Abstract
The herpes simplex virus 1 ICP0 is a regulatory protein. Early in infection ICP0 localizes in ND10 bodies and performs two functions: As an E3 ligase in conjunction with E2 UbcH5a conjugating enzyme, it degrades the ND10 components PML and SP100. Concurrently, it suppresses the silencing of viral DNA by dispersing the HDAC1/CoREST/REST/LSD1 repressor complex. Subsequently, ICP0 is exported to the cytoplasm. In cells treated with HDAC inhibitors or transfected with irrelevant DNA, the export is delayed in a DNA dose-dependent fashion. Here, we follow up an observation that ICP0 binds cyclin D3 and that ICP0 mutants unable to bind cyclin D3 are not exported. Moreover, in infected cells cdk4 is activated, but cdk2 is not. We report that (i) cyclin D1, D2, or D3 colocalize with ND10 bodies and ICP0 early in infection and ultimately become incorporated into viral replication compartments, (ii) each of the D cyclins partially rescues ΔICP0 mutants, and (iii) inhibition of cdk4 by inhibitor I sequesters ICP0 in the nucleus. A key finding is that overexpression of cyclin D3 enables the transport of ICP0 to the cytoplasm. We conclude that (i) ICP0 facilitates the recruitment of cyclin D3 to the sites of viral DNA synthesis, (ii) until its functions are completed, ICP0 is retained in the nucleus, and (iii) a common signal that results in the export of ICP0 to the cytoplasm is the accumulation of a viral DNA-synthesis-dependent late protein.
Keywords: ND10 bodies, nuclear–cytoplasmic translocation, PML
Infected cell protein no. 0 (ICP0) is a major multifunctional herpes simplex virus 1 (HSV-1) regulatory protein made immediately after infection. ICP0 is a promiscuous transactivator, and in cells infected at low multiplicity with ΔICP0 mutants, the transition from α (immediate early) to genes expressed later in infection does not ensue (1). In wild-type virus-infected cells, it accumulates initially at ND10 bodies, where it performs at least two functions. As an E3 ligase, it degrades PML and SP100 in conjunction with the UbcH5a ubiquitin-conjugating enzyme (2, 3). At the same time, it blocks the silencing of the viral DNA by a host repressor complex consisting of several proteins, including HDACs 1 or 2, CoREST, REST, and LSD1 (4, 5). Indeed, a dominant negative CoREST retaining the binding site of ICP0 but lacking the HDAC binding site significantly increases the yields of ΔICP0 mutant in cells that do not support its replication (4).
The interaction of ICP0 with cyclin D3 but not with cyclin D1 was observed initially in a yeast two-hybrid system and confirmed in pull-down experiments (6). The binding site was mapped to Glu199. ICP0 carrying the substitution D199A was not translocated to the cytoplasm. Furthermore, the virus carrying this mutation failed to replicate as well as wild-type in contact-inhibited, resting primary human fibroblasts and was less pathogenic in mice when administered i.p (7). Moreover, in cells infected with this mutant, cyclin D3 was less stable than in wild-type virus-infected cells (7). Earlier studies also have shown that cyclin D3 colocalizes with or at ND10 structures and that the presence of ICP0 does not interfere with the phosphorylation of cdk4 (6, 8). In infected cells, however, cdk2 was not activated (8, 9), suggesting that HSV-1 brings the infected cells to but not into cellular DNA synthesis, consistent with the observation that incorporation of radioactive precursors into cellular DNA ceases with the onset of viral DNA synthesis (10, 11). Relevant to this report are the following: In wild-type virus-infected cells, ICP0 is translocated into the cytoplasm between 5 and 7 h after infection (12). The translocation is accelerated in cells infected with a mutant (R7801) in which the cyclin D3 gene was inserted into a wild-type viral genome but remained unaffected in cells infected with D199A mutant carrying cyclin D3 (8). Recent studies have shown that in cells transfected with DNA and then infected ICP0 is retained in the nucleus in a DNA-dose- and size- but not in a DNA-type-dependent fashion (13). The implication of these studies is that ICP0 is retained in the nucleus until its nuclear functions are completed. These functions include disaggregation of ND10 bodies and “processing” of DNA (13).
