Abstract
The role of Notch signaling in cartilage differentiation and maturation in vivo was examined. Conditional Notch pathway gain and loss of function was achieved using a Cre/loxP approach to manipulate Notch signaling in cartilage precursors and chondrocytes of the developing mouse embryo. Conditional overexpression of activated Notch intracellular domain (NICD) in the chondrocyte lineage results in skeletal malformations with decreased cartilage precursor proliferation and inhibited hypertrophic chondrocyte differentiation. Likewise, expression of NICD in cartilage precursors inhibits sclerotome differentiation, resulting in severe axial skeleton abnormalities. Furthermore, conditional loss of Notch signaling via RBP-J gene deletion in the chondrocyte lineage results in increased chondrocyte proliferation and skeletal malformations consistent with the observed increase in hypertrophic chondrocytes. In addition, the Notch pathway inhibits expression of Sox9 and its target genes required for normal chondrogenic cell proliferation and differentiation. Together, our results demonstrate that appropriate Notch pathway signaling is essential for proper chondrocyte progenitor proliferation and for the normal progression of hypertrophic chondrocyte differentiation into bone in the developing appendicular and axial skeletal elements.
Keywords: cartilage development, endochondral ossification, hypertrophic chondrocyte, Sox9
Chondrogenesis is an essential process in which sequential aggregation, proliferation, differentiation, and hypertrophy of chondrocytes provide the initial patterning of the vertebrate skeleton. The appendicular skeleton comprises the limbs and their attachments to the axial skeleton, which comprises the skull, rib cage, and vertebral column (1). The vertebral column and rib cage develop from the mesenchymal sclerotome of the somites, whereas the appendicular skeleton develops from osteochondroprogenitor cells in the limb buds. During embryonic development, mesenchymal progenitor cells condense, differentiate into chondrocytes, and form a cartilaginous preskeleton, which subsequently is replaced by ossified bone, a process termed endochondral ossification (2). Endochondral ossification, which forms most bones in the body, including the vertebral column of the axial skeleton and the limbs of the appendicular skeleton, is a complex regulatory process during which chondrocyte proliferation, differentiation, and maturation are tightly controlled. The role of the Notch signaling pathway in chondrogenic proliferation, differentiation, and maturation is largely unknown.
The Notch signaling pathway, implicated in skeletal development and disease, consists of 5 ligands (Dll1, 3, and 4, and Jagged1 and 2) that interact with 4 Notch receptors (Notch1–4) (3). Upon ligand binding, the Notch receptor is cleaved, and the Notch intracellular domain (NICD) translocates to the nucleus, where it binds to the transcription factor recombination signal binding protein for immunoglobulin kappa J (RBP-J, also termed RBPJk, CBF1, RBPSUH, or SUH) and ultimately activates expression of downstream genes to affect many cellular processes, including cell proliferation and differentiation (3–6). Several Notch signaling pathway components regulate somite segmentation, which underlies axial skeleton patterning (7). In addition, in vivo Notch pathway overexpression or underexpression in the osteogenic lineage demonstrates that Notch inhibits osteogenic differentiation and growth in the axial and appendicular skeleton (8–10). Although the role of Notch signaling in bone development has been investigated, the role of Notch signaling in prenatal cartilage development has not yet been demonstrated. Cell culture experiments suggest that Notch1 might play an inhibitory role during chondrocyte differentiation; however, in vivo effects of Notch manipulation in the chondrocyte lineage have not been reported (11–13).
To examine the role of Notch in chondrocyte development and differentiation, the Notch signaling pathway was manipulated throughout chondrocyte lineage development using pro-alpha1(II) collagen-Cre (Col2α1Cre) mice (14). Notch gain of function was achieved with RosaNotch mice expressing NICD, the activated form of the Notch1 receptor, in the chondrocyte lineage in the presence of Col2α1Cre (15). Furthermore, Notch signaling was ablated in the chondrocyte lineage using RBP-Jf/f mice (16). Together, these studies demonstrate that Sox9 and its target genes are subject to regulation by Notch signaling that ultimately affects chondrogenic proliferation and differentiation. The cartilage defects resulting from both Notch pathway activation and inactivation in the chondrogenic lineage demonstrate that appropriate Notch signaling is required for proper cartilage progenitor proliferation and hypertrophic chondrocyte differentiation in appendicular and axial skeleton development.
Results
Conditional NICD Expression in the Chondrocyte Lineage Results in Skeletal Malformations and Perinatal Lethality.
