Abstract
The Nup84 complex constitutes a key building block in the nuclear pore complex (NPC). Here we present the crystal structure of one of its 7 components, Nup120, which reveals a β propeller and an α-helical domain representing a novel fold. We discovered a previously unidentified interaction of Nup120 with Nup133 and confirmed the physiological relevance in vivo. As mapping of the individual components in the Nup84 complex places Nup120 and Nup133 at opposite ends of the heptamer, our findings indicate a head-to-tail arrangement of elongated Nup84 complexes into a ring structure, consistent with a fence-like coat for the nuclear pore membrane. The attachment site for Nup133 lies at the very end of an extended unstructured region, which allows for flexibility in the diameter of the Nup84 complex ring. These results illuminate important roles of terminal unstructured segments in nucleoporins for the architecture, function, and assembly of the NPC.
Keywords: crystal structure, fluorescence localization, mRNA export, nuclear pore complex, site-directed mutagenesis
The nuclear pore complex (NPC) mediates the selective exchange of macromolecules between the nucleus and cytoplasm and represents one of the largest proteinaceous assemblies in the eukaryotic cell (1–4). Electron microscopic studies revealed that the NPC is a cylindrical structure consisting of a central core with an 8-fold rotational symmetry across a nucleocytoplasmic axis and a 2-fold symmetry in the plane of the nuclear envelope (5–8). This symmetric core is linked to the asymmetric cytoplasmic filaments and a nuclear basket structure (9). The NPC comprises ≈30 different nucleoporins (nups), organized into several subcomplexes (10–12). Over the last decade, a detailed mechanistic understanding of some of the mobile transport factors and their involvement in nucleocytoplasmic transport has been gained at the atomic level (13, 14); however, a similar level of understanding regarding the structure and assembly of the NPC remains poorly understood, despite its central importance in eukaryotic life.
In cells with open mitosis, the NPC is disassembled either into individual nups or into various subcomplexes from which the NPC reassembles at the completion of the cell cycle (15–17). Similar subcomplexes also were obtained by dissecting intact NPCs, using nonionic detergents and a range of salt concentrations (12). In yeast, a well-characterized heptameric subcomplex consists of Nup84, Nup85, Nup120, Nup133, Nup145C, Sec13, and Seh1 (18–20). Negative-stain electron microscopy of this heptamer assembled from recombinant proteins has revealed a ≈400-Å-long, Y-shaped assembly and established the relative position of its members (19). Nup133 and Nup84 are located at the base, the Sec13·Nup145C pair is in the center, and Nup120 and the Seh1·Nup85 pair form the 2 upper arms of the Y.
All members of the yeast heptamer are well conserved; however, the vertebrate complex contains 2 additional members, Nup37 and Nup43, forming a nonamer (16, 17, 21, 22). The deletion or immunodepletion of any nup from these complexes has dramatic consequences on the architecture and function of the NPC, as well as on the organization of the nuclear envelope (17, 20, 23–26). In particular, Nup120 is involved in nuclear poly(A)+ mRNA and preribosome export, NPC assembly and distribution, and nuclear envelope organization (26–28).
To date, partial atomic structures of several components of the heptameric complex have been determined: the β propeller of Nup133 (29), the Sec13·Nup145C pair (30), the mammalian Nup84 homolog Nup107 in complex with Nup133 (31), and the Seh1·Nup85 pair (32, 33). But atomic-level knowledge of how these modules are precisely arranged and interact with one another in the heptameric complex remains largely elusive. Moreover, the higher-order arrangement of the heptamers in the symmetric NPC core has not yet been established. Notably, the hetero-octameric, rod-shaped assemblies of the Sec13·Nup145C and Seh1·Nup85 nucleoporin pairs support a plausible model for a coat for the nuclear pore membrane (30, 32). According to this model, 32 heptamers cluster into a cylindrical, fence-like coat that bears a striking resemblance to other membrane coat systems, such as COPII (34).
We have previously proposed that this coat cylinder conceptually represents 1 of 4 concentric and interdigitated cylinders in the symmetric core of the NPC (30). The coat cylinder is sandwiched between the nucleoporins of the pore membrane (POM) cylinder and the adapter cylinder. None of the POM nups has been structurally characterized to date; Nic96 represents the only known structure of the adapter cylinder (35, 36). Finally, the innermost cylinder harbors the channel nups, whose natively unfolded phenylalanine-glycine repeats serve as a transport barrier and docking sites for mobile transport factors, such as karyopherins (14). Crystal structures of a mammalian channel nup suggest circumferential sliding as a mechanism to adjust the channel diameter in response to cargo translocation (37).