The functions of D cyclins have been reviewed extensively (14, 15). Noteworthy here are that several members of the herpesvirus family encode orthologs of cyclin D2 and that although each D cyclin appears to have specific functions, the basic functions of the D cyclins with respect to initiation of S phase, cell growth, and differentiation appear to be interchangeable (15–19).
In this work, we show that the sequence targeting cyclin D3 to ND10 bodies maps between residues 69 and 129, that cyclins D1 and D2 also colocalize early in infection with ICP0, and that ΔICP0 mutant viruses carrying cyclin D1, D2, or D3 replicated significantly better than the ΔICP0 mutant virus. Last, exposure of wild-type virus-infected cells to cdk4 inhibitor I resulted in retention of ICP0 in the nucleus. This effect was not seen in cells in which cyclin D3 was overexpressed by a recombinant virus carrying wild-type ICP0 and the cyclin D3 gene.
Results
Cyclin D3 Aggregates at ND10 Bodies Before The Incorporation into the Viral Replication Compartments.
Elsewhere, our laboratory has reported that ICP0 binds cyclin D3 and early in infection both proteins are found in structures similar to those containing PML (6–8). The fate of cyclin D3 after the dispersion and degradation of ND10 was unknown. The objective of this series of experiments was to identify the domain of cyclin D3 that is critical for its recruitment to ND10 bodies. In this series of experiments, we took advantage of special features of cells that were first transfected with DNA and then infected as described elsewhere (13). Specifically, in cell lines resistant to expression of transfected DNA, ICP0 is retained in ND10 structures, and degradation of PML is delayed in a transfected DNA dose-dependent manner (13). This system facilitates studies of the distribution of transgene products at early times after infection. Accordingly, the DNA constructs shown in Fig. 1 were transfected into HEp-2 cells grown in 4× well slides. At 24 h after transfection, the cells were infected with wild-type virus. The cultures were fixed and immunostained 9 h after infection. The results, Fig. 2, were as follows:
Fig. 1.
DNA sequence arrangements of cyclin D3 components used to map the sequence required for colocalization of the protein with ND10 bodies. The procedures for generating plasmids containing Myc-tagged intact or truncated cyclin D3 are described in Materials and Methods.
Fig. 2.
Subcellular localization of D-type cyclins and cdk4. (A) Design of experiments involving transfection or infection of HEp-2 (B–H, J–M): HEp-2 cells were transfected with plasmids encoding either the full-length wild-type cyclin D3 (B, F), cyclin D1 (J, K), cyclin D2 (L, M), or truncated forms of cyclin D3, residues 1–155 (C, G), residues 156–292 (D, H), or residues 1–129 (E). At 24 h after transfection the cells were exposed to 10 pfu of HSV-1(F) per cell. The cells were fixed at 9 h after infection and doubly reacted with either the rabbit polyclonal antibody to ICP0 (B–E, J–M) or PML (F–H) and the Myc-tagged mouse monoclonal antibody (B–H, J–M). (I) HEp-2 cells were exposed to 10 pfu of the recombinant virus R7801 (Fig. 1) per cell. The cells were fixed at 6 h after infection and reacted with mouse monoclonal antibody to cyclin D3 and the rabbit polyclonal antibody to ICP8. (N–P) HEp-2 cells were mock-infected (N) or exposed to 10 pfu of HSV-1(F) per cell. The cells were fixed at 4 h (O) or 9 h (P) after infection and reacted with the mouse monoclonal antibody to ICP0 and the rabbit polyclonal antibody to cdk4. In (B–J, L, M), ICP0, PML, and ICP8 demarcate the nucleus. Except for the cells shown in G, overexpressed cyclin D3 accumulated in both nucleus and cytoplasm. ICP0 is partially cytoplasmic in K and P.
