Abstract
The transcription factor NF-E2-related nuclear factor 2 (Nrf2) regulates expression of genes that protect cells from oxidative damage. Here, we characterized nitric oxide (•NO)-induced Nrf2–Kelch-like ECH-associated protein 1 (Keap1) signaling and its role in counteracting •NO-induced apoptosis of human colon cancer HCT116 cells. Nrf2 was localized in the cytoplasm in control cells; •NO triggered its rapid nuclear accumulation, transcriptional activation, and up-regulation of HO-1, NQO1, and GCL, but not GST A4 and P1 subunits. Nrf2 accumulation in the nucleus was also associated with enhanced transcription and posttranscriptional modifications. (S)-nitrosation of Keap1 may contribute to nuclear accumulation of Nrf2 by facilitating its dissociation from Keap1, thus initiating •NO-mediated Nrf2–Keap1 signaling. •NO-mediated induction of ARE-dependent genes occurred well before apoptosis, as judged by caspase 3 activation. Collectively, these results show that the Nrf2–Keap1 signaling pathway mediates protective cellular responses to mitigate •NO-induced damage and may contribute to the relative resistance of HCT116 to •NO-induced cytotoxicity.
Keywords: inflammation, oxidative stress, reactive nitrogen species, (S)-nitrosation
At low concentrations, nitric oxide (•NO) serves as a signaling molecule, whereas excessive levels, such as those produced by inflammatory cells, trigger downstream signaling pathways, eliciting necrosis or apoptosis (1). Protection against these effects involves enhanced expression of stress-responsive genes through tightly regulated and precisely coordinated processes (2, 3). The pathway mediated by NF-E2-related nuclear factor 2 (Nrf2)-Kelch-like ECH-associated protein 1 (Keap1) is of central importance in regulating expression of detoxifying and antioxidant genes and protecting against carcinogenesis and xenobiotic toxicity (3–5).
Keap1 serves as a sensor for electrophile-induced activation of Nrf2-regulated genes and acts as a negative regulator of Nrf2 signaling in quiescent cells by retaining Nrf2 in the cytoplasm through simultaneous interactions with it and the actin cytoskeleton. Modifications of critical thiol groups in Keap1 result in dissociation of the Keap1–Nrf2 complex and translocation of Nrf2 into the nucleus, where it functions as a strong transcriptional activator of antioxidant response element (ARE)-responsive genes, such as heme oxygenase 1 (HO-1), NAD(P)H:quinone oxidoreductase 1 (NQO1), glutamate cysteine ligase (GCL), and GST (6).
Keap1-mediated proteasomal degradation of Nrf2 also plays an important role in regulating its transcriptional activity (7, 8), and Keap1 serves as an adaptor for Cul3-dependent Nrf2 ubiquitination (9, 10). This suggests that Keap1 actively induces Nrf2 degradation by facilitating its ubiquitination, in addition to passively sequestering it in the cytoplasm (11, 12). In some cells, Nrf2 is reportedly localized primarily in the nucleus, where its activation is repressed by transient Keap1 shuttling into the nucleus to promote its ubiquitination (13). Additionally, up-regulation of Nrf2 gene expression has been shown to increase Nrf2 protein levels in response to ARE inducers (14), and its partition between cytoplasm and nucleus may be controlled by both nuclear import and export (15, 16).
We and others have been investigating mechanisms involved in •NO-induced cellular stress, DNA damage, mutagenesis, and apoptotic signaling pathways (17, 18). We have defined steady-state concentration and cumulative-dose thresholds for •NO-induced toxicity (19) and evaluated biological roles of glutathione in these processes (20). However, precise biochemical and molecular mechanisms by which cells protect themselves from •NO-induced damage remain incompletely understood. •NO donor drugs reportedly stimulated Nrf2 translocation in bovine vascular endothelial cells (21) and induced Nrf2-mediated transcriptional up-regulation of protective genes in human neuroblastoma cells (22), underscoring the possible importance of Nrf2-mediated antioxidant response signaling in cellular defense against •NO-induced damage.
Because inflammation is an established risk factor for colon cancer in humans, we sought to determine whether •NO activates Nrf2–Keap1 signaling and affects induction of apoptosis in HCT116 human colon carcinoma cells. Cells were exposed to •NO via a system that tightly controls steady-state •NO concentrations, physiologically relevant dose rates, and oxygen levels in culture media, mimicking the chemical environment of inflamed tissues (19, 20, 23). Under nonstressed conditions, Nrf2 was sequestered in the cytoplasm along with Keap1. •NO treatment resulted in Nrf2 release and translocation into the nucleus, transcriptional activation, and up-regulation of representative protective genes. •NO also caused (S)-nitrosation of cysteine residues in Keap1, which may represent a mechanism triggering Nrf2 disassociation from Keap1, thus initiating •NO-mediated Nrf2–Keap1 signaling. •NO treatment also induced binding of nuclear extracts to the NQO1 ARE and transcriptional up-regulation of Nrf2 and Keap1 genes, which was associated with reduced sensitivity to •NO-induced apoptosis.