To gain further evidence for and new insight into the architecture of a coat for the nuclear pore membrane, we determined the crystal structure of Nup120, the only component of the Nup84 complex presently lacking structural characterization. The Nup120 structure reveals a 7-bladed β propeller domain and an α-helical domain that represents a novel fold. Importantly, we identified a previously unknown interaction between Nup120 and the N-terminal domain (NTD) of Nup133 that suggests a head-to-tail arrangement of Nup84 complexes into an 8-membered ring. The unstructured, N-terminal extension of Nup133 acts like an anchor, mediating the interaction with Nup120. Because unstructured terminal regions are common in nucleoporins, these findings suggest important, general roles of extended, unstructured regions for the architecture, function, and assembly of the NPC.
Results
Structure Determination.
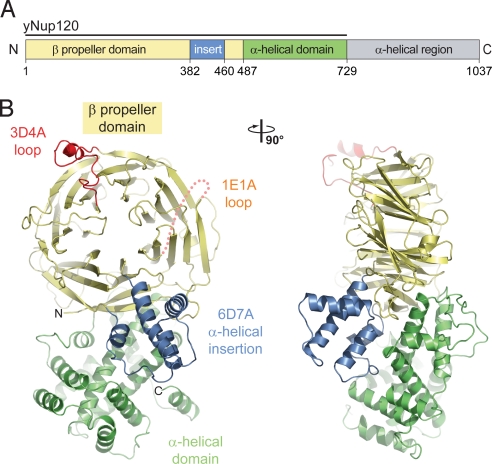
Secondary structure predictions of Saccharomyces cerevisiae Nup120 revealed 2 distinct domains: an N-terminal, ≈500-residue β strand–rich region followedby a C-terminal, ≈550-residue α-helical region (Fig. 1A). Guided by this analysis, we designed a series of expression constructs and identified a stable fragment composed of residues 1–729 and lacking the ≈300-residue C-terminal region. We call this fragment the Nup120 NTD.
Fig. 1.
Structure of the S. cerevisiae Nup120 NTD. (A) Domain structure. Yellow, β propeller domain; blue, α-helical insertion in the 6D7A loop; green, α-helical domain; gray, α-helical region. The bar above the domain structure denotes the crystallized fragment. (B) Structure of the Nup120 NTD in ribbon representation, color-coded as in (A). A 90°-rotated view is shown on the right.
Crystals of the ≈80-kDa Nup120 NTD appeared in the orthorhombic and triclinic space groups P21212 and P1, with 1 and 4 molecules, respectively, in the asymmetric unit. The structure was solved by single anomalous dispersion (SAD), using x-ray diffraction data from a mercury derivative of the S207C mutant in the orthorhombic space group. The structure was refined to 2.6-Å resolution with an Rcryst of 23.2% and an Rfree of 25.4%. Crystals of the wild-type protein were of significantly lower quality, and the structure was refined to 3.0-Å resolution (Rcryst, 25.5%; Rfree, 27.4%). For details of the crystallographic analysis, see Table S1AQ: C.
Architectural Overview.
The polypeptide chain of the Nup120 NTD folds into 2 distinct domains, an N-terminal β propeller domain and a C-terminal α-helical domain with a novel fold (Fig. 1B; Movies S1 and S2). The 7-bladed β propeller domain contains several insertions, most notably a 4-helix bundle that is inserted between strands 6D and 7A, forming a small subdomain. The C-terminal α-helical domain packs against the side of the β propeller, predominantly interacting with blade 7 and the 4-helix bundle. The protein dimensions are ≈90 Å × 50 Å × 35 Å.
The wild-type and mutant Nup120 NTD structures are essentially identical, with a root mean square (RMS) deviation of 0.7 Å over 678 Cα atoms. The only significant structural difference between the 2 proteins pertains to the 3D4A insertion that undergoes a rigid body movement of ≈5 Å (Fig. S1A). Based on the 2 crystal structures, introducing the S207C mutation into helix αB appears to decrease its mobility with respect to the wild-type protein, consistent with the higher diffraction quality of the mutant. Because of the higher quality of the S207C Nup120 structure, the analysis and the figures of Nup120 NTD refer to the mutant structure.
The Nup120 β Propeller Domain.
