Abstract
Slit regulates migration of not only neurons, but also nonneuronal cells, such as leukocytes and cancer cells. Slit effect on cancer cell migration has not been well-characterized. In this study, we used several different assays to examine Slit effect on breast cancer cell migration in vitro. We show that ubiquitin-specific protease 33 (USP33)/VDU1, originally identified as a von Hippel–Lindau tumor suppressor (VHL) protein-interacting deubiquitinating enzyme, binds to the Robo1 receptor, and that USP33 is required for Slit responsiveness in breast cancer cells. Slit induces redistribution of Robo1 from intracellular compartments to the plasma membrane in a USP33-dependent manner. Slit impairs directional migration of breast cancer cells without affecting their migration speed. This inhibitory effect is Robo-mediated and USP33-dependent. These data uncover a previously unknown function of USP33 and reveal a new player in Slit-Robo signaling in cancer cell migration.
Keywords: cell migration and motility, metastasis, Slit-Robo signaling
Cell migration is a fundamental process critical for not only embryonic development but also homeostasis in adult animals. A number of molecular cues guide axons and migrating neurons (1–4). Recent studies suggest that molecular mechanisms modulating migration of cells in different tissues/organs are conserved. For example, guidance cues, receptors, and the intracellular signaling pathways for neuronal migration are also used for cells outside of the nervous system, ranging from immune cells, myoblasts, and endothelial cells to tumor cells (4–7). The Slit gene was first identified in Drosophila, and subsequent studies indicated that secreted proteins of the Slit family and their receptors of the Roundabout (Robo) family play important roles in neuronal guidance (8–13). The Slit genes are frequently inactivated in cancer (14–19).
Accumulating evidence supports that chemokines and their receptors play important roles in tumorigenesis and cancer metastasis, including chemotactic invasion and migration of cancer cells (20, 21). In some cases, cancer cells show increased expression of chemokine receptors not expressed in normal nontumor cells, providing a plausible explanation for distant metastasis to organs that secrete corresponding chemokine ligands (22). Studies from our group and other groups have shown that Slit suppresses chemokine-directed chemotaxis of leukocytes and breast cancer cells (23–25) and inhibits medulloblastoma cell invasion (26). These observations suggest a potential therapeutic strategy in controlling aberrant cell migration during cancer metastasis. Cellular and molecular mechanisms underlying Slit signaling in cancer cells remain to be elucidated.
To dissect the Slit-Robo signaling pathway, we carried out yeast two-hybrid screens by using the intracellular domain of Robo1 as bait (27). From such screening, we have identified ubiquitin-specific protease 33 (USP33)/von Hippel–Lindau tumor suppressor protein (pVHL)-interacting deubiquitinating enzyme 1 (VDU1) as a protein interacting with Robo1 intracellular domain. USP33 was originally discovered as a deubiquitinating enzyme (DUB) binding to pVHL-containing E3 ligase complex targeted for ubiquitin (Ub)-mediated degradation (28). The substrates identified so far for USP33 and/or its homolog USP20/VDU2 are type 2 iodothyronine deiodinase (D2) and hypoxia-inducible factor-1α (HIF-1α) (29–31). The interaction between Robo1 and USP33 suggests that Robo1 is a previously unknown substrate for USP33. Our experiments demonstrate that in breast cancer cells, Slit inhibits chemokine-directed chemotaxis by affecting the directionality of cell migration, and that Slit responsiveness requires USP33. Our study reveals a functional link between USP33 and Robo1 and uncovers a previously uncharacterized role for USP33 in cancer cell migration.
Results
Slit Inhibits Breast Cancer Cell Migration Induced by SDF1 in a Robo-Dependent Manner.
Our previous studies indicate that Slit inhibits the chemokine SDF1-induced leukocyte chemotaxis (23, 25). SDF1 activation has been implicated in breast cancer metastasis (22). To investigate the involvement of Slit in breast cancer metastasis, we examined expression of its receptor Robo1 and CXCR4 in tissue samples from primary breast cancer patients and two metastatic breast cancer cell lines: MDA-MB-231 and MDA-MB-435 (abbreviated as MDA231 and MDA435, respectively). Consistent with previous reports (22, 24), CXCR4 and Robo1 were expressed in cancer samples (Fig. 1A, lanes 1–6) and both of the cell lines (Fig. 1A, lanes 7 and 8). Robo1 expression was also confirmed by Western blotting using a specific anti-Robo1 antibody (Fig. 1B).
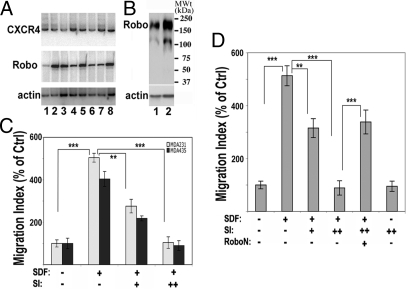
Fig. 1.