As expected on the basis of earlier studies (13), in virtually all transfected or infected cells, ICP0 was retained in the nucleus. In addition, in virtually all experiments, the major part of wild-type cyclin D3 localized largely but not exclusively in the nucleus, initially at ND10 bodies (Fig. 2 B and F) and in cells in more advanced stages of infection in larger bodies (e.g., Fig. 2I). Overexpression of cyclin D3 resulted in accumulation of significant amounts of the protein in the cytoplasm (e.g., Fig. 2B)
The products of truncated cyclin D3 constructs that included residues 1–155 or 1–129 colocalized with ICP0 and PML (Fig. 2 C, E, and G). In contrast, cyclin D3 polypeptides containing residues 156–292 (Fig. 2 D and H) or 1–69 were dispersed throughout the nucleus and did not specifically colocalize with ICP0 or PML.
We conclude from these studies that the sequences required for the colocalization of cyclin D3 with ND10 structures are located between residues 69 and 129.
Localization of Cyclin D3 in Replication Compartments.
ICP8 is a marker of replication compartments. Earlier studies have shown that as replication progresses ICP8 initially localizes at ND10 bodies and then spreads throughout the nucleus (20). In cells exhibiting more advanced stages of replication and reacted with antibody to ICP8 and cyclin D3, the latter appears to localize within the compartment containing ICP8, but it does not colocalize precisely with ICP8 (Fig. 2I)
Cyclins D1 and D2 also Colocalize with ICP0.
Because the functions of cyclins D1 and D2 are similar to those of cyclin D3, determining whether their localization differs from that of cyclin D3 in transfected or wild-type virus-infected cells was of interest. The results of the experiment performed as outlined in Fig. 2A and shown in Fig. 2 J–M indicate that both cyclins D1 and D2 colocalize with ICP0 in transfected or infected cells.
Subcellular Distribution of cdk4 in Wild-Type Virus-Infected Cells.
In these experiments, we examined 4± well cultures of HEp-2 cells fixed and reacted with antibody after mock infection or at 4 and 9 h after infection with wild-type virus, respectively. As illustrated in Fig. 2 N–P, cdk4 was dispersed throughout the nucleus and cytoplasm of mock-infected cells. At relatively early times after infection, the distribution of cdk4 was similar to that of ICP0. Unlike ICP0, cdk4 remained in the nucleus after translocation of ICP0 to the cytoplasm. Its distribution in the nucleus was similar in appearance to that of ICP8 in that it filled most but not the entire nucleus (e.g., Fig. 2P).
Overexpression of D-Type Cyclins Partially Rescue ΔICP0 Mutant Virus.
Relevant to this series of experiments are three observations: (i) ICP0 physically interacts with cyclin D3 but not with cyclin D1. Binding was abolished by the substitution of Glu199 with alanine (6–8). (ii) The ICP0 mutant carrying the D199A substitution was sequestered in the nucleus (6–8). (iii) Overexpression of cyclin D3 by insertion of the gene into the genome of a wild-type virus accelerated the translocation of ICP0 to the cytoplasm, whereas overexpression of cyclin D3 by a mutant carrying the D199A substitution did not enable the export of ICP0 (6–8). In light of the recent studies suggesting that the duration of the nuclear sojourn of ICP0 is related to an activity involving processing of DNA (13), the question arose whether cyclin D3 “assists” in this process and hence could complement the ΔICP0 mutant.
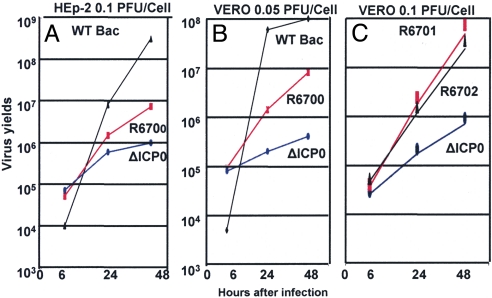
To test this hypothesis, we constructed a ΔICP0 mutant virus in which we inserted cyclin D3 between the UL3 and UL4 ORFs as illustrated in Fig. 3(line 3). The growth properties of the parent and mutant viruses were measured in HEp-2 cells exposed to 0.1 pfu/cell (Fig. 4A) or Vero cells exposed to 0.05 pfu/cell (Fig. 4B). As shown in Fig. 4, the yields of ΔICP0 mutant virus carrying cyclin D3 were >10-fold higher in all assays.