Results
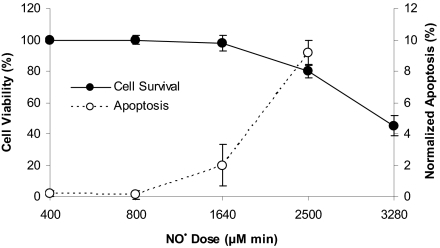
Exposure of HCT116 cells to •NO at steady-state concentrations of 0.6 or 1.8 μM for 24 h caused no cell death, whereas exposure to 7 μM for 1 to 8 h, producing cumulative doses of 400 to 3,200 μM·min, caused loss of cell viability 24 h after treatment (Fig. 1). Cell killing was related to cumulative •NO dose, with a threshold of ≈1,000 μM·min, below which no cell death was observed. Frequency of apoptosis was also related to cumulative dose, with a threshold between 800 and 1,640 μM·min (Fig. 1). Cell cycle analysis 24 h after treatment revealed S-phase arrest in cells treated with 1,600 μM·min. Treatment with argon, the carrier for •NO, had no effect on cell survival, apoptosis, or cell cycle.
Fig. 1.
Cell viability and normalized percent of apoptosis in HCT116 cells 24 h after •NO treatment. Cell viability, determined by trypan blue exclusion, was expressed as the percentage of argon-treated cells. Data represent the mean ± SD of three to four experiments done in duplicate. The threshold dose of •NO-induced apoptosis was ≈1,600 μM·min. Data represent the mean ± SD of three independent experiments, each done in duplicate.
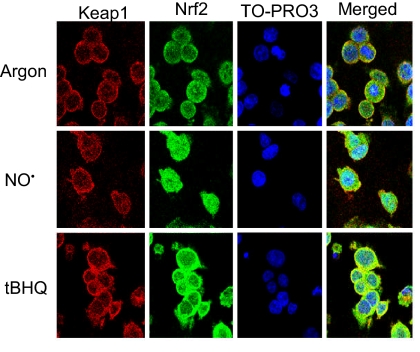
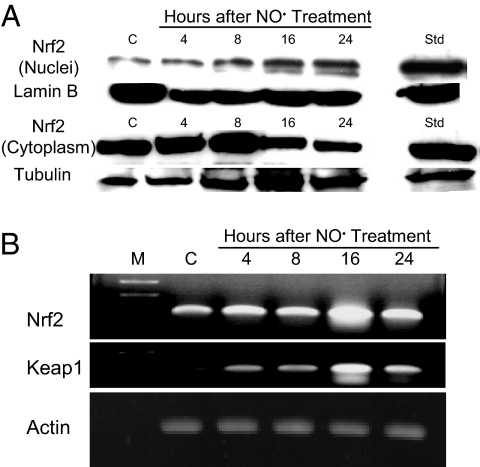
To assess the role of Nrf2-mediated processes in these responses, we examined its localization in cells treated with 1,640 μM·min •NO, which caused 2% apoptosis. Immunofluorescence and confocal microscopy (Fig. 2) showed that Nrf2 and Keap1 were localized in the cytoplasm in control cells. •NO exposure resulted in Nrf2 translocation to the nucleus 4 h after treatment, whereas Keap1 remained localized in the cytoplasm. Similar results were obtained in positive control cells treated with tert-butyl hydroquinone (tBHQ), a potent inducer of Nrf2-dependent gene expression (13). Western blot analysis revealed that Nrf2 protein was present at much higher levels in the cytosolic than in the nuclear fraction after •NO exposure (Fig. 3A). Nrf2 protein accumulated in the nucleus as early as 4 h after exposure, reaching 115–131% (P < 0.01) over control cells during the 8- to 24-h period. Cytoplasmic Nrf2 protein was significantly reduced by 85% (P < 0.05) and 79% (P < 0.01) 16 and 24 h after treatment (Fig. 3A), whereas Keap1 protein remained unchanged.
Fig. 2.
•NO-induced Nrf2 translocation from cytoplasm to nucleus in HCT116 cells 4 h after treatment with 1,640 μM·min •NO, as detected by immunofluorescent staining and confocal microscopy. HCT116 cells treated with 50 μM tBHQ for 4 h were used as positive controls. (Magnification: 200×.)
Fig. 3.