Slit inhibits SDF1-induced chemotaxis of breast cancer cells. (A) Expression of CXCR4 and Robo1. Primary breast cancer samples (lanes 1–6) as well as cell lines (MDA231 and MDA435, lanes 7 and 8, respectively) were examined by RT-PCR with primers specific for CXCR4 or Robo1. Actin was used as a control. (B) Western blotting was used to detect Robo1 protein in MDA231 and MDA435 cells (lanes 1 and 2, respectively). A band of ≈210 kDa (the full-length Robo1 protein) and a lower band (a possible proteolytic product) were detected in these cells. (C and D) Slit2 inhibits cell migration in a Robo-dependent manner. In C, MDA231 and MDA435 cells were tested in a transwell migration assay in the presence of the vehicle control, SDF1 (10 nM), Slit2 (+, 30 pM; ++, 100 pM), or combinations. The cell migration index was measured as the ratio of cells that migrated into the lower well in the test group to those in the control group with only vehicle treatment. In D, similar transwell experiments were carried out by using MDA231 cells in the presence or absence of RoboN (10 nM). In C and D, data are presented as the mean ± SEM, representing the results from three independent experiments, and the differences between the marked groups are statistically significant (**, P < 0.01; ***, P < 0.001 by Mann–Whitney test).
To examine the effect of Slit in cancer cell migration, we set up a transwell migration assay by using either MDA231 or MDA435 cells. Both types of breast cancer cells exhibited SDF1-induced chemotaxis in a similar manner (Fig. 1C). We therefore used MDA231 cells for subsequent experiments. SDF1 induced the migration of MDA231 cells, and this SDF1-induced chemotaxis was inhibited by Slit in a dose-dependent manner, although Slit did not significantly affect the baseline cell migration (Fig. 1D), consistent with a previous study (24). To test the involvement of Robo in Slit signaling in cancer cells, RoboN, a soluble extracellular domain of Robo blocking Slit-Robo signaling (12, 13, 23), was used together with Slit. Addition of RoboN effectively blocked the inhibitory effect of Slit on SDF1-induced chemotaxis (Fig. 1D), indicating that Slit signaling in MDA231 cells is mediated by Robo.
Slit Affects the Directionality of Cancer Cell Migration.
Slit treatment inhibits SDF1-induced chemotaxis of cancer cells in the transwell assay (Fig. 1 C and D). However, this assay does not allow direct visualization or detailed characterization of cell migratory behaviors. We then established a wound-healing assay by using MDA231 cells grown to confluence on fibronectin-coated coverslips and tested Slit effect on wound-induced cancer cell migration. Immediately after wound formation by scratching the cell monolayer using a fine tip, cells were cultured in the presence of the control, SDF1, Slit, or combinations, and then subjected to time-lapse imaging to monitor cell migration. Wound scratching induced cell migration in the direction toward the center of the wound area. Directional cell migration was measured as the forward distance that individual cells migrated from the wound edge line at the zero time point. A uniform application of SDF1 did not show an additive effect on wound-induced directional cell migration. The treatment with Slit alone significantly reduced the wound-induced cell migration (Fig. 2 A and C; see also Movie S1 and Movie S2). Similar to that observed in the transwell assay (Fig. 1C), addition of RoboN blocked the inhibitory effect of Slit on wound-induced directional migration (Fig. 2 A and C), indicating that Slit effect is Robo-dependent. Slit treatment did not significantly affect the cell migration speed (Fig. S1).
Fig. 2.
Slit impairs wound-induced migration of cancer cells. (A) Time-lapse imaging in a wound-healing assay of MDA231 cells was performed in the presence of mock control, Slit2 (25 pM), RoboN, or SDF1 (15 nM). Wound scratches were made so that the cells at the wound edge were migrating toward the right side in each panel. Representative images taken at 0 and 18 hr after wound scratching are shown. The black dotted lines mark the wound edge at 0 hr. (Scale bar: 200 μm.) (B) Cells were fixed after 13 hr of incubation, and MTOC was visualized by immunostaining with an anti-pericentrin antibody followed by Alexa488-conjugated anti-mouse secondary antibody (green). Cell nuclei were stained with Hoechst 33342 (blue). Cells in the first and second rows at the wound edge that had their MTOC in the forward-facing quadrant toward the wound edge were counted as having polarized MTOC. Arrowheads in each panel mark the cells with nonpolarized MTOC. (Scale bar: 20 μm.) (C) Quantification of the forward migration distance of individual cells during the 18-hr imaging period. Dividing cells were excluded from the analysis. Data are presented as the mean ± SEM (n = 30 from three independent experiments; 10 cells per experiment). ***, P < 0.0001 by Mann–Whitney test. (D) Time course of the ratio of cells with polarized MTOC among the total number of cells examined in the first and the second rows of the wound edge quantified from four independent experiments. The difference between the mock control and Slit-treated groups is significant at 8, 13, and 18 hr (*, P < 0.05, Mann–Whitney test).
In a migrating cell, microtubules (MTs) are polarized, with the MT-organizing center (MTOC) positioned between the nucleus and the leading edge of cell migration (32, 33). We examined the effect of Slit treatment on MTOC orientation in the wound-healing assay by pericentrin immunostaining, as described previously (33, 34). At different time points, cells were fixed and stained by using an anti-pericentrin antibody to visualize MTOCs. In this experiment, the percentage of mock-treated cells exhibiting MTOCs redirected toward the wound edge increased during the first 3 h, reaching a plateau of ≈60% (Fig. 2 B and D). The percentage of cells with the polarized MTOCs in the Slit-treated group similarly increased during the first 3 h but reduced to less than 40% afterward, with the maximal inhibitory effect ≈13 h after Slit treatment (Fig. 2 B and D).
