Abstract
Prototrophic strains of budding yeast exhibit robust metabolic cycles during continuous growth under nutrient-limiting conditions. Previous studies revealed periodic fluctuations of aminolevulinic acid, a precursor of heme, indicating that heme biosynthesis is temporally regulated during these metabolic cycles. The enzyme that catabolizes heme, heme oxygenase, was found to be expressed in a highly periodic manner at both the mRNA and protein level. Heme oxygenase generates the biological gas, carbon monoxide (CO), as a product of heme catabolism. It is shown that pulsed administration of CO induces a phase advancement into the oxidative, respiratory phase of the metabolic cycles. This CO-mediated phase advancement takes place only if the gas is administered during the temporal window when it is predicted to be generated. It is further shown that a yeast strain bearing a targeted deletion of the gene encoding heme oxygenase displays protracted metabolic cycles. These observations provide evidence that gaseous CO may function as a cellular signaling molecule that helps cue metabolic cycling.
Keywords: biological gas, circadian, oscillation, heme, metabolic state
Yeast cells have long been known to exhibit various modes of oscillatory behavior. Among the first observed of such phenomena were reduced pyridine nucleotide oscillations that occurred with a period of <1 min in intact yeast cells grown under anaerobic conditions (1–3). Subsequently, oscillations in oxygen consumption and other growth parameters were observed on continuous culture of yeast cells in a chemostat (4–8). These oscillations displayed periods ranging anywhere from ≈40 min to >4 h, depending on the yeast strain and growth conditions. Such studies demonstrated that under appropriate growth conditions, synchronous changes in cellular metabolism could be observed that repeat over time.
Cyclic changes in metabolism also occur during the course of the circadian cycle. Oscillatory changes in metabolism are particularly evident in plants and other photosynthetic organisms, which must harness the energy of sunlight to produce glucose during the day. Microarray studies of gene expression have shown that many genes encoding metabolic enzymes are periodically expressed as a function of the circadian cycle in various species (9–12). The anticipatory expression of particular genes during specified temporal windows enables organisms to optimally coordinate their physiology and metabolism with alternating day and night cycles.
In turn, is it possible that metabolism and metabolic cues might have a reciprocal role in the control and entrainment of such cycles? It is well-known that restricted feeding can readily reset the phase of circadian rhythm in the liver (13, 14). Also, certain species of primitive cyanobacteria, which have robust circadian rhythms, must perform both photosynthesis and nitrogen fixation, two essential, but chemically incompatible processes, for growth. The solution is that these two processes are temporally segregated to different times of the day-night cycle (15). These observations collectively indicate that such biological cycles are fundamentally linked to cellular metabolism (16, 17).
We have studied a system that displays long-period, ≈4- to 5-h oscillations in the budding yeast Saccharomyces cerevisiae, in hopes of uncovering metabolic cues that might drive biological cycles. When yeast cells are grown to a high density, starved for a short period, and then continuously fed low concentrations of glucose in a chemostat, the cell population becomes highly synchronized and undergoes robust oscillations in oxygen consumption. We have reported that under these conditions over half of all yeast genes are periodically expressed, and termed the underlying oscillations yeast metabolic cycles (YMC) (18). These cycles enable cells to temporally compartmentalize various cellular and metabolic processes about three phases: oxidative (OX), reductive building (RB), and reductive charging (RC) (17, 18). Subsequent metabolite profiling has revealed that many common cellular metabolites oscillate in abundance during the YMC, indicating that the metabolic state of a yeast cell is dynamic in time (19). Analytical studies of cyclic changes in gene expression and metabolite concentration have begun to show how and why cells optimally perform specific tasks at different times of this metabolic cycle (17–20). Also, examination of the transcripts and metabolites that cycle has led to predictions of the biosynthetic and regulatory pathways that might contribute to cycling. However, the metabolic cues that drive the YMC and the transitions between its three phases remain poorly understood. Here, we describe how oscillations in the abundance of aminolevulinic acid (ALA), a precursor of heme biosynthesis, led to the finding that yeast cells can respond to the biological gas carbon monoxide (CO). We hypothesize that transient production of CO may function as one such endogenous cue for metabolic cycling.
