated (NP_727938.1) proteins and of the unique disco protein of the louse (PED: Pediculus humanus corporis), the flour beetle (TRI: Tribolium castaneum), the honey bee (API: Apis mellifera), and the jewel wasp (NAS: Nasonia vitripennis). The non-Drosophila disco sequences were deduced from genomic sequences identified by similarity searches using the human bnc2 amino acid sequence (Pediculus: AAZO01002822.1 and AAZO01000000, Tribolium: XP_971658.1, Apis: NW_001253429 and Nasonia: NW_001817459.1). The cysteines and histidines of the zinc fingers are in bold type and underlined. Conservative amino acid replacements are in blue type. Numbers are relative to the first methionine. (B) Comparison of intron boundaries of bnc2, bnc1, and disco. (C) Structure of the basonuclins and disco proteins. Basonuclins contain 3 pairs of zinc fingers (vertical bars) and an NLS (red dot) located between fingers 2 and 3. Disco proteins contain from 1 to 3 pairs of zinc fingers and an NLS placed as in the basonuclins. NLS predicted with PredictNLS software.
Earlier studies have shown that the disco gene is expressed in the gnathal buds and transiently in a sheet of visceral mesoderm around the gut (21); disco-driven lacZ expression is also found in the joints of the adult leg (22). Bnc2-driven lacZ expression was similarly found predominantly in the gut of the mouse embryo (Fig. 6D), in the mesodermal cells that surrounded the submucosa and around the joints (Fig. S2). Therefore the Drosophila disco genes have a pattern of expression similar to that of the bnc2 gene, and unlike that of the bnc1 gene whose expression is confined to keratinocytes and reproductive germ cells (5, 6). By its sequence, its tissue distribution, and its function, bnc2 appears to be the vertebrate homolog of the disco proteins.
Discussion
From results presented here, it is clear that basonuclin 2 is essential for the growth of a number of craniofacial bones, all of which are of neural crest origin (23). Although expression of the basonuclin 2 gene is also observed in mesenchymal cells not derived from the neural crest (Fig. S2), these cells do not seem to require basonuclin 2 for proper multiplication. Cleft of the secondary palate can be caused by mutations in genes encoding transcription factors, growth and signaling molecules, and their receptors (17). Basonuclin 2 might regulate the expression of some of these genes.
The relation between the basonuclins and the disco proteins is of great interest. We provide here incontrovertible evidence that the basonuclins are the vertebrate orthologs of the insect disco proteins (Fig. 6). The homology between basonuclins and disco proteins was not recognized earlier because only Drosophila disco sequences were available. The disco proteins of Drosophila have diverged considerably more from the basonuclin sequences than those of nondipterans, such as Tribolium. It has been suggested (24) that Tribolium is better suited for comparisons between phyla than the widely used dipterans and that when attempting to link human genes to their Drosophila homologs, data from Tribolium should help to provide a more conservative sequence of reference. Such an approach was indeed necessary to resolve the relationship between the Drosophila zen gene and the human Hox3 genes (25).
It has been postulated that the gene for bnc2 is the older of the 2 basonuclin genes and after its duplication to produce bnc1, the latter was free to evolve in other directions, while the gene for bnc2 remained largely invariant and retained its original function (12). We believe that this interpretation is correct and that bnc2 carries out in vertebrates a function similar to that of disco in insects. Embryos lacking both disco and discorelated fail to hatch and show absence of the cephalopharyngeal skeletal structures that develop from the gnathal segments (4, 26). This phenotype is strikingly similar to that observed in mice lacking bnc2, whose most pronounced defects lie in the pharyngeal region and particularly in the gnathal bones (Fig. S1). Loss of disco in Tribolium also affects the development of appendages derived from the head segments. Disco RNA interference in larvae leads to a failure of the mandibular, maxillary and labial segments to extend from the head. RNA interference in adults causes severe truncation of the maxillary and labial appendages in both Drosophila and Tribolium (27, 28). The truncation of the head appendages observed in the disco mutants of these species resembles the truncation of the palatine and pterygoid processes observed in mice lacking bnc2 (Fig. 3A). It would be of interest to determine whether nondipteran disco genes can rescue the phenotype of bnc2−/− mice.
Bnc1 is associated with the ability of keratinocytes to multiply (7). We show here that bnc2 also has a function in cell multiplication, albeit in a different cell type. It has similarly been suggested that the disco proteins promote cell proliferation in the appendage primordia (28). Conservation of the zinc fingers by bnc1, bnc2, and the disco proteins might be related to the function in cell multiplication shared by these proteins and suggests that their molecular targets are similar or identical.
Human monosomy 9p associates various craniofacial abnormalities, which include an excessively arched palate and defects of the nasal septum. The genomic region involved has been narrowed down to 4.7 Mb in close proximity to the bnc2 gene (29). Duplications of the 9p22–24 region associated with breakpoints in the region of the bnc2 gene are also associated with craniofacial abnormalities including the palate (30). These human developmental defects may be the consequence of bnc2 haploinsufficiency caused by disruption of the gene.
Materials and Methods
Inverse Genomic and Multiplex PCR.