Although the wound-healing assay allowed direct visualization of cell migration, it is difficult to monitor individual migrating cells and measure their trajectories. To examine the migrating behavior of isolated individual cells, we established a Dunn chamber chemotaxis time-lapse microscopy assay (35) using MDA231 cells (Fig. 3A). We first tested whether Slit acted as a repellent for cancer cells, as was reported for neurons (13, 36). When placed under a Slit concentration gradient, MDA231 cells did not show repulsive response or change in migration speed (Slit, 0.40 ± 0.03 μm/min vs. control, 0.42 ± 0.03 μm/min; n = 30; not statistically significant). Thus, Slit treatment alone did not affect the migration direction or speed of MDA231 cells.
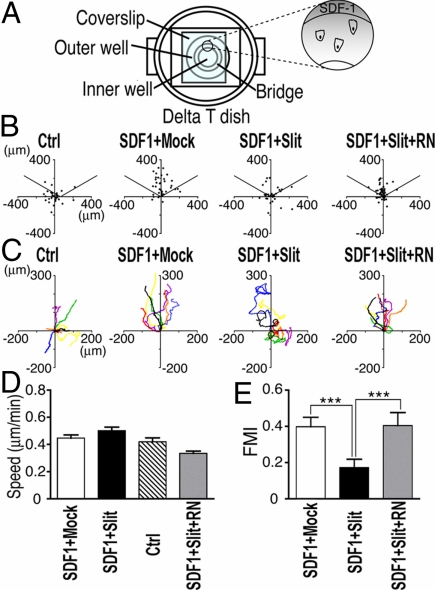
Fig. 3.
Slit affects SDF1-induced chemotaxis, reducing directionality but not migration speed in the Dunn chamber assay. MDA231 cells were monitored by time-lapse microscopy in SDF1 gradients in the presence of mock control, Slit2 (25 pM), or RoboN in combinations. (A) A schematic drawing of the Dunn chamber. The outer well contained the SDF1 source (15 nM) located at the top of the vertical axis so that an SDF1 gradient was created along the axis, with the higher concentration toward the top. Cell migration was recorded and tracked for 18 hr. (B) Distribution of mock- or Slit2-treated cells that migrated under SDF1 gradients. The dots represent final positions of cells relative to initial positions within the imaging field. In the absence of SDF1 gradients, cells migrated randomly without directionality, with their final positions randomly distributed. SDF1 induced chemotaxis toward the source located at the top. Addition of Slit2 reduced this SDF1-directed chemotaxis. The data are representative of at least three independent experiments. (C) Representative trajectories of seven migrating cells in each group during the imaging period, treated with SDF1 gradients and the mock control, Slit2, or RoboN in combinations. The initial positions of individual cells are fixed at the origin (0, 0). (D and E) Quantitative analyses of migration speed (D) and FMI (E) of the mock- and Slit-treated cells under SDF1 gradients. FMI is calculated as the ratio of the net distance that the cell progressed in the forward direction to the total distance that the cell traveled. Data are presented as the mean ± SEM (n = 30 from three independent experiments; 10 cells per experiment). ***, P < 0.001 by Mann–Whitney test.
We next tested MDA231 cells in SDF1 gradients in the presence of mock or Slit protein preparation (Fig. 3A). Fig. 3B shows the distribution of cells in the absence or presence of Slit. It is important to note that if cells display completely random migration, 33% of cells should distribute in each 120° arc. The cells treated with the mock preparation migrated toward the SDF1 gradient with relatively straight paths, such that 77% ± 1% of the cells were in the 120° arc facing the SDF1 source at the top of the y axis, compared with randomly distributed cells in the absence of the SDF1 gradient (Fig. 3 B and C, Control vs. SDF1 + Mock; see also Movie S3). By contrast, Slit-treated cells migrated at a wide angle toward the SDF1 gradient, or even away from the source of SDF1, with much less consistent migration trajectories. Only 55% ± 5% of the cells distributed within the top 120° arc (Fig. 3 B and C, SDF1 + Slit; see also Movie S4). Remarkably, Slit treatment did not reduce the migration speed (SDF1 plus Slit, 0.50 ± 0.03 μm/min vs. SDF1 plus mock, 0.45 ± 0.02 μm/min; n = 30; not statistically significant; Fig. 3D). To investigate whether Robo mediated the effect of Slit on SDF1-induced chemotaxis, we examined the migration behavior of cells treated with SDF1 plus Slit plus RoboN combination. These cells showed slightly reduced migration speed compared with cells in SDF1 plus mock, SDF1 plus Slit, or no SDF1 groups. However, the difference was not statistically significant (Fig. 3D). To test Slit effect on migration directionality, we examined the forward migration index (FMI) (37) during the 12-h imaging period. The FMI is defined as the ratio of the net distance that the cell progressed in the forward direction to the total migration length that the cell traveled, similar to the chemotropism index as described by McCutcheon (38) and Zigmond (39). Slit-treated cells showed significantly lower FMI than the mock-treated cells, indicating that Slit reduced chemotaxis efficiency of these cancer cells in response to SDF1 (Slit-treated group, 0.17 ± 0.05 vs. mock control group, 0.40 ± 0.05; P < 0.001; Fig. 3E).