Exposure to •NO-induced nuclear translocation and increased expression of Nrf2. (A) Nrf2 protein was constitutively expressed in both cytoplasm and nucleus and accumulated in the nucleus upon •NO treatment, as measured by Western blot analysis. HCT116 cells treated with 50 μM tBHQ for 4 h were used as positive controls (Std). (B) •NO treatment induced transcriptional up-regulation of Nrf2 and Keap1 genes, as measured by RT-PCR.
Expression of the Nrf2 and Keap1 genes measured by RT-PCR was maximally increased at 16 h, subsequently declining 24 h after •NO treatment (Fig. 3B). Lamin B and tubulin served as markers for nuclear and cytoplasmic proteins, respectively (22). Lamin B was undetectable in the cytoplasmic extract and tubulin undetectable in the nuclear extract when the same amounts of proteins were analyzed by Western blotting. Hence, •NO treatment not only triggered translocation of Nrf2 to the nucleus, but it also induced transcriptional up-regulation of the Nrf2 and Keap1 genes in HCT116 cells.
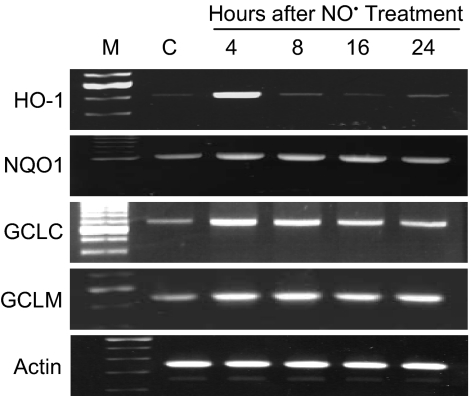
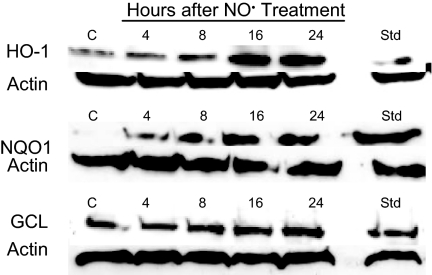
Semiquantitative RT-PCR analyses revealed that •NO treatment induced up-regulation of a battery of detoxifying and antioxidant genes (Fig. 4). Transcript levels of the HO-1 gene were sharply increased 4 h after treatment, returning to control levels by 8 h and later. Substantial increases in NQO1 and GCL gene transcripts were also observed as early as 4 h after exposure, whereas levels of the GST A4 and GST P1 genes were unaffected. •NO treatment also resulted in elevated levels of the HO-1, NQO1, and GCL proteins (Fig. 5); HO-1 levels increased to 125% at 16 h and 132% at 24 h; NQO1 levels reached a maximum of 142% at 24 h; and GCL protein rose to 114–116% over a period of 8–24 h. All increases were statistically significant (P < 0.05).
Fig. 4.
•NO-induced transcriptional up-regulation of detoxifying enzymes and antioxidant genes in HCT116 cells treated with 1640 μM·min •NO, as measured by RT-PCR.
Fig. 5.
Increased levels of HO-1, NQO1, and GCL proteins were observed in HCT116 cells after treatment with 1,640 μM·min •NO, as measured by Western blot analysis. HCT116 cells treated with 50 μM tBHQ for 4 h were used as positive controls (Std).
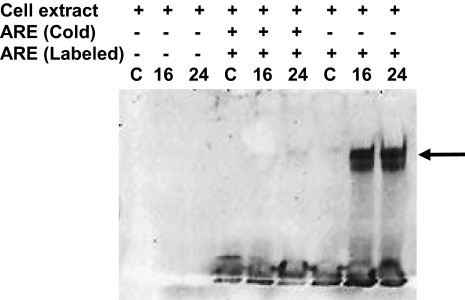
To assess transcription factors mediating the ARE response, nuclear extracts from cells treated with 1,600 μM·min •NO were incubated with digoxigenin (DIG)-labeled NQO1 ARE oligonucleotide, and a gel-shift assay was performed (Fig. 6). •NO treatment resulted in increased binding of nuclear factors to the NQO1 ARE. The specificity of enhanced binding was verified by a competition experiment, which showed that excess unlabeled NQO1 oligonucleotides completely blocked formation of the DNA–protein complex (Fig. 6).
Fig. 6.
•NO-enhanced binding of nuclear factors to the ARE of the NQO1 gene. DIG-end-labeled ARE of the human NQO1 gene was incubated with nuclear extract from •NO-treated HCT116 cells as described in Materials and Methods. Mobility-shifted ARE–nuclear protein complex is indicated by the arrow. To verify specificity of binding, unlabeled ARE and 125-fold excess of unlabeled ARE plus labeled ARE were added in a competition assay.