DNA was prepared from mouse tails as previously described (31). For inverse genomic PCR, DNA was digested with PstI, self-ligated, and amplified according to (16). For multiplex PCR, 2 pairs of primers (one around the insertion site and 1 in pU21) were mixed in the same PCR (30 cycles of amplification at 95° for 1 min, 56° for 1 min, and 72° for 1 min).
Southern Blot Analysis.
Southern blots of mouse tail DNA were performed as previously described (32). The probe was an 861-bp fragment of the bnc2 intron located immediately 3′ to the vector.
Northern Blot.
Embryos were pulverized under liquid nitrogen and RNA was prepared using the TRIzol reagent (Invitrogen). Total RNA (40 μg) was resolved by electrophoresis on a formaldehyde/1% agarose gel, partially hydrolyzed in 50 mM NaOH to facilitate transfer of large RNA, and transferred onto a nylon membrane by vacuum blotting. The probe was a 900-bp fragment in exon 5 of bnc2. Hybridization was carried out in ExpressHyb (Clontech) using the manufacturer's protocol except that the last washes were at 65° instead of 50°.
Expression of Genes Located in the Vicinity of the Gene-Trap Insertion Site.
Genes located within 2 Mb of the insertion site and analyzed by RT-PCR were the following (GenBank): 1810054D0, Snapc3, Psip1, LOC100040611, 4930473A06, LOC668010, LOC100040660, LOC100040284, D530005L17, Sh3gl2, A930009N24, and Adamtsl1.
Microarray analysis was performed with RNA isolated from mouse embryonic heads at E16.5 using Qiagen columns and Affymetrix mouse gene 1.0 ST arrays. Three wt and 3 homozygous mutants were analyzed. Gene-level expression values were derived from the CEL file probe-level hybridization intensities using the model-based Robust Multichip Average algorithm (RMA). RMA performs normalization, background correction, and data summarization. Differential analysis was performed using the local-pooled-error test, and a P value threshold of P < 0.05 was used as the criterion for expression. The estimated false discovery rate of this analysis was calculated using the Benjamini and Hochberg approach in order to correct for multiple comparisons.
Histological Analysis.
For sections stained with hematoxylin and eosin, embryos were fixed in either Bouin's solution or 4% paraformaldehyde and then embedded in paraffin and stained.
For indirect immunofluorescence, embryos were snap frozen in OCT (Miles) and isopentane and cut with a cryomicrotome at 7 μm. Sections were fixed in acetone/methanol (1:1) at −20° and stained with primary antibodies as described earlier (33). See SI Text for antibodies used.
Phosphohistone H3-positive cells were counted using the ImageJ software. Over 40,000 cells were counted in a total of thirteen E14.5, E16.5, and E18.5 embryos. For each embryo, cells were counted in 3 widely spaced coronal sections.
Method for staining of cartilage and bone with alizarin red/alcian blue is available as SI Text.
Supplementary Material
Acknowledgments.
We thank Bruno Passet (INRA, Jouy-en-Josas) for implanting Ayu21–18 embryos into foster mothers. This work was supported by the Centre National de la Recherche Scientifique, the Association pour la Recherche sur le Cancer, the Ligue contre le Cancer, fellowships from the Fondation pour la Recherche Médicale and the Fondation Bettencourt-Schueller (A.V.), and fellowships from the Ministère de l'Enseignement Supérieur et de la Recherche (A.M.-D. and C.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0905840106/DCSupplemental.
References
- 1.Romano R, Li H, Tummala R, Maul R, Sinha S. Identification of Basonuclin2, a DNA-binding zinc-finger protein expressed in germ tissues and skin keratinocytes. Genomics. 2004;83:821–833. doi: 10.1016/j.ygeno.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Vanhoutteghem A, Djian P. Basonuclin 2: An extremely conserved homolog of the zinc finger protein basonuclin. Proc Natl Acad Sci USA. 2004;101:3468–3473. doi: 10.1073/pnas.0400268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steller H, Fischbach K, Rubin G. Disconnected: A locus required for neuronal pathway formation in the visual system of Drosophila. Cell. 1987;50:1139–1153. doi: 10.1016/0092-8674(87)90180-2. [DOI] [PubMed] [Google Scholar]
- 4.Mahaffey J, Griswold C, Cao Q. The Drosophila genes disconnected and disco-related are redundant with respect to larval head development and accumulation of mRNAs from deformed target genes. Genetics. 2001;157:225–236. doi: 10.1093/genetics/157.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tseng H, Green H. Basonuclin: A keratinocyte protein with multiple paired zinc fingers. Proc Natl Acad Sci USA. 1992;89:10311–10315. doi: 10.1073/pnas.89.21.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang Z, Gallicano G, Yu Q, Fuchs E. An unexpected localization of basonuclin in the centrosome, mitochondria, and acrosome of developing spermatids. J Cell Biol. 1997;137:657–669. doi: 10.1083/jcb.137.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tseng H, Green H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Tseng H. Basonuclin-null mutation impairs homeostasis and wound repair in mouse corneal epithelium. PLoS ONE. 2007;2:e1087. doi: 10.1371/journal.pone.0001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tian Q, Kopf G, Brown R, Tseng H. Function of basonuclin in increasing transcription of the ribosomal RNA genes during mouse oogenesis. Development. 2001;128:407–416. doi: 10.1242/dev.128.3.407. [DOI] [PubMed] [Google Scholar]