Fig. 3.
Sequence arrangements of recombinant viruses used in these studies. (1) Sequence arrangement of the ΔICP0 BAC (4) used as the parental virus for the construction of recombinants R6700, R6701, and R6702. (2) Organization and direction of transcription of UL2, UL3, and UL4 ORFs. (3-5) Sequence arrangements of transcriptional cassettes composed of the Egr-1 promoter, the Myc-tagged cyclin D3, D2, or D1, and the bidirectional UL21/UL22 poly(A) sequence inserted into the unique BamHI site between the UL3 and UL4 ORFs for construction of the recombinant viruses R6700, R6701, and R6702, respectively. (6) Sequence arrangement of the HSV-1(F). (7) Sequence arrangement of the R7801, that carries wild-type cyclin D3 in the thymidine kinase gene (6). B, BamHI; Hp, HpaI; X, XbaI; R, EcoRV; M, MluI; H, HindIII; Pm, PmeI.
Fig. 4.
Enhanced replication of ΔICP0 mutant viruses encoding cyclin D1, D2, or D3. Replicate cultures of HEp-2 (A) or Vero cells (B) were exposed to 0.1 or 0.05 pfu/cell of wild-type virus derived from a wild-type BAC, ΔICP0 recombinant virus derived from a ΔICP0 BAC (4), or ΔICP0 mutant virus carrying cyclin D3 (R6700). (C) Vero cells infected with 0.1 pfu/cell of wild-type virus, ΔICP0 virus, or ΔICP0 mutant virus carrying the gene for cyclin D2 (R6701) or cyclin D1 (R6702) respectively. The infected cells were harvested at 6, 24, or 48 h after infection and titered on U2OS cells.
In light of the observation that cyclins D1 and D2 colocalized with ICP0 early in infection, determining whether overexpression of these cyclins also is able to complement ΔICP0 mutant virus was of interest. To answer this question, we constructed two additional ΔICP0 mutant viruses into which we inserted cyclins D1 and D2, respectively, between the open frames of UL3 and UL4 (Fig. 3). In Vero cells, the recombinant viruses overexpressing cyclin D1 or D2 yielded >10-fold higher virus than the parent, ΔICP0 mutant virus (Fig. 4C).
We conclude from these studies that, consistent with the interactions of ICP0 with cyclin D3, the replicative processes require specific functions expressed by cyclin D3 and that within the range of functions tested cyclin D1 or D2 can substitute for cyclin D3.
Overexpression of Cyclin D3 Reverses the Retention of ICP0 in the Nucleus Induced by Inhibition of cdk4.
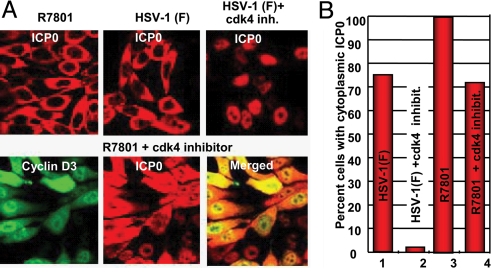
Earlier studies have shown that in infected cells cdk4 is phosphorylated but cdk2 is not (8, 9). Moreover, within a short interval after infection, DNA synthesis shifts from cellular to viral (10, 11). Inasmuch as one function of D cyclins is activation of cdk4 (14), the question arose as to the requirement of cdk4 activation for viral gene expression. In this series of experiments, we examined the localization of ICP0 in cells treated with the cdk4 inhibitor I. Immunofluorescence studies showed that ICP0 was retained in the nucleus in cells infected 18 h after exposure to the inhibitor and incubated for an additional 9 h (Fig. 5 A and B). To determine whether overexpression of cyclin D3 compensated for the inhibition of cdk4, in parallel experiments the cells were exposed to the inhibitor, infected with the recombinant virus R7801 (Fig. 3), and then incubated for an additional 9 h. As shown in Fig. 5 A and B, ICP0 was translocated to the cytoplasm in the vast majority of the cells infected with R7801 recombinant virus.