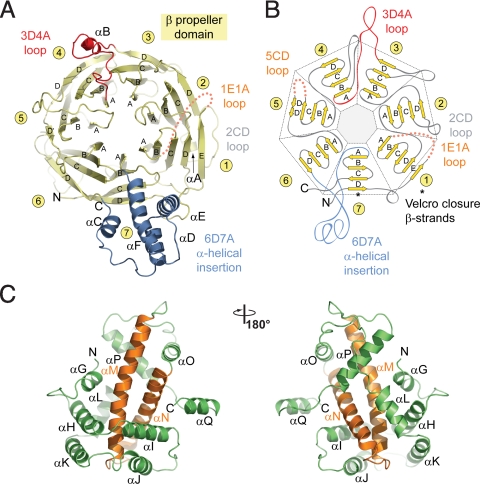
The β propeller domain generally conforms to the canonical β propeller fold (38, 39) (Figs. 2A and 2B). In contrast to the canonical fold, the Nup120 β propeller contains an additional β strand in blade 1 that is inserted into the 7D1A connector (the loop between strand D of blade 7 and strand A of blade 1), providing a fifth strand to the first blade. Moreover, the Nup120 β propeller features a 78-residue 4-helix bundle inserted into the 6D7A loop. Because the major part of the surface-exposed 1E1A and 5CD loops (residues 30–52 and 306–310) are invisible in the electron density, presumably due to disorder, these 2 regions were omitted from the final model. The bottom face of the Nup120 β propeller domain contains a deep cavity with a small opening on the top face of the β propeller, resulting in a hole through the domain (Fig. S1B).
Fig. 2.
Structural analysis of the Nup120 NTD domains. (A) Ribbon representation of the Nup120 β propeller domain. The 7 blades of the β propeller core (yellow), the location of the disordered 1E1A loop (orange), the 3D4A loop (red), the α-helical insertion in the 6D7A loop (blue), and their secondary structure elements are indicated. (B) Schematic representation of the Nup120 β propeller domain and the locations of its various insertions. (C) Ribbon representation of the Nup120 α-helical domain. The leucine zipper–like core (orange) and the 9 surrounding α-helices (green) are indicated. A 180°-rotated view is shown on the right.
The Nup120 α-Helical Domain.
The Nup120 α-helical domain comprises 11 α helices, forming a compact, novel fold (Fig. 2C). Helices αN and αM, at the core of the domain, form a leucine zipper–like α-helical hairpin surrounded by 6 helices (αG–αL) at its tip and 3 helices (αO–αQ) at its base. The hydrophobic interface between the 2 domains is formed by helices, αG, αI, αM, αN, αO, and αP of the α-helical domain and helices αC and αD, β strands 1B, 7A, 7B, and 7D, and loops 1CD, 6AB, 7AB, and 7D1E of the β propeller domain. A total of 93 residues participate in the interaction between the 2 domains, burying a surface area of ≈4,000 Å2.
Surface Properties.
The surface of Nup120 NTD features 5 essentially invariant patches primarily on one side of the protein (Figs. S1B–D and S2), with 3 regions located in the β propeller domain and 2 regions located in the α-helical domain. Region 1, located at the side of the β propeller, is formed by blades 3 and 4. Region 2, at the top face of the β propeller domain, is formed by blades 6 and 7, and region 3 localizes to the α-helical insertion in the 6D7A connector. Regions 4 and 5, in the α-helical domain, are formed by helices αH and αK and by helices αJ, αK, αL, αM, αN, and their connecting loops, respectively. Although the cavities found in β propeller domains are often used for ligand binding (39, 40), the surface of the cavity in Nup120 is not evolutionarily conserved, suggesting that this region is not likely to be used as a binding site. Whereas the surface of Nup120 NTD is predominantly negatively charged, the conserved region 1 is primarily hydrophobic. Interestingly, similar electrostatic surface properties have been identified in the structures of the other members of the Nup84 complex.
Nup120 Interacts With Nup133.
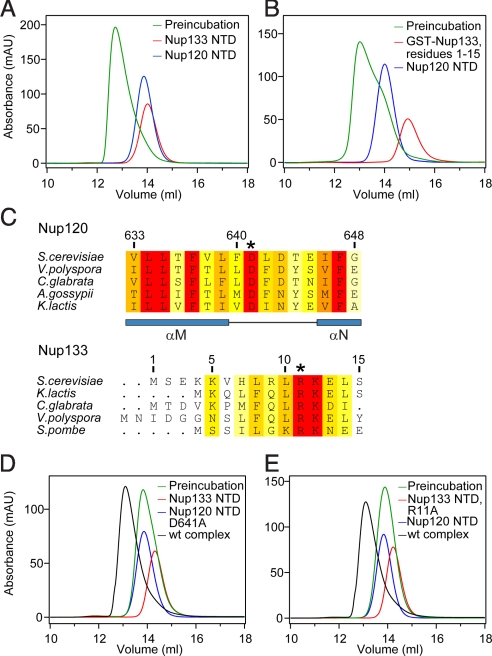
We hypothesized that the conserved surface patches on Nup120 NTD are harnessed for the interaction with adjacent nucleoporins in the NPC, with members of the Nup84 complex being prime candidates. Thus, we systematically probed the remaining proteins of the heptameric Nup84 complex for their ability to bind to Nup120. Although full-length Nup120 is capable of forming trimeric complexes with the nucleoporin pairs Seh1·Nup85 and Sec13·Nup145C (19), the corresponding complex formations with Nup120 NTD could not be detected by size-exclusion chromatography. This finding suggests that the C-terminal domain of Nup120 mediates the interaction with Seh1·Nup85 and also with Sec13·Nup145C.