Slit effect on chemotaxis of the breast cancer cells was abolished by RoboN addition, such that 75% ± 1% of the cells ended up within the top arc (FMI: RoboN plus Slit, 0.40 ± 0.07 vs. Slit, 0.17 ± 0.05; P < 0.0001; Fig. 3 B and C, SDF1 + Slit + RN, and E). The FMI of cells in the Slit plus RoboN group was comparable to that of the mock-treated cells, indicating that Slit treatment caused a loss of migration directionality by a Robo-dependent mechanism. In contrast to the previous flow cytometry study showing that only small populations (21–35%) of MDA231 cells express Robo1 and Robo2 (24), our immunostaining demonstrated that Robo1 was expressed in more than 95% of MDA231 cells. Consistently, more than 95% of the cells exhibited Slit responsiveness in our migration assays. Taken together, Slit-Robo signaling inhibits SDF1-induced chemotaxis by affecting the directionality rather than the migration speed in the cancer cells.
Identification of USP33 as a Robo1-Interacting Protein.
To identify components in the Slit-Robo signaling pathway, we performed yeast two-hybrid screens of mouse cDNA libraries by using Robo1 intracellular domain as bait (27). In addition to the GTPase-activating proteins srGAPs, we isolated 18 cDNA clones encoding mouse USP33/VDU1, a deubiquitinating enzyme of the USP family (28). RT-PCR analysis showed that USP33 was expressed in human breast cancer cells, including MDA231, and immunoblotting confirmed that USP33 protein was expressed in MDA231 cells (Fig. 4C).
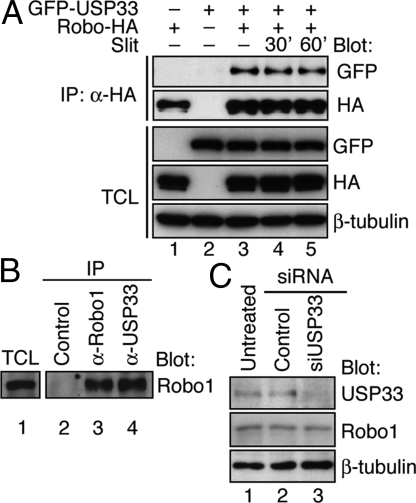
Fig. 4.
UPS33 interacts with Robo1. (A) Coimmunoprecipitation was carried out to detect the interaction of Robo1-HA with GFP-USP33 in transfected HEK293 cells. Antibodies used in immunoprecipitation (IP) and immunoblotting (Blot) are indicated. Total cell lysate (TCL) panels show the expression of corresponding proteins in the total cell lysates. The cells were stimulated with mock control or Slit2 protein (25 pM) for the indicated time periods. Interaction between Robo1 and USP33 was not dramatically affected by Slit2 treatment. (B) Interaction of endogenous Robo1 and USP33 proteins in MDA231 cells. Coimmunoprecipitation was done with control IgG, anti-Robo1, or anti-USP33 antibodies. Immunoprecipitated proteins were detected by Western blotting with anti-Robo1. (C) In MDA231 cells, siUSP33 no. 1, but not control siRNA, suppressed expression of endogenous USP33 but not Robo1, as detected by Western blotting.
To examine Robo1–USP33 interaction in mammalian cells, we carried out coimmunoprecipitation experiments. Plasmids expressing Robo1 tagged with a hemagglutinin (HA) epitope (Robo1-HA) and/or USP33 fused to green fluorescent protein (GFP-USP33), or control vector plasmids were cotransfected in combinations into human embryonic kidney (HEK) 293 cells. After immunoprecipitation using an anti-HA antibody, GFP-USP33 was detected only in the immunoprecipitates of the cells expressing both Robo1 and USP33 proteins, not of the control cells (Fig. 4A, lanes 1–3). Slit treatment did not affect Robo1–USP33 interaction (Fig. 4A, compare lanes 4 and 5 to lane 3). To test whether the endogenously expressed Robo1 and USP33 interacted with each other, we used MDA231 cell lysates for immunoprecipitation by using the control IgG, anti-Robo1, or anti-USP33 antibodies (Fig. 4B, lanes 2–4). Anti-USP33 antibody did not cross-react with Robo protein, but it coimmunoprecipitated Robo1 (Fig. 4B, lane 4), whereas control IgG did not precipitate Robo1 (lane 2), indicating that Robo1 interacts with USP33 in MDA231 cells. Detailed characterization of the domains involved in Robo1–USP33 interaction will be described elsewhere.
USP33 Is Required for Slit Signaling in Breast Cancer Cells.