Fig. 5.
Effects of cdk4 inhibitor I are reversed by overexpression of cyclin D3. Hep-2 cells were either left untreated or pretreated with 2.5 μM of the cdk4 inhibitor I for 18 h, in their culture medium, before infection with either HSV-1(F) or R7801, at 10 pfu/cell, as indicated in (A). The cells were fixed at 9 h after infection and reacted with the rabbit polyclonal antibody to ICP0 and the mouse monoclonal antibody to cyclin D3. The images were captured with the same settings of a Zeiss confocal microscope (A). In (B), ≈450 cells from sequential fields were counted to determine the percentage of cells with cytoplasmic ICP0.
Discussion
Earlier studies from our laboratory have shown that ICP0 performs at least two key functions during its sojourn in the nucleus of the infected cells. These are: (i) degradation of PML and SP100 that at the very least are preemptive acts to block exogenous IFN from interfering with viral replication (2, 21) and (ii) suppression of silencing of viral DNA by the dissociation of the HDAC/CoREST/REST/LSD1 complex (4, 5). Mutation in the ICP0 RING domain responsible for the E3 ligase function resulted in retention of ICP0 in the nucleus. In the same vein, ICP0 was retained in the nucleus of cells transfected with irrelevant, coding or noncoding DNA or by exposing cells to HDAC inhibitors (13). ICP0 also was retained in the nucleus of cells infected with a virus carrying a mutation in the binding site of CoREST in ICP0 (4). These studies suggest the following: (i) ICP0 performs specific functions in the nucleus of infected cells. To date, two functions have been identified and both map in specific, separate domains of ICP0. (ii) Completion of these functions results in export of ICP0 to the cytoplasm. Export arising from completion of two distinct functions, albeit performed concurrently, suggests a common export signal. Earlier studies have shown that inhibition of viral DNA synthesis results in a significant delay in the translocation of ICP0. These studies suggested that a late viral gene product is primarily responsible for the export of ICP0 (12). In this work, we have added an additional function that must be performed by ICP0 in order for its export to the cytoplasm. This function involves the activation of cdk4 by cyclin D3. The results presented here raise two questions: First, what is the function of cyclin D3 and, second, why has HSV-1 evolved the recruitment of cyclin D3 rather than the acquisition or recruitment of cyclin D2, as have other herpes viruses (16–19)?
Relevant to the problems posed by the questions are the following: Cyclin D3 accumulates at ND10 bodies in cells infected with the D199A mutant. Because ND10 structures serve as the nucleation sites for the formation of the HSV-1 replication compartments, those observations suggest that recruitment of cyclin D3 to the sites of formation of the replication compartments is reinforced by ICP0 but does not depend on it. Another role of wild-type ICP0 is to stabilize cyclin D3 (6–8). The partial complementation of ΔICP0 mutants by overexpressed cyclin D3 suggests that the interaction of ICP0 with cyclin D3 involves the stabilization and recruitment of cyclin D3 ultimately to the replication compartment and that this function plays an important role in meeting the goal of the virus to maximize the amounts of progeny produced by infected cells. The interchangeability of the D cyclins suggests that this function is shared by all three D cyclins. The key function shared by all three D cyclins involves activation of the pathway to the S phase. Finally, cells infected by HSV-1 in vivo are nondividing, most likely in G1 phase. The studies on the D199A substitution mutant predict that recruitment of D cyclin is particularly important in these cells and in cells in which HSV-1 reactivates from a latent state (7).