However, Nup120 NTD formed a stable complex with the N-terminal domain of Nup133 in size-exclusion chromatography and by analytical ultracentrifugation (Figs. 3A and S3). In contrast, the remaining proteins of the heptamer, including the C-terminal domain (CTD) of Nup133, were incapable of interacting with the Nup120 NTD (Fig. S3B). A smaller Nup120 fragment containing only the N-terminal β propeller domain, residues 1–486, failed to interact with the Nup133 NTD (Fig. S3F). Because the C-terminal α-helical domain of Nup120 (residues 487–729) was insoluble in our bacterial expression system, most likely due to the large hydrophobic surface exposed in the absence of the β propeller domain, we were unable to directly test whether the α-helical domain would be sufficient for Nup133 binding.
Fig. 3.
Nup120 NTD interacts with the 15 N-terminal residues of Nup133. (A) Gel filtration profiles of Nup120 NTD (blue), Nup133 NTD (red), and the Nup120 NTD·Nup133 NTD complex (green). (B) Gel filtration profiles of Nup120 NTD (blue), the 15 N-terminal residues of Nup133 fused to GST (red), and their complex (green). All proteins were injected at approximately the same concentrations. (C) The invariant Asp-641 of Nup120 and Arg-11 of Nup133 are key residues for complex formation. The location of Asp-641 and Arg-11 are indicated by asterisks in multispecies sequence alignments. (D) Gel filtration profiles of Nup120 NTD D641A mutant (blue) and the Nup133 NTD (red), and the elution profile resulting from incubation of the 2 proteins before injection (green). (E) Gel filtration profiles of Nup120 NTD (blue) and the Nup133 NTD R11A mutant (red), and the elution profile resulting from incubation of the 2 proteins before injection (green). As a reference, the gel filtration profile of the wild-type Nup120 NTD·Nup133 NTD is indicated in black.
A 15-Residue Segment in Nup133 Mediates Binding to Nup120.
The human Nup133 NTD structure revealed a 7-bladed β propeller domain with a presumably unstructured N-terminal segment of ≈75 residues (29). Secondary structure predictions and limited proteolysis experiments suggest that the ≈55 N-terminal residues also are unstructured in the yeast Nup133 NTD. We refer to this 55-residue fragment (residues 1–55) as the N-terminal extension (NTE). Using circular dichroism spectroscopy, we confirmed a random coil conformation of the Nup133 NTE (Fig. S3G). Although the Nup133 NTE fragment was capable of interacting with Nup120, the β propeller domain of Nup133 comprising residues 56–520 failed to bind to Nup120 (Figs. S3C and D). Further analysis revealed that an N-terminal Nup133 fragment containing only residues 1–15 was sufficient for complex formation with Nup120 (Figs. 3B and S3E). To quantitate the affinity between Nup120 and Nup133, we determined their binding constant to be ≈0.7 μM (Fig. S4A) by isothermal titration calorimetry (ITC). The first 15 N-terminal residues are primarily responsible for this interaction, because the affinity of this peptide for Nup120 was decreased only slightly (≈2 μM) (Fig. S4B). These data demonstrate that the very N-terminal segment of Nup133 is necessary and sufficient for the interaction with Nup120.
The Interaction of Nup120 and Nup133 Is Electrostatic in Nature.
Given the strongly negative electrostatic potential of the Nup120 surface and the presence of several conserved lysine and arginine residues in the Nup133 NTE, we examined the influence of high-salt buffer conditions on the stability of the Nup120·Nup133 complex. We found that the apparent molecular weight of the complex decreased with increasing salt concentration (Fig. S4C). Moreover, the Nup120·Nup133 complex exhibited substantial protein concentration–dependent mobility, as judged by size exclusion chromatography (Fig. S4D). Thus, the association between Nup120 and Nup133 is dynamic and appears to be governed by electrostatic interactions that can be substantially weakened in high-salt conditions.