To test the role of USP33 in Slit signaling in cancer cells, we transfected MDA231 cells with siRNAs targeted against either luciferase (used as a control siRNA; ref. 40) or USP33 (siUSP33). Two different siRNAs specifically against USP33, siUSP33 nos. 1 and 2, were efficient for knocking down USP33 expression (Figs. 4C and 6C and Fig. S2C). Data obtained by using siUSP33 no. 2 are presented in Fig. S2. In the subsequent experiments, siUSP33 no. 1 was used. Immunoblotting experiments confirmed that siUSP33 efficiently and specifically inhibited USP33 expression in both HEK293 cells and MDA231 cells (Fig. 4C and Fig. S2C), reducing USP33 expression to 20.7% ± 0.6% of the level in control siRNA-transfected MDA231 cells. In the Dunn chamber chemotaxis assays, 67% ± 1% or 49% ± 2% of control siRNA-transfected cells treated with mock or Slit preparation, respectively, ended up within the 120° arc facing the SDF1 source, similar to the parental cells (Fig. 5A). By contrast, 78% ± 4% or 74% ± 3% of USP33 knockdown cells that were treated with mock or Slit preparation, respectively, distributed within the arc facing the SDF1 source, indicating that siUSP33, but not control siRNA, eliminated Slit effect in inhibiting SDF1-induced chemotaxis (Fig. 5A). Moreover, tracking the centroids of the migrating cells showed significantly more straight trajectories of the USP33 knockdown cells treated with Slit compared with the control siRNA-treated cells exposed to Slit (FMI: siUSP33, 0.52 ± 0.05 vs. control, 0.17 ± 0.04, P < 0.001; Fig. 5 B and D). Therefore, siUSP33, but not control siRNA, specifically abolished Slit activity in inhibiting SDF1-induced chemotaxis. The siUSP33-transfected cells showed higher FMI than the control siRNA-treated cells in the absence of Slit (SDF1 plus mock) (siUSP33, 0.56 ± 0.03 vs. control siRNA, 0.40 ± 0.06; P < 0.05; Fig. 5 B and D). Knockdown of USP33 did not significantly affect the migration speed of these cancer cells (Fig. 5C). In addition, wound-healing experiments with MDA231 cells transfected with two independent siRNAs directed against USP33 revealed that the inhibitory effect of Slit on wound-induced directional migration depends on USP33 (Fig. S2). Therefore, USP33 is required for Slit signaling in the cancer cells.
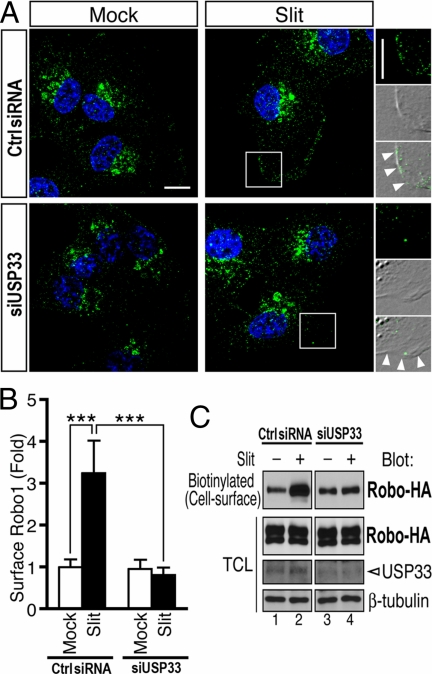
Fig. 6.
USP33 is required for Slit-induced Robo1 relocation to the plasma membrane. (A) Robo1 immunostaining of MDA231 cells transfected with either control or USP33 siRNA at 10 min after Slit stimulation (25 pM), with Robo1 signals in green and nuclear staining in blue (Hoechst 33342). Images represent two-dimensional images of three-dimensional reconstruction. (Right) High-power views of the boxed areas, merged with differential interference contrast (DIC) images, showing Robo1 signals distributed at the plasma membrane (arrowheads). (Scale bars: 10 μm.) (B) Quantification of surface levels of Robo1 in MDA231 cells. The ratio of Robo1 signal intensity at the plasma membrane to its total signal intensity in the entire cell was normalized to that in mock-stimulated control cells. To quantify cell surface-localized versus intracellularly localized Robo1, the plasma membrane area was manually marked by tracing the cell circumference. Twenty cells per group were analyzed. Data are presented as the mean ± SEM. ***, P < 0.001 by Mann–Whitney test. (C) Cell surface biotin labeling shows that Slit stimulation significantly increases cell surface expression of Robo1. Robo1-HA-expressing HEK cells transfected with control siRNA or siUSP33 were stimulated with Slit for 5 min. Extracellular domains were probed by cell surface biotinylation. Biotinylated proteins were then harvested by avidin-agarose pulldown and examined by Western blotting using anti-HA.
Fig. 5.
Knocking down USP33 eliminates Slit activity in suppressing SDF1-induced chemotaxis. Dunn chamber chemotaxis assay was performed as described for Fig. 3, except that MDA231 cells were transfected with control or USP33-specific siRNAs. (A) Distribution of mock- or Slit2-treated cells under SDF1 gradients after transfection with the corresponding siRNA. (B) Representative trajectories of seven migrating cells expressing control siRNA or siUSP33, in the presence of the control or Slit2 under SDF1 gradients. (C and D) Quantitative analyses are shown in C (migration speed) and D (FMI). Data are presented as the mean ± SEM (n = 30 from three independent experiments; 10 cells per experiment). ***, P < 0.001 by Mann–Whitney test.
Slit Stimulates the Redistribution of Robo to the Plasma Membrane in a USP33-Dependent Manner.