Consistent with these conclusions is the observation that inhibition of cdk4 by inhibitor I, which interacts with its ATP binding site, resulted in retention of ICP0 in the nucleus. This observation suggests that the function performed by ICP0 with respect to cyclin D3 ultimately leads to the activation of cdk4 and that the sensor that determines the translocation of ICP0 to the cytoplasm requires the function of cdk4. Current literature suggests that cdk4 may have multiple functions but the one common to the D cyclins is activation of transcription of genes that enable the initiation of the S phase of the cell cycle (14, 15). One hypothesis that is consistent with all available data is that ICP0 recruits cyclin D3 to augment or ensure availability of activated cdk4 in the replication compartment, that the function of cdk4 is to facilitate the onset and maintenance of viral DNA synthesis, and that the signal for translocation of ICP0 to the cytoplasm is the accumulation of a viral gene product dependent on the synthesis of viral DNA. Consistent with this hypothesis, ICP0 is retained in the nucleus in cells exposed to phosphonoacetate, a powerful inhibitor of viral DNA synthesis, and blocks the accumulation of γ2 (late) gene products (12). However, HSV-1 blocks cellular DNA synthesis (10, 11).
The question regarding the selective recruitment of cyclin D3 poses an interesting challenge. In principle, a fundamental strategy of HSV-1 that is different from those of EBV or human herpes virus no. 8 (HHV8) is that HSV-1 has evolved proteins that recruit and redirect cellular proteins rather than incorporate into its genome and subsequently modify the gene whose product that it needs. Hence, we may assume that HSV-1 specifically evolved a function to recruit cyclin D3 even though both cyclins D1 and D2 perform similar functions. One hypothesis that remains to be explored is based on the observation that cyclin D3 interacts with lamina A/C (22). The formation of replication compartments at the ND10 structures has been reported to occur at nuclear lamina, and disruption of the lamina has been reported to interfere with the development of these compartments (23). Conceivably, the additional function performed by cyclin D3 played a role in the evolution of its recruitment by ICP0. This potential role of cyclin D3 has to be verified. If this were the case, then determining whether herpes virus saimiri or HHV8 have evolved separate functions to bring their replication compartments to nuclear lamina or whether cyclin D2 encoded by these viruses has acquired this function would be of interest.
Materials and Methods
Cells and Viruses.
The sources, properties, and propagation of HEp-2, U2OS, and Vero cells were reported elsewhere (13, 24). The properties of the parent HSV-1(F) strain, the R7801 mutant carrying cyclin D3 inserted into the thymidine kinase gene, and the BAC encoding either the wild-type HSV-1(F) DNA or the ΔICP0 DNA (Fig. 3) were described elsewhere (4, 6, 25–27). The wild-type and mutant viruses were titered on U2OS cells.
Construction of Recombinant Viruses.
Wild-type or mutated forms of Myc-tagged D-type cyclins under control of the Egr-1 promoter followed by the bidirectional UL21/UL22 poly(A) were inserted into the unique BamHI site between UL3 and UL4 of ΔICP0 with the aid of the BAC system. For the development of a pKO5 plasmid with UL3/UL4 flanking sequences, the steps were as follows: Initially, a HindIII fragment from pRB209 (28) was inserted into the HindIII site of pRB442 (29) to generate pRB6716. Then, the SphI–PvuII fragment from pRB6716 that contains the sequence between nucleotides 9598 and 13679 of the viral genome was inserted into the NotI–BglII sites of pKO5 to generate pRB6717. The cassettes containing the Myc-tagged D-type cyclins under control of the Egr-1 promoter followed by the UL21/22 poly(A) were amplified from pRB6709, pRB6714, and pRB6715, described below, digested with PmeI, and inserted into the BamHI site of pRB6717, generating pRB6718, pRB6719, and pRB6720, respectively.