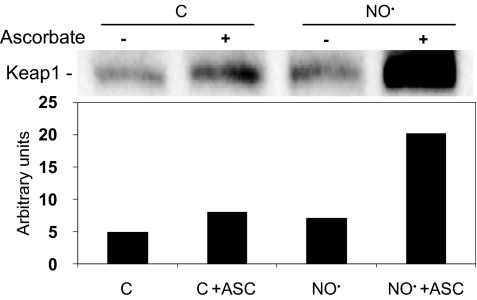
To determine whether Nrf2 release was associated with (S)-nitrosation of Keap1, cells were exposed to 1,600 μM·min •NO, and Keap1 was analyzed by the biotin switch assay. As shown in Fig. 7, a significant increase in Keap1 (S)-nitrosation was detected in cells exposed to •NO compared with control cells, in which a low basal level of (S)-nitrosation was also observed. Thus, increased (S)-nitrosation of Keap1, leading to release of Nrf2 and its nuclear translocation, may represent an important triggering event in cellular responses to •NO.
Fig. 7.
(S)-nitrosation of Keap1. HCT116 cells were exposed to 1,600 μM·min •NO and submitted to biotin switch assay of protein (S)-nitrosation as described in Materials and Methods. Biotinylated proteins were isolated and analyzed by Western blot with anti-Keap1 antibody. An increased signal in the presence of ascorbate (ASC), which displaces •NO from nitrosothiols, enhancing biotin labeling, verifies specificity of (S)-nitrosation. Cells exposed to argon were used as controls. The bar graph represents the densitometric analysis of Keap1 bands in the blot.
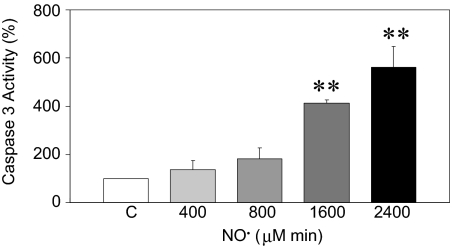
Caspase 3 activity was monitored by using the caspase 3 substrate (DEVD-pNA) to study the temporal relationship between initiation of •NO-induced Nrf2–Keap1 signaling and induction of apoptosis. •NO treatment resulted in a dose-dependent induction of caspase 3 activity. Significant (P < 0.01) increases—455% ± 13% and 560% ± 88% of control values—were evidenced 24 h after treatment with 1,600 and 2,400 μM·min •NO, respectively (Fig. 8). Activation of caspase 3 and increased frequency of apoptosis (Fig. 1) both required the same threshold cumulative dose; namely, 1,600 μM·min. No increase in caspase activity was observed in cells treated with 400 and 800 μM·min •NO, nor was it present 4 h after •NO exposure to the higher doses.
Fig. 8.
Caspase 3 activity in HCT116 cells 24 h after treatment with 400 to 2,400 μM·min •NO. Increases in caspase 3 activity were calculated as percentage of A405 nm in •NO-treated versus argon-treated control cells. Data are mean ± SE from three independent experiments. **, P < 0.01 compared with argon control.
Discussion
In unstressed conditions, expression of Nrf2 target genes is minimized by Nrf2 sequestration in the cytoplasm by Keap1 binding, ubiquitinylation, and proteosomal degradation (7, 8). The Nrf2–Keap1 interaction has been suggested to be a sensor of oxidative stressors (9, 25), and the involvement of Keap1 cysteine residues 151, 273, and 288 in Nrf2 degradation supports such a role (26). •NO modulates the cellular redox state and has been shown to have an impact on functionally important cysteines (27). Our findings indicate that •NO modifies Keap1 cysteine residues, and may thus interfere with Nrf2 binding and enable Nrf2 to accumulate in the nucleus (Figs. 2 and 3A) (13). We found that the level of Keap1 (S)-nitrosation was strongly increased in HCT116 cells exposed to a dose of •NO that induced Nrf2 nuclear translocation (Fig. 7). In the nucleus, Nrf2 forms a complex with the small Maf protein (28) that in turn induces transcriptional up-regulation of detoxifying and antioxidant genes. Our results are consistent with that mechanism, in that •NO-induced Nrf2 accumulation in the nucleus (Fig. 3A) was associated with increased expression of HO-1, NQO1, and GCL (Fig. 4), and gel-shift assay showed that nuclear extracts of •NO-treated cells stimulated ARE-mediated NQO1 gene expression (Fig. 6). (S)-nitrosation of Keap1 in HEK293H cells treated with 500 μM CSNO has been reported to precede nuclear translocation of the Keap1–Nrf2 complex (29). Our results show that exposure of cells to •NO at a steady-state concentration of 7 μM causes (S)-nitrosation of Keap1 and nuclear translocation of Nrf2. •NO also promotes generation of 8-nitro-cGMP, a compound with high affinity for sulfhydryl groups (30), suggesting that Keap1 may be highly susceptible to (S)-guanylation. Possible functional effects of this modification require further investigation.