To understand the role of USP33 in Slit signaling, we examined whether USP33 affected the Robo protein level. USP33 overexpression or knockdown did not affect the total Robo1 protein level as determined by immunoblotting (Fig. 4C and Fig. S2C), suggesting that USP33 may not regulate Robo1 protein stability. We then examined whether USP33 affected Slit responsiveness by altering the subcellular distribution of Robo1. In nontreated MDA231 cells, a significant portion of Robo1 immunostaining signal was in the perinuclear compartment, and only a low level of Robo1 signal was detected at the plasma membrane. Slit treatment stimulated redistribution of Robo1 immunostaining signals from the perinuclear region to the plasma membrane. Robo1 redistribution was not affected by the control siRNA (Fig. 6A Top). In the control siRNA-transfected group, Slit treatment induced a ≈3-fold increase in the Robo1 signal at the plasma membrane, whereas in the siUSP33-treated cells, there was no significant change in Robo1 immunostaining signal at the plasma membrane upon Slit stimulation (Fig. 6 A and B). Thus, siUSP33, but not control siRNA, abolished Slit-induced Robo1 redistribution process, with the majority of Robo1 signals remaining in the perinuclear region (Fig. 6A Bottom).
By extracellular biotinylation, we examined the surface levels of Robo1 in control and USP33-depleted HEK cells. After Slit stimulation, cell surface proteins were immediately biotinylated at 4 °C and were collected by using avidin-immobilized agarose beads. As shown in Fig. 6C, lanes 1 and 2, in control cells, the surface level of Robo1 increased by ≈6-fold 5 min after Slit stimulation. In contrast, Slit-induced up-regulation of Robo1 surface presentation was dramatically inhibited by siUSP33 (Fig. 6C, lanes 3 and 4). Together, these results reveal an essential role for USP33 in Slit-induced Robo redistribution from the intracellular compartments to the plasma membrane.
Discussion
Slit Signaling in Breast Cancer Cells.
In this study, we used several assays to characterize Slit effect on cancer cell migration in vitro. Consistent with a previous study using a transwell assay (24), Slit inhibited SDF1-induced cancer cell migration. The transwell assay allowed examination of Slit effect on a large number of cells at the same time. The wound-healing assay enabled us to examine Slit effect in cancer cells in monolayer cultures. The Dunn chamber time-lapse microscopy allowed us to directly visualize migration of isolated cells and investigate Slit effects on chemotactic behaviors of the cells, with quantitative measurements of migration speed and directionality. Slit clearly impairs directional migration induced by SDF1 or by wounding without affecting the migration speed. The loss of directionality in cell migration provides an explanation for a Slit inhibitory effect on cancer cells in the transwell assays (ref. 24 and the present study). It is interesting to note that a recent study suggested that N-terminal fragment of Slit may act as a chemoattractant for breast cancer cells in transwell assays (41). By using the full-length Slit protein, we did not observe such activity in the transwell assay.
Microarray studies implicate high levels of Robo1 expression in breast and liver cancers (42, 43) and suggest that the role of Slit-Robo signaling in cancer cells is complex and multifold, similar to netrin-deleted in colorectal cancer (DCC) signaling in cancer (7). Our data suggest that Slit plays a role in blocking chemokine-induced metastasis toward tissues expressing high levels of chemokines, such as SDF1. Consistent with this is the observation that Slit is frequently inactivated by methylation in cancer cells (15–19). These data support the role of Slit in tumor suppression. Recent evidence further indicates that Slit exerts the tumor-suppressive effect by regulating β-catenin (44).
The Role of USP33 in Slit Signaling in Breast Cancer Cells.
The critical role of Ub-mediated modification in regulating protein stability/function has been well-established (45). Endocytic trafficking of receptors is also regulated by ubiquitination (46). DUBs have important regulatory roles in receptor trafficking and degradation of oncoproteins or tumor suppressor proteins (47–49). For example, USP7/HAUSP interacts with the tumor suppressor p53 and influences its activity (50, 51). Mutations in a tumor suppressor gene encoding the deubiquitinating enzyme CYLD have been identified in familial cylindromatosis. CYLD interacts with proteins regulating NF-κB and TNF receptors (52–54). DUBs often act to protect target proteins from degradation. For example, AMSH inhibits down-regulation of EGF receptor signaling via deubiquitination in endosomes (55), although it is not clear whether EGF responses are completely abolished by AMSH depletion. In our study, USP33 interacts with Robo1 and is required for Slit-induced Robo1 redistribution to the plasma membrane without affecting the total Robo1 protein level. In contrast to the previously reported role of DUBs in preventing down-regulation of receptors and other substrate proteins by the Ub-mediated pathway (30, 31, 55), our data reveal a previously unknown activity of USP33 in modulating subcellular distribution of Robo.
Our understanding of functional relationships between the Slit-Robo and other signal transduction pathways in cancer cells remains limited. Together with previous studies (23, 24), our study supports a role for the Slit-Robo pathway in regulating cancer cell responses to chemokines. USP33 is a downstream target for ubiquitination by pVHL E3 ligase and is degraded via the Ub-proteasome system (28). It will be important to test in future studies whether the Slit-Robo pathway cross-communicates with those involving pVHL and its substrates, such as HIFs, and to address what role Slit signaling plays in tumorigenesis and tumor angiogenesis.
Materials and Methods
Cell Migration Assays.