- 10.Iuchi S, Green H. Basonuclin, a zinc finger protein of keratinocytes and reproductive germ cells, binds to the rRNA gene promoter. Proc Natl Acad Sci USA. 1999;96:9628–9632. doi: 10.1073/pnas.96.17.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Zhang S, Schultz R, Tseng H. Search for basonuclin target genes. Biochem Biophys Res Commun. 2006;348:1261–1271. doi: 10.1016/j.bbrc.2006.07.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green H, Tseng H. In: Basonuclin: A Zinc Finger Protein of Epithelial Cells and Reproductive Germ Cells. Iuchi S, Kuldell N, editors. Georgetown, Texas: Landes Bioscience; 2005. [Google Scholar]
- 13.Ma J, Zeng FY, Schultz RM, Tseng H. Basonuclin: A novel mammalian maternal-effect gene. Development. 2006;133:2053–2062. doi: 10.1242/dev.02371. [DOI] [PubMed] [Google Scholar]
- 14.Vanhoutteghem A, Djian P. Basonuclins 1 and 2, whose genes share a common origin, are proteins with widely different properties and functions. Proc Natl Acad Sci USA. 2006;103:12423–12428. doi: 10.1073/pnas.0605086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniwaki T, et al. Characterization of an exchangeable gene trap using pU-17 carrying a stop codon-β geo cassette. Dev Growth Differ. 2005;47:163–172. doi: 10.1111/j.1440-169X.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- 16.Silver J, Keerikatte V. Novel use of polymerase chain reaction to amplify cellular DNA adjacent to an integrated provirus. J Virol. 1989;63:1924–1928. doi: 10.1128/jvi.63.5.1924-1928.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Gurley L, D'Anna J, Barham S, Deaven L, Tobey R. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- 19.Goto H, et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- 20.Richards S, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 21.Lee K, Freeman M, Steller H. Expression of the disconnected gene during development of Drosophila melanogaster. EMBO J. 1991;10:817–826. doi: 10.1002/j.1460-2075.1991.tb08014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bishop S, Klein T, Arias A, Couso J. Composite signaling from Serrate and Delta establishes leg segments in Drosophila through Notch. Development. 1999;126:2993–3003. doi: 10.1242/dev.126.13.2993. [DOI] [PubMed] [Google Scholar]
- 23.Le Douarin N, Creuzet S, Couly G, Dupin E. Neural crest cell plasticity and its limits. Development. 2004;131:4637–4650. doi: 10.1242/dev.01350. [DOI] [PubMed] [Google Scholar]
- 24.Savard J, Tautz D, Lercher M. Genome-wide acceleration of protein evolution in flies (Diptera) BMC Evol Biol. 2006;6:7. doi: 10.1186/1471-2148-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falciani F, et al. Class 3 Hox genes in insects and the origin of zen. Proc Natl Acad Sci USA. 1996;93:8479–8484. doi: 10.1073/pnas.93.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson L, Bowling D, Mahaffey J, Imiolczyk B, Mahaffey J. An interactive network of zinc-finger proteins contributes to regionalization of the Drosophila embryo and establishes the domains of HOM-C protein function. Development. 2004;131:2781–2789. doi: 10.1242/dev.01159. [DOI] [PubMed] [Google Scholar]
- 27.Robertson L. Raleigh: University of North Carolina; 2005. A developmental genetic analysis of the C2H2 zinc finger encoding gene, disconnected, in Drosophila melanogaster and Tribolium castaneum. PhD. [Google Scholar]
- 28.Patel M, et al. The appendage role of insect disco genes and possible implications on the evolution of the maggot larval form. Dev Biol. 2007;309:56–69. doi: 10.1016/j.ydbio.2007.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Kawara H, et al. Narrowing candidate region for monosomy 9p syndrome to a 4.7-Mb segment at 9p22.2-p23. Am J Med Genet A. 2006;140:373–377. doi: 10.1002/ajmg.a.31094. [DOI] [PubMed] [Google Scholar]
- 30.Fujimoto A, Lin M, Schwartz S. Direct duplication of 9p22–>p24 in a child with duplication 9p syndrome. Am J Med Genet. 1998;77:268–271. [PubMed] [Google Scholar]
- 31.Laird P, et al. Simplified mammalian DNA isolation procedure. Nucleic Acids Res. 1991;19:4293. doi: 10.1093/nar/19.15.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tseng H, Green H. Remodeling of the involucrin gene during primate evolution. Cell. 1988;54:491–496. doi: 10.1016/0092-8674(88)90070-0. [DOI] [PubMed] [Google Scholar]
- 33.Vanhoutteghem A, Djian P. The human basonuclin 2 gene has the potential to generate nearly 90,000 mRNA isoforms encoding over 2,000 different proteins. Genomics. 2007;89:44–58. doi: 10.1016/j.ygeno.2006.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.