To fine-map the interaction between Nup120 and Nup133, we performed alanine-scanning mutagenesis of residues within the 5 conserved surface patches and tested for complex formation by size-exclusion chromatography (Figs. 3D and E, Fig. S5, and Table S2). All mutants were purified to homogeneity in milligram amounts; their behavior on a gel filtration column was indistinguishable from that of the wild-type protein. For Nup120, mutation of the invariant residue Asp-641, located in the αM–αN connector of conserved region 5, had the strongest effect and abolished complex formation with Nup133 NTD (Fig. 3D). In contrast, mutations of Asp-578, Ile-646, or Phe-647 had only moderate effects on the interaction between the 2 proteins, as indicated by a substantial shift toward lower molecular weight (Fig. S5). Interestingly, the combination of 2 of these mutants (T551A and I579A) aggravated their individual effects and completely abolished the interaction (Fig. S5F). For Nup133, we identified the invariant residue Arg-11 as essential for complex formation (Fig. 3E), whereas other residues in the Nup133 NTE had no detectable effect (Table S2). To test whether Nup120 Asp-641 and Nup133 Arg-11 form a direct salt bridge in the interface between the 2 proteins, we created residue substitutions that reversed the charges at these 2 key residues (Nup120 Asp641Arg and Nup133 Arg11Asp). The 2 mutants failed to rescue the binding between the 2 proteins, however (Fig. S5G).
Altogether, these findings are consistent with an electrostatic interface between Nup120 and Nup133, with Nup133 Arg-11 and Nup120 Asp-641 playing key roles in mediating this interaction.
In Vivo Analysis of the Nup120–Nup133 Interaction.
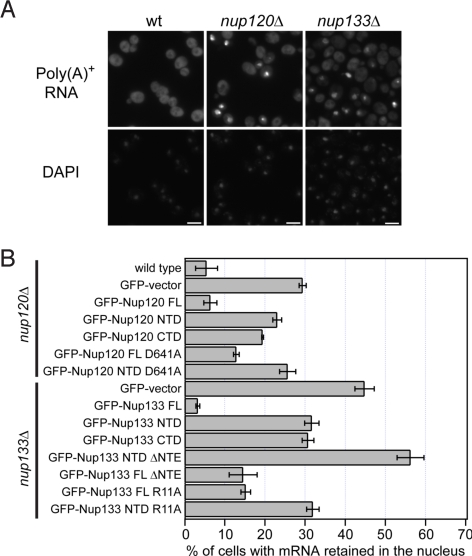
To characterize the physiological relevance of the Nup120–Nup133 interaction, we analyzed the nuclear retention of poly(A)+ mRNA, the localization, and the growth phenotype of various Nup120 and Nup133 GFP fusion proteins and mutants (Figs. 4, S6, and S7). For Nup120, we analyzed the full-length protein, the NTD (residues 1–729), the CTD (residues 730–1037), and the mutant full-length protein and the NTD carrying the D641A mutation. For Nup133, we analyzed the full-length protein, the NTD (residues 1–520), the NTD lacking the unstructured NTE (residues 56–520), the CTD (residues 521–1157), and the mutant full-length protein and the NTD harboring the R11A mutation.
Fig. 4.
Physiological relevance of the Nup120–Nup133 interaction. (A) Detection of poly(A)+ mRNA using an Alexa-647 labeled 50-mer oligo dT FISH probe (Top). Wild-type cells (Left) display a diffuse FISH signal, while nup120Δ (Middle) and nup133Δ (Right) cells yield strong nuclear signals that coincide with DAPI staining, consistent with poly(A)+ mRNA retention inside the nucleus. (B) Quantitation of nuclear poly(A)+ mRNA retention in nup120Δ and nup133Δ yeast strains complemented with various Nup120 and Nup133 variants. The percentages refer to the fraction of cells that displayed marked nuclear staining and are derived from 3 independent experiments.
The nuclear retention of poly(A)+ mRNA was determined by oligo dT FISH using single-knockout Nup120 and Nup133 strains complemented with Nup120 and Nup133 variants, respectively (Fig. 4). Whereas in a typical wild-type population, only 5% of the cells displayed a nuclear signal with the oligo dT FISH probe, 29% of the Nup120-deficient cells exhibited mRNA accumulation in the nucleus (Fig. 4), consistent with the previously reported mRNA export defect of nup120Δ cells (27, 28). This mRNA retention phenotype could be completely rescued by complementation with full-length Nup120. In contrast, cells complemented with full-length Nup120 harboring the point mutation D641A showed a marked increase in their mRNA retention phenotype (13%). For comparison, the NTD (23%) or the CTD of Nup120 (19%) rescued the deletion phenotype only moderately. Similarly, wild-type Nup133 (3%) rescued the mRNA export defect in nup133Δ cells (44%), whereas a fragment lacking the NTE or even the full-length protein harboring the single R11A mutation rescued the phenotype only partially (15%). For comparison, the NTD (32%), the CTD (31%), and the NTD containing the R11A mutant (32%) restore the wild type only to some extent. Strikingly, removal of the NTE from the Nup133 NTD caused a severe defect in mRNA export, with 56% of cells accumulating mRNA in the nucleus (Fig. 4). Together, these data suggest that the interaction between Nup120 and Nup133 is physiologically relevant and required for proper mRNA export. Further in vivo analyses are described in SI Text.