Our results also show that •NO up-regulated expression of HO-1, NQO1, and GCL genes and increased their protein levels (Figs. 4 and 5). Clearly, activation of these and other detoxifying enzymes and protective proteins through the Nrf2–Keap1 system could counteract •NO-mediated toxicity. In contrast to a previous report that •NO donor drugs up-regulated GST A4 and GST P1 genes in human neuroblastoma cells (22), we did not observe this in HCT116 cells, suggesting that regulation of these genes may depend on cell type and/or inducer level. HCT116 cells were relatively resistant to killing by •NO, compared with lymphoblastoid TK6 cells, in which the threshold steady-state concentration for cell death was ≈0.5 μM (31), whereas HCT116 cells tolerated 0.6 μM and 1.8 μM •NO without evidence of cytotoxicity (Fig. 1). Whereas the threshold •NO dose for both lethality and apoptosis was 150 μM·min in TK6 cells (19), these values were 1,000 and 1,600 μM·min, respectively, in HCT116 cells (Fig. 1). As in TK6 cells, •NO-induced apoptosis in HCT116 cells was mediated via caspase 3 activation (Fig. 8). We found that the 1,600 μM·min threshold •NO dose that induced apoptosis also increased caspase 3 activity, evident at 24 h, but not 4 h, after •NO treatment. Thus, because up-regulation of the Nrf2 gene and nuclear accumulation of its protein were detected as early as 4 h after •NO exposure (Fig. 3), activation of Nrf2–Keap signaling by •NO took place before caspase 3 activation and induction of apoptosis. This result is supported by a study in neuroblastoma cells demonstrating that ARE activation mediated by •NO donors also occurred before caspase 3 activation (22).
Our data reveal that the Nrf2–Keap1 signaling pathway is activated by •NO, participates in responses to mitigate •NO damage, and may contribute to the relative resistance of HCT116 to •NO toxicity. These findings provide new insights into protective mechanisms used by cells responding to oxidative and nitrosative stresses caused by excessive •NO exposure as created by inflammatory cells in inflamed tissues.
Materials and Methods
Cell Culture and •NO Treatment.
HCT116 human colon carcinoma cells (32, 33) (from C. C. Harris, National Cancer Institute, Bethesda, MD) were maintained at 37 °C, 5% CO2 in McCoy's 5A medium containing 10% FBS, 100 units/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine. Cells (1 × 106) were plated in 60-mm dishes 24 h or 48 h before exposure to •NO at steady-state concentrations of 0.6, 1.8, or 7 μM (19, 20, 23, 31). An O2 (50%)/CO2 (5%) mixture maintained O2 at air saturation. Cumulative total •NO dose, expressed as micromolar·minute, was controlled by varying exposure time. Cells exposed to argon gas, the carrier for •NO, delivered under identical conditions served as negative controls in all experiments. Cell-culture reagents were purchased from Lonza, and gases were provided by Airgas.
Cell Viability and Apoptosis Analysis.
Cell viability 24 h after treatment was determined by trypan blue exclusion, which produced results comparable to plating efficiency and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay (17–20). Apoptosis 24 h after treatment was quantitatively measured with a Becton Dickinson FACScan equipped with CellQuest software after annexin V-FITC and propidium iodide staining (Clontech) (17, 18).
Cell Cycle Analysis.
Cells collected 24 h after treatment were washed twice in PBS, fixed in 70% cold ethanol overnight, resuspended in 1% BSA-PBS solution containing 50 μg/mL propidium iodide and 50 μg/mL RNase, incubated at 37 °C for 30 min, then analyzed with a Becton Dickinson FACScan. Cell-fit analysis determined the percentage of cells in a specific stage of the cell cycle (19).
Immunofluorescence and Confocal Microscopy.
Cells (1 × 105) were grown in poly-d-lysine-coated glass dishes (MatTek) for 24 h, then treated with •NO as above. Cells treated with 50 μM tBHQ for 4 h served as positive controls. After treatment, cells were fixed with 4% formaldehyde, permeabilized with 100% cold methanol and 0.2% Triton X-100, treated with RNase, and blocked in 100 mM glycine and 0.5% IgG-free BSA. Rabbit anti-Nrf2 polyclonal antibody C-20 and goat anti-Keap1 polyclonal antibody E20 (Santa Cruz Biotechnology) (22), 1 μg per 100 μL in blocking buffer (0.5% BSA in PBS), were added and incubated for 1 h. Rabbit anti-Nrf2 antibody αNrf2-NTR (dilution 1:100; from C. B. Pickett and T. Nguyen, Schering-Plough Research Institute, Kenilworth, NJ) (13) was also used in immunofluorescent staining. FITC-conjugated donkey anti-rabbit IgG (1:200) and rhodamine-conjugated AffiniPure donkey anti-goat IgG (1:200; Jackson Laboratories) were used to label Nrf2 and Keap1, respectively. Cells were stained with the nuclear stain TO-PRO3 (Invitrogen). Confocal images were obtained by using a Zeiss laser scanning confocal microscope and processed by using Zeiss LSM 510 Image Browser software.