Escherichia coli RR1 strain harboring ΔICP0 BAC (4) was electroporated with either pRB6718, pRB6719, or pRB6720 and incubated at 43 °C on LB plates containing 25 μg/mL zeosin (Zeo) and 20 μg/mL chloramphenicol. The colonies were diluted and plated on LB plates containing chloramphenicol (20 μg/mL) and 5% sucrose. Colonies grown on sucrose plates were screened by colony hybridization. Plasmids isolated from positive colonies were transfected into U2OS cells. The incorporation of cyclin D at the desired location (Fig. 3) was verified in recombinant viruses R6700, R6701, and R6702 by PCR.
Plasmid Construction and Site-Directed Mutagenesis.
The PCR product of wild-type cyclin D3 from pRB5162 was digested with EcoRV and inserted into the EcoRV site of pcDNA3.1(+)ZeoMyc to generate pRB6700. For the Myc-tagged deletion forms of cyclin D3, PCR products of either the amino or the carboxy terminus from pRB5162 (6) were digested with EcoRV and inserted into the EcoRV site of pcDNA3.1(+)ZeoMyc to generate pRB6701 (1–155 aa), pRB6702 (156–292 aa), pRB6703 (1–129 aa), pRB6704 (1–100 aa), pRB6705 (1–69 aa), and pRB6706 (1–41 aa). The PmeI fragment from pRB6700, containing the Myc-tagged full-length cyclin D3, was inserted into the SpeI–XhoI sites of pPRB5162 to generate pRB6709. The PCR products of full-length wild-type cyclin D2 and wild-type cyclin D1 (kindly provided by K. Riabiowol, University of Calgary Canada) were digested with NruI and EcoRV, respectively, and inserted into the EcoRV of pcDNA3.1(+)Myc to generate pRB6712 and pRB6713, respectively. The PmeI fragments from pRB6712 and pRB6713, containing Myc-tagged cyclin D2 and Myc-tagged cyclin D1, were inserted into the SpeI–XhoI sites of pRB5162 to generate pRB6714 and pRB6715, respectively.
Transfection and Infections.
Cells grown in four-well slides (Erie Scientific) were transfected when 60 to 70% confluent with 250 ng per well of DNA in mixtures of 1 μL of Lipofectamine and 1.5 μL of Plus reagents per well, as specified by the supplier (Invitrogen). At 3 h after transfection, the medium was removed, and the cells were rinsed extensively with DMEM supplemented with 10% FBS and further incubated for 24 h. The cells were exposed to 10 pfu of virus per cell in medium 199V, consisting of mixture 199 (Sigma) supplemented with 1% calf serum 24 h after transfection.
Immunofluorescence Studies.
The procedures were described elsewhere (13, 24). Briefly, the cells were fixed in 4% paraformaldehyde, at times indicated in the Results section, permeabilized, blocked with PBS-TBH solution consisting of 0.1% Triton X-100 in PBS, 10% human serum, and 1% BSA, and reacted with primary antibodies diluted in PBS-TBH. The ICP0 mouse monoclonal antibody (Goodwin Institute for Cancer Research, Plantation, FL) was used at a dilution of 1:1,000. The ICP0 exon 2 rabbit polyclonal antibody was used at a dilution of 1:2,000. The ICP8 rabbit polyclonal antibody was kindly provided by D. Knipe (Harvard Medical School, Cambridge, MA) and used at a dilution of 1:2,000. The cdk4 and PML rabbit polyclonal antibodies and the c-Myc mouse monoclonal antibody (Santa Cruz Biotechnology) were used at a dilution of 1:500. The cyclin D3 mouse monoclonal antibody (BD Pharmigen) was used at a dilution of 1:500. The four-well cultures were then rinsed several times with PBS-TBH and reacted with Alexa-Fluor-594-conjugated goat anti-rabbit or Alexa-Fluor-488-conjugated goat anti-mouse, diluted 1:1,000 in PBS-TBH. After several rinses, first with PBS-TBH and then with PBS, the samples were mounted and examined with a Zeiss confocal microscope equipped with software provided by Zeiss. For the quantification studies, ≈450 cells were counted. The cdk4 inhibitor I (Calbiochem) was used as indicated in the figure captions.
Acknowledgments.