Results
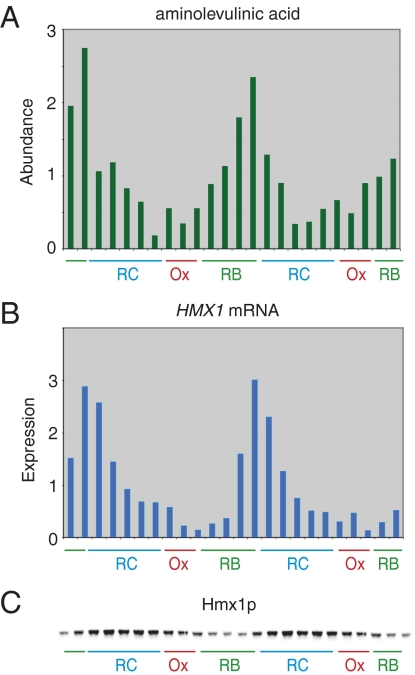
One of the most acutely regulated metabolites fluctuating in abundance over the YMC is ALA, the first and rate-limiting intermediate in the biosynthesis of heme (Fig. 1A). That heme synthesis might fluctuate during the YMC is predictable from DNA microarray studies showing that the genes encoding anabolic enzymes for heme biosynthesis are expressed in a cyclic manner preceding the observed accumulation of ALA (19). ALA levels increase as cells exit the OX phase and enter the RB phase, suggesting that heme synthesis may be required after a period of intense respiration.
Fig. 1.
Cycles of heme synthesis and breakdown during the YMC. (A) ALA levels fluctuate in a periodic fashion. The intracellular levels of ALA, a precursor of heme biosynthesis, were quantitated by LC-MS/MS over two consecutive metabolic cycles (12 time points per cycle) as described previously (19). Abundance refers to the relative quantity of a 114-Da daughter fragment specific to ALA (132 Da). The x axis tick marks denote ≈20- to 25-min time intervals. (B) Heme oxygenase transcript levels are highly periodic as a function of the YMC. Transcript levels of the gene encoding heme oxygenase (HMX1), which breaks down heme, were determined by microarray analysis over two consecutive cycles as described previously (18). (C) Heme oxygenase protein levels are highly periodic as a function of the YMC. Hmx1p protein levels were determined at 12 time intervals over one cycle by Western blot analysis using a strain expressing a Flag-tagged version of Hmx1p from its native chromosomal locus. The data are double-plotted to reveal cyclic changes in protein level.
Further inspection of the transcript profiles of heme pathway genes led to the prediction that heme breakdown might also be regulated temporally during the YMC. The gene encoding the rate-limiting enzyme in heme catabolism, heme oxygenase (HMX1 in yeast) (21, 22), is tightly regulated as a function of the YMC (Fig. 1B). Western blot studies confirm that levels of the heme oxygenase enzyme itself fluctuate acutely as well (Fig. 1C), peaking in the midst of the RC phase. It is predictable from these data that maximal rates of heme degradation should occur during the RC phase of the YMC.
Yeast cells synthesize heme during the RB and RC phases of the YMC, presumably such that mitochondrial hemoproteins can be loaded and optimally functional in anticipation of the ensuing OX phase. Having observed peak levels of heme oxygenase gene expression and enzyme abundance during the RC phase of the YMC, we hypothesized that yeast cells catabolize free heme that might transiently exceed cellular demand during this phase of the YMC. A second possibility is that heme oxygenase degrades aged or damaged heme as hemoproteins are turned over during this temporal window. The heme oxygenase enzyme converts heme into biliverdin, evolving the gas CO as a byproduct (23, 24). Heme oxygenase-mediated catabolism of heme represents the only known biological source of CO. Having observed tight regulation of ALA accumulation, heme oxygenase gene expression, and heme oxygenase enzyme abundance as a function of the YMC, it is predictable that CO is transiently produced specifically during the RC phase of the YMC.