Cell Lysate and Nuclear and Cytoplasmic Extract Preparation.
Cells were lysed by using a kit from Cell Signaling Technology. Nuclear and cytoplasmic proteins were extracted by using a kit from Panomics following the manufacturer's instructions. Immunoblotting analysis of tubulin and lamin B indicated no contamination between nuclear and cytoplasmic fractions.
Western Blot Analysis.
Western blot analyses were performed as described previously (18). Primary antibodies used include: goat anti-Keap1 polyclonal antibody E20 and rabbit anti-Nrf2 polyclonal antibody C-20 (Santa Cruz Biotechnology); mouse anti-HO-1 monoclonal antibody (Stressgen); goat anti-NQO1 polyclonal antibody (Novus Biologicals); and rabbit anti-GCL polyclonal antibody Ab-1 (LabVision). To control for protein loading, membranes were stripped and reprobed with mouse anti-actin monoclonal antibody, mouse anti-lamin B monoclonal antibody (Oncogene), or mouse anti-α-tubulin monoclonal antibody (Upstate Biotechnology).
EMSA.
EMSA was performed by using a DIG gel-shift kit (Roche). A double-stranded DNA probe containing the NQO1 gene ARE sequence 5′-CAGTCACAGTGACTCAGCAGAATCT-3′ was used (34). DNA probes were labeled at 3′ ends with DIG-11-ddUTP by using terminal transferase. Nuclear extract (5 μg) was incubated with 1 μg of poly[d(I-C)], 1 μg of poly-l-lysine, and 5× binding buffer [100 mM Hepes, pH 7.6; 5 mM EDTA; 50 mM (NH4)2SO4; 5 mM DTT; 1% Tween 20; and 150 mM KCl] at room temperature for 15 min, then mixed with loading buffer (0.25× 90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3, 40% glycerol, and 0.2% bromophenol blue). The mixture was loaded on a native 5% polyacrylamide gel and separated in 0.5× TBE buffer at 120 V at 4 °C for 1 h. The gel was transferred in 0.5× TBE onto a positively charged nylon membrane at 400 mA for 30 min at 4 °C. After transfer, the sample was fixed on the membrane by UV cross-linking. DIG detection was performed as described by the manufacturer and detection of enhanced chemiluminescence by exposure to Hyperfilm ECL (GE Life Sciences). For binding competition experiments, unlabeled DNA and excess unlabeled competitor DNA (100 ng) were included in the reaction mixture.
Semiquantitative RT-PCR Analysis.
Total RNA was extracted by using the Tri Reagent kit (Sigma), and semiquantitative RT-PCR analysis was performed by using the SuperScript one-step RT-PCR kit (Invitrogen) following the manufacturer's protocol. A total of 2 μg of total RNA was used in each reaction.
Immunoprecipitation of Keap1.
For Keap1–Nrf2-binding analysis, 500 μg of cytoplasmic protein isolated from cells treated with either •NO or argon gas was immunoprecipitated with goat anti-Keap1 polyclonal antibody E20 following a protocol reported previously (33). Samples were then resolved on 15% SDS/PAGE gels under reducing conditions and were transferred to polyvinylidene difluoride membrane (Bio-Rad) for immunodetection with rabbit anti-Nrf2 polyclonal antibody C-20 (Santa Cruz Biotechnology) and secondary horseradish peroxidase-conjugated anti-rabbit IgG antibody (Bio-Rad).
Keap1 (S)-Nitrosation Analysis.