We thank David Knipe (Harvard Medical School, Boston) for the rabbit polyclonal antibody to ICP8 and Karl Riabowol (University of Calgary, Calgary, AB, Canada) for the plasmids encoding cyclins D1 and D2.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sacks WR, Schaffer PA. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100:8963–8968. doi: 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76:841–850. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST–REST complex. Proc Natl Acad Sci USA. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol. 2009;83:4376–4385. doi: 10.1128/JVI.02515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Sant C, Lopez P, Advani SJ, Roizman B. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J Virol. 2001;75:1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehmann GL, McLean TI, Bachenheimer SL. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevents G1 entry in quiescent cells. Virology. 2000;267:335–349. doi: 10.1006/viro.1999.0147. [DOI] [PubMed] [Google Scholar]
- 10.Roizman B, Aurelian L, Roane PR., Jr The multiplication of herpes simplex virus. I. The programming of viral DNA duplication in HEp-2 cells. Virology. 1963;21:482–498. doi: 10.1016/0042-6822(63)90209-5. [DOI] [PubMed] [Google Scholar]
- 11.Roizman B, Roane PR., Jr The multiplication of herpes simplex virus. II. The relation between protein synthesis and the duplication of viral DNA in infected HEp-2 cells. Virology. 1964;22:262–269. doi: 10.1016/0042-6822(64)90011-x. [DOI] [PubMed] [Google Scholar]
- 12.Lopez P, Van Sant C, Roizman B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol. 2001;75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalamvoki M, Roizman B. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci USA. 2008;105:20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187–190. doi: 10.1016/s0968-0004(00)89005-2. [DOI] [PubMed] [Google Scholar]
- 15.Sherr CJ, Roberts JM. Living with or without cyclins and cyclin-dependent kinases. Genes Dev. 2004;18:2699–2711. doi: 10.1101/gad.1256504. [DOI] [PubMed] [Google Scholar]
- 16.Chang Y, et al. Cyclin encoded by KS herpesvirus. Nature. 1996;382:410. doi: 10.1038/382410a0. [DOI] [PubMed] [Google Scholar]
- 17.Jung JU, Stager M, Desrosiers RC. Virus-encoded cyclin. Mol Cell Biol. 1994;14:7235–7244. doi: 10.1128/mcb.14.11.7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, et al. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholas J, Cameron KR, Honess RW. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 20.Uprichard SL, Knipe DM. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology. 1997;229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 21.Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101–7105. doi: 10.1128/JVI.77.12.7101-7105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mariappan I, Gurung R, Thanumalayan S, Parnaik VK. Identification of cyclin D3 as a new interaction partner of lamin A/C. Biochem Biophys Res Commun. 2007;355:981–985. doi: 10.1016/j.bbrc.2007.02.060. [DOI] [PubMed] [Google Scholar]
- 23.Silva L, Cliffe A, Chang L, Knipe DM. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008;4:e1000071. doi: 10.1371/journal.ppat.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalamvoki M, Qu J, Roizman B. Translocation and colocalization of ICP4 and ICP0 in cells infected with herpes simplex virus 1 mutants lacking glycoprotein E, glycoprotein I, or the virion host shutoff product of the UL41 gene. J Virol. 2008;82:1701–1713. doi: 10.1128/JVI.02157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 26.Horsburgh BC, Hubinette MM, Tufaro F. Genetic manipulation of herpes simplex virus using bacterial artificial chromosomes. Methods Enzymol. 1999;306:337–352. doi: 10.1016/s0076-6879(99)06022-x. [DOI] [PubMed] [Google Scholar]
- 27.Ye GJ, Roizman B. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc Natl Acad Sci USA. 2000;97:11002–11007. doi: 10.1073/pnas.97.20.11002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poffenberger KL, Tabares E, Roizman B. Characterization of a viable, noninverting herpes simplex virus 1 genome derived by insertion and deletion of sequences at the junction of components L and S. Proc Natl Acad Sci USA. 1983;80:2690–2694. doi: 10.1073/pnas.80.9.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baines JD, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]