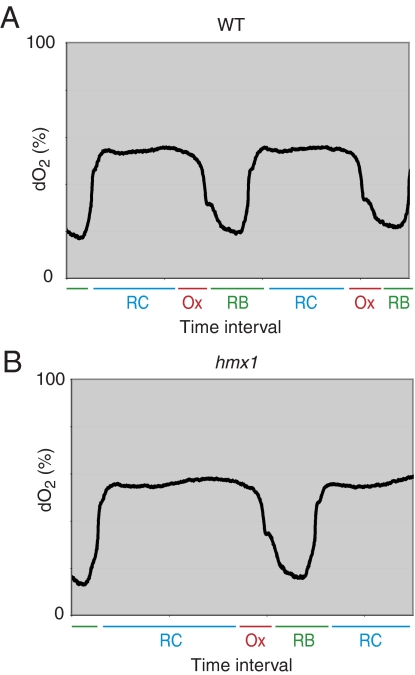
If the CO produced by yeast cells has any biological role, it is possible that cells lacking the heme oxygenase enzyme might display an altered phenotype during continuous growth in the chemostat. We used homologous recombination to delete the heme oxygenase gene and studied the growth of the resulting Δhmx1 mutant under glucose-rich, log growth conditions, as compared with sustained growth in a chemostat. Yeast cells lacking the heme oxygenase gene were indistinguishable from WT parental cells when grown in high glucose. Heme oxygenase-deficient cells were fully capable of chemostat growth, yet displayed a metabolic cycle protracted in length by up to ≈1 h or ≈25% of the period length of the cycle observed for WT cells (Fig. 2 A and B).
Fig. 2.
Cells lacking heme oxygenase exhibit longer metabolic cycles. (A) Shown are two consecutive metabolic cycles exhibited by the CEN.PK MATa strain during continuous growth. Under these conditions the cycles have a period length of ≈3.5 h. The y axis denotes dissolved oxygen (dO2) levels. The x axis tick marks denote 2-h time intervals. (B) Δhmx1 cells were subject to the same continuous growth conditions as in A. The cycles exhibited by the Δhmx1 strain are ≈4.5 h, ≈25% longer than WT.
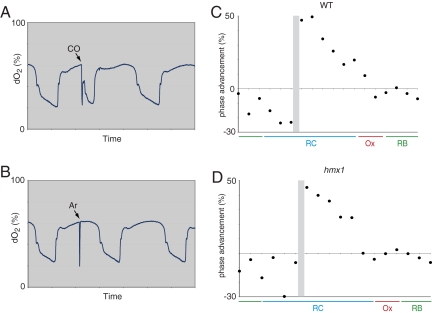
The patterns of steep decline in dissolved oxygen during the OX phase of the YMC, and subsequent increase in oxygen levels during the ensuing RB phase, were virtually indistinguishable between parental and heme oxygenase-deficient strains. Therefore, we hypothesize that heme oxygenase deficiency does not affect either of these phases, but instead, affects overall metabolic cycle length by extending the duration of the RC phase of the YMC. This interpretation raises the possibility that transient production of CO might assist movement of cells from the RC phase of the YMC to OX. In the absence of the heme oxygenase enzyme, cells should be incapable of producing CO. Because the absence of heme oxygenase activity lengthens the duration of the RC phase of the YMC, it is possible that prolongation of the RC phase can be attributed to the absence of CO. To test this hypothesis, we established a phase response curve (PRC). To this end, we administered brief pulses of CO to chemostat cultures at 18 equally spaced time intervals that span the three phases of the YMC. As deduced from macroscopic patterns of oxygen consumption, it was possible to observe CO-mediated phase-advancement of the YMC (Fig. 3A). When brief pulses of CO were applied to cells existing in either the OX or RB phases of the YMC, no significant effects on cycling were observed. By contrast, when applied to cells existing in the RC phase, CO triggered premature advancement into the OX phase and respiratory metabolism (Fig. 3C). Thereafter, robust metabolic cycles of normal temporal dimensions resumed. Dosing of the inert gas argon had no effect on cycling other than a transient dip in dO2 levels (Fig. 3B), suggesting that phase advancement is mediated specifically by CO and not by transient reduction in dissolved oxygen concentration.
Fig. 3.
CO generated from heme breakdown can function as a metabolic cue. (A) Transient administration of CO induces a phase advance into OX phase. Cycling WT cells were exposed to CO applied at ≈5 psi for 2 min in the midst of RC phase. Note that CO induced phase advancement into OX phase. The x axis tick marks denote 2-h time intervals. (B) Transient administration of argon has no apparent effect on metabolic cycling. Cycling WT cells were exposed to argon gas applied at ≈5 psi for 2 min in the midst of RC phase. (C) CO can induce phase advancement only during the RC phase. A PRC was constructed by dosing CO at each of 18 equally spaced time intervals across one metabolic cycle of a WT haploid strain. The y axis denotes quantitation of phase advancement as a percentage of the immediately previous cycle length. The competency gate that defines the point at which phase advancement becomes possible is denoted by the shaded area. The x axis tick marks denote ≈15-min time intervals. (D) CO can induce phase advancement of cells lacking heme oxygenase. CO was administered at each of 18 equally spaced time intervals to cycling Δhmx1 cells. The competency gate for phase advancement is denoted by the shaded area.