Transwell assay was carried out as described previously (23), except that 8-μm pore polycarbonate filter membranes coated with fibronectin (5 μg/mL) were used to separate the upper and lower wells. In wound-healing assay, MDA231 cells were seeded on fibronectin-coated coverslips and grown to confluence. A wound scratch was made on the cell monolayer by using a micropipette tip. After removing the dislodged cells by extensive washing with culture media, cells were treated with mock control or Slit. Cell migration was monitored by time-lapse microscopy, and the behaviors of individual, randomly selected cells were tracked by using ImageJ software (National Institutes of Health, Bethesda, MD). These data were processed with Microsoft Excel software and were subjected to statistical analysis using Prism 4.0b software (GraphPad). Dunn chamber chemotaxis assay was performed with modification of published protocols (35, 56).
MTOC Orientation Assay.
Wound-healing assay was carried out as described above. After incubation at 37 °C for different periods of time, the cells were fixed in methanol (−20 °C) for 5 min. MTOC was visualized by immunostaining using anti-pericentrin and Alexa488-conjugated anti-mouse secondary antibody. The cells in the first and second rows at the wound edge were scored for MTOC orientation (at least 150 cells in each group).
Immunoprecipitation Assay and Immunostaining.
For a detailed description of experimental procedures, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Fujio Murakami (Osaka University, Japan) for generously providing anti-Robo1 antibody. We are grateful to Drs. John Wikswo and Yuxin Liu for the early stages of chemotaxis analyses, Dr. James Monypenny for advice on the Dunn chamber assay, and Xiaoping Chen and Bo Zheng for excellent technical assistance. We thank Masanori Katakura and Dr. Shigeru Yanagi for their support, Yasutomo Kubota for instructions on Metamorph software and deconvolution analyses, Kazunori Hakozaki (Leica Microsystems) for providing Leica Deblur software and anonymous reviewers for helpful suggestions to improve this paper. This work was supported by National Institutes of Health Grants CA114197, CA107193 (to J.Y.W.), and DA0176404 (to Y.R.), and James S. McDonnell Foundation Grant JSMF220o20180 (to J.Y.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801262106/DCSupplemental.
References
- 1.Dickson BJ. Molecular mechanisms of axon guidance. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 2.Guan KL, Rao Y. Signalling mechanisms mediating neuronal responses to guidance cues. Nat Rev Neurosci. 2003;4:941–956. doi: 10.1038/nrn1254. [DOI] [PubMed] [Google Scholar]
- 3.Huber AB, Kolodkin AL, Ginty DD, Cloutier JF. Signaling at the growth cone: Ligand-receptor complexes and the control of axon growth and guidance. Annu Rev Neurosci. 2003;26:509–563. doi: 10.1146/annurev.neuro.26.010302.081139. [DOI] [PubMed] [Google Scholar]
- 4.Carmeliet P, Tessier-Lavigne M. Common mechanisms of nerve and blood vessel wiring. Nature. 2005;436:193–200. doi: 10.1038/nature03875. [DOI] [PubMed] [Google Scholar]
- 5.Rao Y, Wong K, Ward M, Jurgensen C, Wu JY. Neuronal migration and molecular conservation with leukocyte chemotaxis. Genes Dev. 2002;16:2973–2984. doi: 10.1101/gad.1005802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong K, Park HT, Wu JY, Rao Y. Slit proteins: Guidance cues for cells ranging from neurons to leukocytes. Curr Opin Genet Dev. 2002;12:583–591. doi: 10.1016/s0959-437x(02)00343-x. [DOI] [PubMed] [Google Scholar]
- 7.Arakawa H. Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer. 2004;4:978–987. doi: 10.1038/nrc1504. [DOI] [PubMed] [Google Scholar]
- 8.Rothberg JM, Hartley DA, Walther Z, Artavanis-Tsakonas S. slit: An EGF-homologous locus of D. melanogaster involved in the development of the embryonic central nervous system. Cell. 1988;55:1047–1059. doi: 10.1016/0092-8674(88)90249-8. [DOI] [PubMed] [Google Scholar]
- 9.Brose K, et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- 10.Kidd T, et al. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- 11.Kidd T, Bland KS, Goodman CS. Slit is the midline repellent for the Robo receptor in Drosophila. Cell. 1999;96:785–794. doi: 10.1016/s0092-8674(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 12.Li HS, et al. Vertebrate Slit, a secreted ligand for the transmembrane protein Roundabout, is a repellent for olfactory bulb axons. Cell. 1999;96:807–818. doi: 10.1016/s0092-8674(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 13.Wu W, et al. Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sundaresan V, et al. Homozygous deletions at 3p12 in breast and lung cancer. Oncogene. 1998;17:1723–1729. doi: 10.1038/sj.onc.1202103. [DOI] [PubMed] [Google Scholar]
- 15.Dallol A, et al. SLIT2, a human homolog of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res. 2002;62:5874–5880. [PubMed] [Google Scholar]
- 16.Dallol A, et al. Frequent epigenetic inactivation of the SLIT2 gene in gliomas. Oncogene. 2003;22:4611–4616. doi: 10.1038/sj.onc.1206687. [DOI] [PubMed] [Google Scholar]
- 17.Dallol A, Morton D, Maher ER, Latif F. SLIT2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Cancer Res. 2003;63:1054–1058. [PubMed] [Google Scholar]
- 18.Latil A, et al. Quantification of expression of netrins, slit and their receptors in human prostate tumors. Int J Cancer. 2003;103:306–315. doi: 10.1002/ijc.10821. [DOI] [PubMed] [Google Scholar]