(S)-nitrosation of Keap1 was assessed by a modified biotin switch assay (35). Unless stated otherwise, all reagents were from Sigma. HCT116 cells exposed to 1600 μM·min •NO were washed in HEN buffer (250 mM Hepes sodium salt, pH 7.7; 1 mM EDTA; and 0.1 mM neocuproine) and lysed for 30 min at 4 °C in HEN buffer containing 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 0.1% SDS, and protease inhibitors. Lysates were centrifuged at 14,000 × g, and 1 mg of total protein from each sample was precipitated with acetone and resuspended in HEN buffer containing 1% SDS (HENS buffer) at a final protein concentration of 0.8 μg/μL. Free thiols were blocked by addition of 20 mM (S)-methyl methanethiosulfonate and incubation at 50 °C for 30 min with vortexing, and proteins were precipitated in acetone and resuspended in 200 μL of HENS buffer. Biotinylation of nitrosothiols was carried out by incubation with 20 mM ascorbic acid and 1 mM N-(6-(biotinamido)hexyl)-3′-(2′-pyridyldithio)-propionamide (biotin-HPDP; Pierce–Thermo Scientific) for 1 h at room temperature. To verify specific labeling of nitrosothiols, an internal control was performed in which samples were not treated with ascorbic acid, which is responsible for displacing •NO from nitrosothiols, allowing biotin to bind. All previous steps were performed in the dark. Excess biotin-HPDP was removed by acetone precipitation, and samples were resuspended in 300 μL of HENS buffer plus 600 μL of neutralization buffer (20 mM Hepes, pH 7.7; 100 mM NaCl; 1 mM EDTA; and 0.5% Triton X-100). Biotinylated proteins were isolated by overnight incubation with neutravidin-coupled agarose beads (Pierce–Thermo Scientific). After incubation, beads were washed five times with neutralization buffer containing 600 mM NaCl and once with PBS. Isolated proteins were recovered from beads by addition of equal parts elution buffer (20 mM Hepes, pH 7.7; 100 mM NaCl; 1 mM EDTA; and 100 mM 2-mercaptoethanol) and reducing SDS/PAGE sample buffer, after heating at 100 °C for 10 min. The amount of (S)-nitrosated Keap1 in the samples was analyzed by Western blot analysis using the E20 antibody anti-Keap1 (Santa Cruz Biotechnology).
Caspase 3 Activity Assay.
Effect of treatment on caspase 3 activation was determined by using ApoAlert caspase colorimetric assay kit from Clontech following the manufacturer's protocol, using 100 μg of whole-cell lysate. Absorbance was read at 405 nm in a microtiter plate reader, and activity was expressed as a percentage of values from control cells.
Statistical Analysis.
All experiments were repeated two to four times after experimental conditions were optimized. Statistical analysis was performed by using a two-tailed Student's t test, and P < 0.05 was considered to be statistically significant.
Acknowledgments.
This work was supported by National Cancer Institute Program Project Grant 5 P01 CA26731 and National Institute of Environmental Health Sciences Center Grant ES02109.
Footnotes
The authors declare no conflict of interest.
References
- 1.Li CQ, Wogan GN. Nitric oxide as a modulator of apoptosis. Cancer Lett. 2005;226:1–15. doi: 10.1016/j.canlet.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 2.Holtzclaw WD, Dinkova-Kostova AT, Talalay P. Protection against electrophile and oxidative stress by induction of phase 2 genes: The quest for the elusive sensor that responds to inducers. Adv Enzyme Regul. 2004;44:335–367. doi: 10.1016/j.advenzreg.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 3.Motohashi H, Yamamoto M. Nrf2–Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10:549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Dinkova-Kostova AT, et al. Extremely potent triterpenoid inducers of the phase 2 response: Correlations of protection against oxidant and inflammatory stress. Proc Natl Acad Sci USA. 2005;102:4584–4589. doi: 10.1073/pnas.0500815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JS, Surh YJ. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005;224:171–184. doi: 10.1016/j.canlet.2004.09.042. [DOI] [PubMed] [Google Scholar]
- 6.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36:1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 7.Stewart D, et al. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem. 2003;278:2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 8.Kwak MK, et al. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23:8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang DD, et al. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang DD, et al. Ubiquitination of Keap1, a BTB-Kelch substrate adaptor protein for Cul3, targets Keap1 for degradation by a proteasome-independent pathway. J Biol Chem. 2005;280:30091–30099. doi: 10.1074/jbc.M501279200. [DOI] [PubMed] [Google Scholar]
- 12.Hong F, Sekhar KR, Freeman ML, Liebler DC. Specific patterns of electrophile adduction trigger Keap1 ubiquitination and Nrf2 activation. J Biol Chem. 2005;280:31768–31775. doi: 10.1074/jbc.M503346200. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T, Sherratt PJ, Nioi P, Yang CS, Pickett CB. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J Biol Chem. 2005;280:32485–32492. doi: 10.1074/jbc.M503074200. [DOI] [PubMed] [Google Scholar]