A specific “competency gate” during early RC phase between time intervals 6 and 7 represents the time at which CO-mediated phase advancement becomes possible. Interestingly, this competency gate coincides with the point at which the population of cells that committed to cell division has just finished the division process (18). The cell population remains susceptible to phase advancement until they enter the time intervals 13–14, which represents initiation of the OX phase (Fig. 3C). We further asked whether yeast cells lacking heme oxygenase and the ability to synthesize endogenous CO could still respond to the gas. Brief pulses of CO were administered to cycling Δhmx1 cell populations over 18 evenly spaced time intervals. The Δhmx1 mutant exhibited a similar PRC to CO as the WT parental strain (Fig. 3D), indicating that heme-oxygenase deficient cells can still respond to CO.
Discussion
Having observed a protracted RC phase in a strain lacking heme oxygenase (Fig. 2B), as well as RC-restricted phase advancement of the YMC in response to pulsed administration of CO, we hypothesize that transient production of CO might have a role in regulating the metabolic state of eukaryotic cells. Heme-containing proteins represent a likely target of CO signaling. It is known that CO can readily bind to hemoproteins, including hemoglobin and mitochondrial enzymes critical to the respiratory chain (25). In these cases, CO functions as an impediment to respiration. As such, it is paradoxical that transient pulses of CO would trigger advancement of the YMC from the RC to OX phase. It is possible that yeast cells are able to sense CO-mediated inhibition of mitochondrial respiration, and respond by activation of OX metabolism. No such phase advancement was observed in response to attenuation of oxygen levels as a function of argon administration (Fig. 3B). As such, we speculate that yeast cells may be endowed with a CO receptor that functions to trigger entry into the OX phase of the YMC by sensing the catabolism of free heme as a marker of the timing of when cytochromes have been fully heme-loaded such that cells are optimally prepared for respiratory metabolism. Clearly, there are other signals besides CO that can mediate entry into OX phase and respiratory metabolism, because although temporarily delayed, heme oxygenase-deficient cells can still enter OX phase.
The Km of heme oxygenase for heme has been estimated to be ≈1 μM, and free heme is kept below micromolar concentrations in normal cells (26). Although it is difficult to determine the effective concentration of CO in our experiments, we estimate that the concentration of CO required to induce phase advancement of the YMC is within an order of magnitude of predicted physiological concentrations of CO.
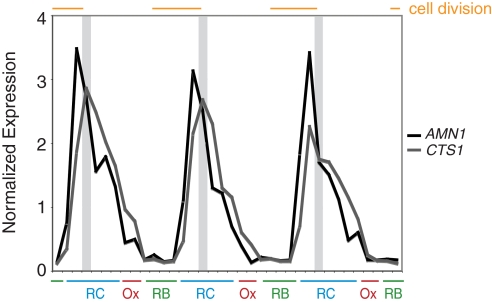
Based on the timing of when cells respond to CO, exit of the cell population from the cell division process can be hypothesized to be a prerequisite for such phase advancement (Fig. 4). The peaks of expression of AMN1 and CTS1, two periodic genes required for separation of daughter cells from mothers, represent temporal markers that denote the time at which cells finish the division process (27, 28). The phase advancement competency gate occurs immediately after the peaks of expression of these two genes (Fig. 4). Therefore, the competency gate defines a new feature of such metabolic cycles, and suggests that cells transition to a different metabolic state during this time; thereby, rendering them susceptible to phase advancement into OX phase. We have observed that other metabolites such as methionine can only trigger phase advancement after the competency gate as well. Together, these observations reveal a specific temporal window during RC phase where the cell population changes in a manner enabling response to metabolic cues capable of triggering transition from the RC to OX phase of the YMC.
Fig. 4.
The competency gate for phase advancement from RC to OX phase coincides with exit from cell division. The previously determined expression profiles of two genes (AMN1 and CTS1) that are induced at the very end of mitosis are shown (18). The competency gates are denoted by the shaded areas, and the time of cell division is denoted by the orange bars. The x axis tick marks denote ≈25-min time intervals.