- 19.Dickinson RE, et al. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer. 2004;91:2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murdoch C, Muthana M, Coffelt SB, Lewis CE. The role of myeloid cells in the promotion of tumor angiogenesis. Nat Rev Cancer. 2008;8:618–631. doi: 10.1038/nrc2444. [DOI] [PubMed] [Google Scholar]
- 21.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 22.Müller A, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 23.Wu JY, et al. The neuronal repellent Slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. 2001;410:948–952. doi: 10.1038/35073616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad A, Fernandis AZ, Rao Y, Ganju RK. Slit protein-mediated inhibition of CXCR4-induced chemotactic and chemoinvasive signaling pathways in breast cancer cells. J Biol Chem. 2004;279:9115–9124. doi: 10.1074/jbc.M308083200. [DOI] [PubMed] [Google Scholar]
- 25.Prasad A, Qamri Z, Wu J, Ganju RK. Slit-2/Robo-1 modulates the CXCL12/CXCR4-induced chemotaxis of T cells. J Leukoc Biol. 2007;82:465–476. doi: 10.1189/jlb.1106678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Werbowetski-Ogilvie TE, et al. Inhibition of medulloblastoma cell invasion by Slit. Oncogene. 2006;25:5103–5112. doi: 10.1038/sj.onc.1209524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong K, et al. Signal transduction in neuronal migration: Roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, et al. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem. 2002;277:4656–4662. doi: 10.1074/jbc.M108269200. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, et al. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Comm. 2002;294:700–709. doi: 10.1016/S0006-291X(02)00534-X. [DOI] [PubMed] [Google Scholar]
- 30.Curcio-Morelli C, et al. Deubiquitination of type 2 iodothyronine deiodinase by von Hippel-Lindau protein-interacting deubiquitinating enzymes regulates thyroid hormone activation. J Clin Invest. 2003;112:189–196. doi: 10.1172/JCI18348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1α. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gotlieb AI, May LM, Subrahmanyan L, Kalnins VI. Distribution of microtubule organizing centers in migrating sheets of endothelial cells. J Cell Biol. 1981;91:589–594. doi: 10.1083/jcb.91.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 34.Michaut MA, Williams CJ, Schultz RM. Phosphorylated MARCKS: A novel centrosome component that also defines a peripheral subdomain of the cortical actin cap in mouse eggs. Dev Biol. 2005;280:26–37. doi: 10.1016/j.ydbio.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 35.Zicha D, Dunn GA, Brown AF. A new direct-viewing chemotaxis chamber. J Cell Sci. 1991;99:769–775. doi: 10.1242/jcs.99.4.769. [DOI] [PubMed] [Google Scholar]
- 36.Ward M, McCann C, DeWulf M, Wu JY, Rao Y. Distinguishing between directional guidance and motility regulation in neuronal migration. J Neurosci. 2003;23:5170–5177. doi: 10.1523/JNEUROSCI.23-12-05170.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foxman EF, Kunkel EJ, Butcher EC. Integrating conflicting chemotactic signals: The role of memory in leukocyte navigation. J Cell Biol. 1999;147:577–587. doi: 10.1083/jcb.147.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCutcheon M. Chemotaxis in leukocytes. Physiol Rev. 1946;26:319–336. doi: 10.1152/physrev.1946.26.3.319. [DOI] [PubMed] [Google Scholar]
- 39.Zigmond SH. Mechanisms of sensing chemical gradients by polymorphonuclear leukocytes. Nature. 1974;249:450–452. doi: 10.1038/249450a0. [DOI] [PubMed] [Google Scholar]
- 40.Elbashir SM, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 41.Schmid BC, et al. The neuronal guidance cue Slit2 induces targeted migration and may play a role in brain metastasis of breast cancer cells. Breast Cancer Res Treat. 2007;106:333–342. doi: 10.1007/s10549-007-9504-0. [DOI] [PubMed] [Google Scholar]
- 42.Minn AJ, et al. Genes that mediates breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito H, et al. Identification of ROBO1 as a novel hepatocellular carcinoma antigen and a potential therapeutic and diagnostic target. Clin Cancer Res. 2006;12:3257–3264. doi: 10.1158/1078-0432.CCR-05-2787. [DOI] [PubMed] [Google Scholar]
- 44.Prasad A, et al. Slit-2 induces a tumor suppressive effect by regulating β-catenin in breast cancer cells. J Biol Chem. 2008;283:26624–26633. doi: 10.1074/jbc.M800679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 46.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 47.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Nijman SMB, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Clague MJ, Urbé S. Endocytosis: The DUB version. Trends Cell Biol. 2006;16:551–559. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 50.Cummins JM, et al. Disruption of HAUSP gene stabilizes p53. Nature. 2004;428:486. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- 51.Li M, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 52.Brummelkamp TR, Nijman SMB, Dirac AMG, Bernards R. Loss of the cylindromatosis tumor suppressor inhibits apoptosis by activating NF-κB. Nature. 2003;424:797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- 53.Kovalenko A, et al. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 54.Trompouki E, et al. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424:793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- 55.McCullough J, Clague MJ, Urbé S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wells CM, Ridley A. Analysis of cell migration using the Dunn chemotaxis chamber and time-lapse microscopy. In: Guan JL, editor. Methods in Molecular Biology, Cell Migration, Developmental Methods and Protocols. Totowa, NJ: Humana Press; 2005. pp. 31–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.