Discussion
We have determined the structure of a large Nup120 fragment and identified a previously undescribed interaction with Nup133, which provides new insight into the structure and assembly of the NPC coat (30, 32). Nup120 consists of an N-terminal β propeller and a C-terminal α-helical domain with a novel fold. These domain types are characteristic structural modules of nucleoporins and constituents of other membrane coats (41, 42). Analysis of the surface properties suggested 5 putative protein–protein interaction sites. We discovered that Nup120 interacts with a 15-residue stretch at the very N terminus of Nup133, which falls into an unstructured region of ≈55 residues. Nup120 and Nup133 variants, which fail to interact with one another in vitro, impair nuclear envelope localization and mRNA export. These results suggest a critical role for the interaction between these 2 proteins in the assembled NPC.
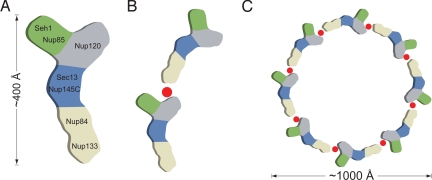
What are the implications of the discovery of this novel interaction on the architecture of the assembled NPC? Electron microscopy studies on the Nup84 complex mapped the approximate position of the 7 nucleoporins that assemble into an overall Y-shaped structure (19). Within the heptamer, Nup120 was mapped as 1 of 2 arms at the top of the Y, while Nup133 was located at the base of the Y, roughly 400 Å away from Nup120 (Fig. 5A). Because a completely extended 55-residue peptide stretch would reach ≈200 Å at best, corresponding to only about half of the length of the Nup84 complex, the interaction of these 2 proteins within a single heptamer seems unlikely. This scenario is even more improbable given the fact that negative-stain electron microscopy revealed only limited conformational flexibility of the heptamer (43), which would not allow the 2 distant proteins to approach each other via conformational changes. Thus, the newly discovered interaction between Nup120 and Nup133 likely depicts a contact between 2 adjacent heptamers in a head-to-tail fashion (Fig. 5B).
Fig. 5.
Model for the ring formation of the Nup84 complex. (A) Schematic representation of the heptameric complex and the approximate localization of its 7 nups (19). (B) The interaction of Nup120 and Nup133 suggests the intermolecular interaction between 2 heptamers in a head-to-tail fashion mediated by a short stretch at the very N terminus of an extended unstructured region of Nup133. (C) Eight heptamers are arranged in a closed ring with a diameter of ≈1,000 Å in accordance with the NPC dimensions determined by cryo-electron microscopy (8).
According to our working hypothesis, 32 copies of the heptameric Nup84 complex associate into a cylindrical, fence-like scaffold that forms the outermost perimeter of the symmetric NPC core, apposed to the POM cylinder (30). We envision that the coat cylinder is composed of 4 rings each harboring 8 heptamers that are stacked on top of each other with alternating directionality. The observed oligomerizations of the Seh1·Nup85 and Sec13·Nup145C nucleoporin pairs would result in 16 poles forming vertical connections among the 4 rings. The interactions within the heptamer would establish the horizontal connectivity in the NPC coat (30, 32). Here, we describe an additional interaction between 2 adjacent heptamers that has not yet been reported. This interaction is consistent with the proposed head-to-tail arrangement of 8 heptamers into a closed ring (Fig. 5C).
A circular arrangement of heptamers into closed, 8-membered rings also has been proposed in 2 alternative NPC models (33, 44). The head-to-tail arrangement that we describe here is principally in accordance with a computational model (44). However, in contrast to our proposed NPC coat, which consists of 4 stacked rings of 32 heptamers, the computational model proposes only 16 heptamers in 2 separated outer rings located at the nucleoplasmic and cytoplasmic periphery. The second alternative model, proposed by Schwartz and coworkers (33), also postulates nucleoplasmic and cytoplasmic rings consisting of 16 heptamers. In this case, however, the long axes of the heptamers are arranged not horizontally but vertically, so that Nup133, at the base of the heptamer, faces outward toward the nucleus and cytoplasm, respectively, while Nup120, at the opposite end of the heptamer, faces inward toward the equatorial plane of the symmetric NPC core. In light of our new finding that Nup120 interacts with Nup133, the proposed arrangement of vertically aligned heptamers seems unlikely. Notably, the present data do not allow us to predict the directionality of the Nup84 complex rings in our model. Similarly, the authors of the computational model were unable to distinguish between the 2 possible mirror-symmetric solutions, resulting in an ambiguity in the protein configurations of the 2 opposing rings (45).