Cycles of heme synthesis, and presumably heme breakdown, also occur as a function of the circadian cycle (29). It may be notable that CO has been shown to be capable of binding to the heme prosthetic group associated with NPAS2, a mammalian transcription factor dedicated to the control of circadian rhythm, and that heme binding by the gas ligand inhibits DNA binding activity of the purified transcription factor (30). Also, heme has also been shown to be bound by the nuclear orphan receptor Rev-Erbα (31, 32), another protein involved in the control of circadian rhythm. Together, these observations indicate that cycles of heme synthesis and regulated production of CO might represent evolutionarily conserved mechanisms involved in the control of metabolic state and biological rhythms.
Methods
Yeast Protocols and Chemostat Growth.
All yeast manipulations and chemostat growth experiments were carried out as described previously (18). Briefly, CEN.PK MATa haploid strains were grown in batch mode to saturation in media containing 5 g/L (NH4)2SO4, 2 g/L KH2PO4, 0.5 g/L MgSO4·7H2O, 0.1 g/L CaCl2·2H2O, 0.02 g/L FeSO4·7H2O, 0.01 g/L ZnSO4·7H2O, 0.005 g/L CuSO4·5H2O, 0.001 g/L MnCl2·4H2O, 1 g/L yeast extract, 10 g/L glucose, 0.5 mL/L 70% H2SO4, and 0.5 mL/L Antifoam 204 (Sigma). After starvation for at least 6 h, the cell population was continuously fed the same media at a dilution rate of ≈0.09 h−1. The CEN.PK Δhmx1 haploid strain was constructed by homologous recombination of a KanR cassette into the coding region of the HMX1 locus (33). For Western blot analysis of Hmx1p, a strain expressing HMX1 tagged at its C terminus with a triple-Flag epitope was constructed using homologous recombination (34). Both the Δhmx1 and HMX1-FL3 strains showed no growth defect in glucose-rich medium. Western blot analysis revealed that the Hmx1p-FL3 epitope-tagged protein was the correct size (≈37 kDa). Three replicates were performed for the Western blot analysis and representative data from one experiment are shown.
CO Administration.
For transient administration of CO and Ar, the gases (>99.9% purity) were flowed into the bottom of the chemostat vessel at a pressure of 5 psi for 2 min while normal aeration and agitation of the chemostat were ongoing (1 L/min, 450 rpm). Such pulses of gas were sufficient to temporarily displace ≈30% (≈72 μM) of air-saturated dissolved oxygen in the growth media. The solubility of CO in water is ≈0.026 g/L (≈930 μM) at 20°. Because our doses of CO were done while the chemostat was actively being aerated at 1 L/min with house air, we estimate the maximal dissolved concentration of CO in the media to be <100 μM. Shorter pulses of CO (<1 min) did not induce phase advancement, and longer pulses of Ar (>3 min), which reduced dO2 levels to only ≈10% of air-saturated levels, had no observable effect on the metabolic cycles. Cells were allowed to proceed through at least one complete metabolic cycle between administrations of gas. Phase advancement is calculated as a percentage of the previous cycle length. There is an estimated ≈5% error in calculation of cycle times due to variables such as feed/harvest pump operation and replenishment of new media to continue the cycles. At least three replicates of the phase advancement experiments were performed, and representative data from one experiment are shown.
Acknowledgments.
We thank Jessica Liu and Benjamin Sutter for technial assistance and Drs. Ueli Schibler, Jay Dunlap, and David Botstein for critical review of the manuscript. This work was supported by a National Institutes of Health Director's Pioneer Award (to S.L.M.), a Helen Hay Whitney Foundation Fellowship (to B.P.T.), the Frank and Sara McKnight Fund for Biochemical Research (to B.P.T.), a Burroughs Wellcome Fund Career Award in Biomedical Sciences (to B.P.T.), a Welch Foundation Research Grant I-1697 (to B.P.T.), and unrestricted funds from an anonymous donor (to S.L.M.).
Footnotes
The authors declare no conflict of interest.