- 14.Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemopreventive agents: Role of antioxidant response element-like sequences in the Nrf2 promoter. Mol Cell Biol. 2002;22:2883–2892. doi: 10.1128/MCB.22.9.2883-2892.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain AK, Bloom DA, Jaiswal AK. Nuclear import and export signals in control of Nrf2. J Biol Chem. 2005;280:29158–29168. doi: 10.1074/jbc.M502083200. [DOI] [PubMed] [Google Scholar]
- 16.Velichkova M, Hasson T. Keap1 regulates the oxidation-sensitive shuttling of Nrf2 into and out of the nucleus via a Crm1-dependent nuclear export mechanism. Mol Cell Biol. 2005;25:4501–4513. doi: 10.1128/MCB.25.11.4501-4513.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CQ, Trudel LJ, Wogan GN. Nitric oxide-induced genotoxicity, mitochondrial damage, and apoptosis in human lymphoblastoid cells expressing wild-type and mutant p53. Proc Natl Acad Sci USA. 2002;99:10364–10369. doi: 10.1073/pnas.162356399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CQ, Trudel LJ, Wogan GN. Genotoxicity, mitochondrial damage, and apoptosis in human lymphoblastoid cells exposed to peroxynitrite generated from SIN-1. Chem Res Toxicol. 2002;15:527–535. doi: 10.1021/tx010171x. [DOI] [PubMed] [Google Scholar]
- 19.Li CQ, et al. Threshold effects of nitric oxide-induced toxicity and cellular responses in wild-type and p53-null human lymphoblastoid cells. Chem Res Toxicol. 2006;19:399–406. doi: 10.1021/tx050283e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li CQ, et al. Biological role of glutathione in nitric oxide-induced toxicity in cell culture and animal models. Free Radic Biol Med. 2005;39:1489–1498. doi: 10.1016/j.freeradbiomed.2005.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Buckley BJ, Marshall ZM, Whorton AR. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun. 2003;307:973–979. doi: 10.1016/s0006-291x(03)01308-1. [DOI] [PubMed] [Google Scholar]
- 22.Dhakshinamoorthy S, Porter AG. Nitric oxide-induced transcriptional up-regulation of protective genes by Nrf2 via the antioxidant response element counteracts apoptosis of neuroblastoma cells. J Biol Chem. 2004;279:20096–20107. doi: 10.1074/jbc.M312492200. [DOI] [PubMed] [Google Scholar]
- 23.Wang C, Deen WM. Nitric oxide delivery system for cell culture studies. Ann Biomed Eng. 2003;31:65–79. doi: 10.1114/1.1533072. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Lee JM, Johnson JA. Microarray analysis reveals an antioxidant responsive element-driven gene set involved in conferring protection from an oxidative stress-induced apoptosis in IMR-32 cells. J Biol Chem. 2002;277:388–394. doi: 10.1074/jbc.M109380200. [DOI] [PubMed] [Google Scholar]
- 25.Wakabayashi N, et al. Protection against electrophile and oxidant stress by induction of the phase 2 response: Fate of cysteines of the Keap1 sensor modified by inducers. Proc Natl Acad Sci USA. 2004;101:2040–2045. doi: 10.1073/pnas.0307301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamler JS. Redox signaling: Nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 28.Motohashi H, Katsuoka F, Engel JD, Yamamoto M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc Natl Acad Sci USA. 2004;101:6379–6384. doi: 10.1073/pnas.0305902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckley BJ, Li S, Worton AR. Keap1 modification and nuclear accumulation in response to S-nitrosocysteine. Free Rad Biol Med. 2008;44:692–698. doi: 10.1016/j.freeradbiomed.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawa T, et al. Protein S-guanylation by the biological signal 8-nitroguanosine 3′,5′-cyclic monophosphate. Nature Chem Biol. 2007;3:727–735. doi: 10.1038/nchembio.2007.33. [DOI] [PubMed] [Google Scholar]
- 31.Wang C, Trudel LJ, Wogan GN, Deen WM. Thresholds of nitric oxide-mediated toxicity in human lymphoblastoid cells. Chem Res Toxicol. 2003;16:1004–1013. doi: 10.1021/tx0340448. [DOI] [PubMed] [Google Scholar]
- 32.Liu L, et al. Pharmacologic disruption of base excision repair sensitizes mismatch repair-deficient and -proficient colon cancer cells to methylating agents. Clin Cancer Res. 1999;5:2908–2917. [PubMed] [Google Scholar]
- 33.Hofseth LJ, et al. Nitric oxide-induced cellular stress and p53 activation in chronic inflammation. Proc Natl Acad Sci USA. 2005;100:143–148. doi: 10.1073/pnas.0237083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhakshinamoorthy S, Jaiswal AK. Small Maf (MafG and MafK) proteins negatively regulate antioxidant response element-mediated expression and antioxidant induction of the NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2000;275:40134–40141. doi: 10.1074/jbc.M003531200. [DOI] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Snyder SH. The biotin switch method for the detection of s-nitrosylated proteins. Sci STKE. 2001;2001:PL1. doi: 10.1126/stke.2001.86.pl1. [DOI] [PubMed] [Google Scholar]