Circular dichroism spectroscopy shows that the 55 N-terminal residues of Nup133, which contain the Nup120 binding region, are unstructured. This result has intriguing implications for the architecture of the NPC coat. The nuclear pore membrane features distinct geometric constraints, with a convex curvature of the membrane from the outside to the inside of the nuclear envelope and a concave curvature in the plane of the pore (45). These constraints result in a cylindrical NPC coat that has a larger diameter at the periphery. The unstructured region that mediates the interaction between adjacent heptamers is well suited to provide a flexible tether of variable length to accommodate rings of varying diameters. In addition to this architectural requirement, the loose tethering of adjacent heptamers may even be functionally important by providing flexibility to the NPC coat. Notably, flexibility of the NPC and in the association of nucleoporins has been reported previously (5, 32, 37, 46).
Many nucleoporins contain a structured core flanked by disordered regions (42). Here we describe a terminal segment that provides the attachment site to an adjacent nucleoporin and that is tethered to the structured core via a long, flexible linker. The common occurrence of such unstructured terminal segments, which are different from the phenylalanine-glycine repeats of nucleoporins, suggests that this anchor-like mechanism may be generally applicable to the assembled NPC. These unstructured regions not only may provide flexibility for the NPC, but also may have important implications for its reversible disassembly in cells that undergo an open mitosis. In fact, the unstructured segments of the nonameric Nup107–Nup160 complex, the human homolog of the heptameric Nup84 complex, are hyperphosphorylated exclusively during mitosis (47). Here we show that an unstructured segment indeed relays the attachment between adjacent nucleoporins. Thus, natively disordered regions harboring terminal anchoring sites would be ideal targets for enzymes that perform posttranslational modifications, which may constitute essential triggers for the reversible disassembly of the NPC.
Conclusion
The detailed molecular architecture of the symmetric core of the NPC is unknown at present; however, much progress has recently been made toward the structural characterization of one of its key building blocks—the heptameric Nup84 complex. The structure and functional analyses of Nup120 that we present here provides further evidence in support of a coat for the nuclear pore membrane (30, 32). Our analyses suggest that a terminal, unstructured linker segment mediates a flexible head-to-tail association between adjacent heptameric Nup84 complexes, consistent with the formation of an 8-membered ring. This novel interaction adds further robustness to the NPC coat, complementing the interactions within the heptamer, as well as the interactions that result from the oligomerization of 2 of its nucleoporin pairs (30, 32). This redundancy suggests that not all interactions need to occur simultaneously. Along with the flexibility of the central channel, rearrangements of the NPC coat may be possible, perhaps even necessary, to facilitate plasticity or enable the nuclear import of integral membrane proteins of the nuclear envelope (48).
In general, significant progress in the structural characterization of nucleoporins, their associations, and their complexes with the transport machinery has been made over the last few years. These advances have already transformed our understanding of the NPC and its associated processes and demonstrate that a reductionist approach is feasible. Ultimately, a mosaic image of the entire NPC constructed from atomic resolution pieces will emerge. The resulting NPC structure will then provide a detailed road map for comprehensive structure–function analyses.
Materials and Methods
The details of molecular cloning, expression, purification, crystallization, x-ray diffraction data collection, structure determination, protein interaction analysis, and in vivo experiments are described in the SI Text published online.
Supplementary Material
Acknowledgments.
We thank K.-C. Hsia, V. Nagy, A. Patke, and P. Stavropoulos for discussions and comments on the manuscript; S. Etherton for help with editing the manuscript; D. King for mass spectrometry analysis; T. Huber for help with CD spectroscopy; V. Nagy for providing material; and A. Davenport for technical assistance. Analytical ultracentrifugation was carried out by the Wadsworth Center Biochemistry Core Facility; isothermal titration calorimetry by the Biophysics Core Facility at the University of Colorado Denver. We also thank W. Shi (NSLS) and M. Becker and R. Fischetti (GM/CA-CAT) for support during data collection. E.W.D. is the Dale F. and Betty Ann Frey Fellow of the Damon Runyon Cancer Research Foundation (DRG-1977–08). D.W. was supported by a fellowship of the German Academic Exchange Service. A.H. was supported by a grant from the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3F7F and 3H7N).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907453106/DCSupplemental.