References
- 1.Chance B, Estabrook RW, Ghosh A. Damped Sinusoidal Oscillations of Cytoplasmic Reduced Pyridine Nucleotide in Yeast Cells. Proc Natl Acad Sci USA. 1964;51:1244–1251. doi: 10.1073/pnas.51.6.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghosh A, Chance B. Oscillations of glycolytic intermediates in yeast cells. Biochem Biophys Res Commun. 1964;16:174–181. doi: 10.1016/0006-291x(64)90357-2. [DOI] [PubMed] [Google Scholar]
- 3.Hommes FA. Oscillatory Reductions of Pyridine Nucleotides during Anaerobic Glycolysis in Brewers' Yeast. Arch Biochem Biophys. 1964;108:36–46. doi: 10.1016/0003-9861(64)90352-2. [DOI] [PubMed] [Google Scholar]
- 4.von Meyenburg HK. Energetics of the budding cycle of Saccharomyces cerevisiae during glucose limited aerobic growth. Arch Microbiol. 1969;66:289–303. doi: 10.1007/BF00414585. [DOI] [PubMed] [Google Scholar]
- 5.Parulekar SJ, Semones GB, Rolf MJ, Lievense JC, Lim HC. Induction and elimination of oscillations in continuous cultures of Saccharomyces cerevisiae. Biotechnol Bioeng. 1986;28:700–710. doi: 10.1002/bit.260280509. [DOI] [PubMed] [Google Scholar]
- 6.Porro D, Martegani E, Ranzi BM, Alberghina L. Oscillations in continuous cultures of budding yeast: A segregated parameter analysis. Biotechnol Bioeng. 1988;32:411–417. doi: 10.1002/bit.260320402. [DOI] [PubMed] [Google Scholar]
- 7.Satroutdinov AD, Kuriyama H, Kobayashi H. Oscillatory metabolism of Saccharomyces cerevisiae in continuous culture. FEMS Microbiol Lett. 1992;77:261–267. doi: 10.1016/0378-1097(92)90167-m. [DOI] [PubMed] [Google Scholar]
- 8.Klevecz RR, Bolen J, Forrest G, Murray DB. A genomewide oscillation in transcription gates DNA replication and cell cycle. Proc Natl Acad Sci USA. 2004;101:1200–1205. doi: 10.1073/pnas.0306490101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmer SL, et al. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 10.Claridge-Chang A, et al. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 11.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 12.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 13.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 15.Mitsui A, et al. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature. 1986;323:720–722. [Google Scholar]
- 16.Rutter J, Reick M, McKnight SL. Metabolism and the control of circadian rhythms. Annu Rev Biochem. 2002;71:307–331. doi: 10.1146/annurev.biochem.71.090501.142857. [DOI] [PubMed] [Google Scholar]
- 17.Tu BP, McKnight SL. Metabolic cycles as an underlying basis of biological oscillations. Nat Rev Mol Cell Biol. 2006;7:696–701. doi: 10.1038/nrm1980. [DOI] [PubMed] [Google Scholar]
- 18.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: Temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 19.Tu BP, et al. Cyclic changes in metabolic state during the life of a yeast cell. Proc Natl Acad Sci USA. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tu BP, McKnight SL. The yeast metabolic cycle: Insights into the life of a eukaryotic cell. Cold Spring Harb Symp Quant Biol. 2007;72:339–343. doi: 10.1101/sqb.2007.72.019. [DOI] [PubMed] [Google Scholar]
- 21.Shibahara S, Muller R, Taguchi H, Yoshida T. Cloning and expression of cDNA for rat heme oxygenase. Proc Natl Acad Sci USA. 1985;82:7865–7869. doi: 10.1073/pnas.82.23.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Protchenko O, Philpott CC. Regulation of intracellular heme levels by HMX1, a homologue of heme oxygenase, in Saccharomyces cerevisiae. J Biol Chem. 2003;278:36582–36587. doi: 10.1074/jbc.M306584200. [DOI] [PubMed] [Google Scholar]
- 23.Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. Carbon monoxide: A putative neural messenger. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 24.Maines MD. Heme oxygenase: Function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J. 1988;2:2557–2568. [PubMed] [Google Scholar]
- 25.Desmard M, Boczkowski J, Poderoso J, Motterlini R. Mitochondrial and cellular heme-dependent proteins as targets for the bioactive function of the heme oxygenase/carbon monoxide system. Antioxid Redox Signal. 2007;9:2139–2155. doi: 10.1089/ars.2007.1803. [DOI] [PubMed] [Google Scholar]
- 26.Sassa S. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide? Antioxid Redox Signal. 2004;6:819–824. doi: 10.1089/ars.2004.6.819. [DOI] [PubMed] [Google Scholar]
- 27.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowicka M, Kudlicki A, Tu BP, Otwinowski Z. High-resolution timing of cell cycle-regulated gene expression. Proc Natl Acad Sci USA. 2007;104:16892–16897. doi: 10.1073/pnas.0706022104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 30.Dioum EM, et al. NPAS2: A gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 31.Yin L, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 32.Reinking J, et al. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 33.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 34.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: Analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]