References
- 1.Hoelz A, Blobel G. Cell biology: Popping out of the nucleus. Nature. 2004;432:815–816. doi: 10.1038/432815a. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 3.Stewart M. Ratcheting mRNA out of the nucleus. Mol Cell. 2007;25:327–330. doi: 10.1016/j.molcel.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 4.Reichelt R, et al. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–894. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- 6.Hinshaw JE, Carragher BO, Milligan RA. Architecture and design of the nuclear pore complex. Cell. 1992;69:1133–1141. doi: 10.1016/0092-8674(92)90635-p. [DOI] [PubMed] [Google Scholar]
- 7.Kiseleva E, et al. A protocol for isolation and visualization of yeast nuclei by scanning electron microscopy (SEM) Nat Protoc. 2007;2:1943–1953. doi: 10.1038/nprot.2007.251. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: Functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 9.Fahrenkrog B, Koser J, Aebi U. The nuclear pore complex: A jack of all trades? Trends Biochem Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rout MP, et al. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suntharalingam M, Wente SR. Peering through the pore: Nuclear pore complex structure, assembly, and function. Dev Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 13.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 14.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 15.Belgareh N, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loïodice I, et al. The entire Nup107–160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasu S, et al. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allen NP, Huang L, Burlingame A, Rexach M. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem. 2001;276:29268–29274. doi: 10.1074/jbc.M102629200. [DOI] [PubMed] [Google Scholar]
- 19.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siniossoglou S, et al. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontoura BM, Blobel G, Matunis MJ. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol. 1999;144:1097–1112. doi: 10.1083/jcb.144.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walther TC, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 23.Baï SW, et al. The fission yeast Nup107–120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24:6379–6392. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci USA. 2003;100:981–985. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN. C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol. 1997;17:906–920. doi: 10.1128/mcb.17.2.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siniossoglou S, et al. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–275. doi: 10.1016/s0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 27.Aitchison JD, Blobel G, Rout MP. Nup120p: A yeast nucleoporin required for NPC distribution and mRNA transport. J Cell Biol. 1995;131:1659–1675. doi: 10.1083/jcb.131.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heath CV, et al. Nuclear pore complex clustering and nuclear accumulation of poly(A)+ RNA associated with mutation of the Saccharomyces cerevisiae RAT2/NUP120 gene. J Cell Biol. 1995;131:1677–1679. doi: 10.1083/jcb.131.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debler EW, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 35.Jeudy S, Schwartz TU. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem. 2007;282:34904–34912. doi: 10.1074/jbc.M705479200. [DOI] [PubMed] [Google Scholar]
- 36.Schrader N, et al. Structural basis of the nic96 subcomplex organization in the nuclear pore channel. Mol Cell. 2008;29:46–55. doi: 10.1016/j.molcel.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 37.Melčák I, Hoelz A, Blobel G. Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science. 2007;315:1729–1732. doi: 10.1126/science.1135730. [DOI] [PubMed] [Google Scholar]
- 38.Napetschnig J, Blobel G, Hoelz A. Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN. Proc Natl Acad Sci USA. 2007;104:1783–1788. doi: 10.1073/pnas.0610828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paoli M. Protein folds propelled by diversity. Prog Biophys Mol Biol. 2001;76:103–130. doi: 10.1016/s0079-6107(01)00007-4. [DOI] [PubMed] [Google Scholar]
- 40.Vellieux FM, et al. Structure of quinoprotein methylamine dehydrogenase at 2.25 A resolution. EMBO J. 1989;8:2171–2178. doi: 10.1002/j.1460-2075.1989.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:2085–2093. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devos D, et al. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci USA. 2006;103:2172–2177. doi: 10.1073/pnas.0506345103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–788. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 45.Antonin W, Mattaj IW. Nuclear pore complexes: Round the bend? Nat Cell Biol. 2005;7:10–12. doi: 10.1038/ncb0105-10. [DOI] [PubMed] [Google Scholar]
- 46.Akey CW. Structural plasticity of the nuclear pore complex. J Mol Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- 47.Glavy JS, et al. Cell-cycle-dependent phosphorylation of the nuclear pore Nup107–160 complex. Proc Natl Acad Sci USA. 2007;104:3811–3816. doi: 10.1073/pnas.0700058104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King MC, Lusk CP, Blobel G. Karyopherin-mediated import of integral inner nuclear membrane proteins. Nature. 2006;442:1003–1007. doi: 10.1038/nature05075. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.