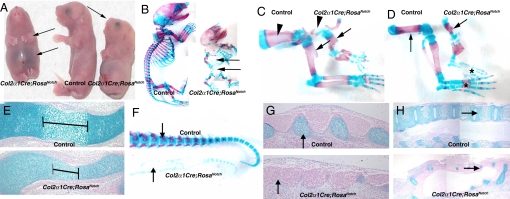
To examine the role of Notch signaling in chondrogenesis, RosaNotch mice with Cre-dependent NICD activation were crossed with Col2α1Cre mice, which express Cre in condensed mesenchymal cartilage progenitor cells and differentiated chondrocytes (14, 15). The resulting Col2α1Cre;RosaNotch mice are perinatal lethal (likely due to respiratory deficiency) and present with a rounded head with a short snout, distended abdomen, shortened trunk and tail, and truncated limbs, among other abnormalities (Fig. 1A; Figs. S1 and S2). Alcian blue (cartilage) and alizarin red (bone) staining of embryonic day (E) 18.5 skeletal preparations showed that skeletal elements formed by endochondral ossification, including vertebrae, ribs, and limbs, are undersized and underdeveloped in Col2α1Cre;RosaNotch embryos compared with controls. Skeletal elements of these mice are characterized by a severe and generalized chondrodysplasia, as demonstrated by significantly decreased alcian blue staining in cartilaginous structures throughout the embryo. The rib cage of Col2α1Cre;RosaNotch mice is smaller than that of controls (an average of 10 ribs versus 13), and the vertebral column and tail are virtually absent (Fig. 1B). In addition, these mice also have a shortened or absent neck, umbilical hernias, and decreased or absent cartilage in the trachea and skull compared with controls (Figs. 1 A and B, Figs. S1 and S2, and data not shown). Overall, the Col2α1Cre;RosaNotch phenotype is consistent with a global defect in cartilage formation and differentiation, resulting in gross malformations of the appendicular and axial skeleton.
Fig. 1.
Col2α1Cre;RosaNotch mice exhibit appendicular and axial skeletal malformations associated with chondrodysplasia. (A) Decreased limb length, distended abdomen, and malformed head (arrows) of Col2α1Cre;RosaNotch pups at birth. (B) Whole-mount E18.5 skeletal preparations stained with alcian blue (cartilage) and alizarin red (bone) exhibit a smaller, abnormal rib cage and lack of vertebral column in Col2α1Cre;RosaNotch mice (arrows). (C and D) Shortened forelimbs and hind limb skeletal elements of E18.5 Col2α1Cre;RosaNotch mice (arrows), and absent scapula (arrowheads in C) and digit ossification centers (asterisks in D). (E) E14.5 alcian blue–stained Col2α1Cre;RosaNotch humerus exhibit decreased bone density, as well as a truncated ossification center (brackets). (F) E18.5 vertebral column of Col2α1Cre;RosaNotch mice exhibit decreased cartilage, as well as an absence of bone (arrows). (G and H) Alcian blue staining at E14.5 (G) and E18.5 (H) shows an absence of cartilage precursors (arrows in G) in Col2α1Cre;RosaNotch vertebral column, resulting in loss of skeletal elements in the axial skeleton (arrows in H).
The appendicular skeleton of Col2α1Cre;RosaNotch mice consists of malformed long bones with reduced limb size and length. The forelimbs and hind limbs of these mice exhibit a shortened humerus and femur, respectively (Fig. 1C–D, arrows). In addition, the scapulae are virtually nonexistent, and the ossification centers of the digits are absent in the limbs (Fig. 1C–D, arrowhead and asterisk). Alcian blue staining of the limbs at E14.5 shows little to no chondrocyte hypertrophy, and the size of the Col2α1Cre;RosaNotch humerus bone is reduced compared with controls, consistent with inhibited chondrogenic differentiation (Fig. 1E).
The axial skeleton of Col2α1Cre;RosaNotch mice is malformed, with a nearly absent vertebral column (Fig. 1 B and F). At E14.5, alcian blue staining demonstrates severely hypomorphic or absent segmental cartilage precursor condensations (which ultimately form the vertebral column), whereas MF20 antibody labeling reveals normal muscle precursor segments (Fig. 1G, arrows and data not shown). At E18.5, control mice have a stereotypic segmented vertebral column, whereas the Col2α1Cre;RosaNotch mice display a disorganized and severely reduced cartilage anlagen (Fig. 1H, arrows). This severe embryonic chondrodysplasia demonstrates that NICD activation inhibits cartilage precursor development and endochondral ossification in the appendicular and axial skeleton.
Endogenous NICD Expression in Cartilage Progenitor Cells Is Increased and Expressed Earlier in Col2α1Cre;RosaNotch Embryos.
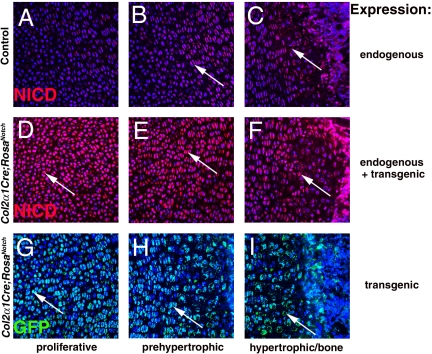
While the localization of Notch pathway activation and NICD expression has been described in osteogenesis, much less is known about Notch signaling during chondrogenesis in vivo (8–10). Localization of endogenous NICD expression indicative of Notch pathway activity, distinct from transgenic GFP-tagged NICD expression, was analyzed in E18.5 limb growth plates. In normal skeletal development, endogenous NICD is first expressed in prehypertrophic chondrocytes and continues to be expressed in hypertrophic chondrocytes and osteoblasts, supporting a role for the Notch pathway in chondrocyte differentiation and endochondral ossification (Fig. 2A–C). Importantly, endogenous NICD is at low to undetectable levels in the earlier, proliferative zone of chondrocyte progenitors (Fig. 2A). In Col2α1Cre;RosaNotch mice, GFP-tagged NICD expression is detected earlier in proliferating chondrocytes and also is expressed in all chondrocytes as well as in bone, with increased NICD immunoreactivity compared with controls (Fig. 2D–I). Furthermore, several Notch pathway components, including Dll4 (proliferative chondrocytes) and Jag1, Notch1, Hey1, Hes1, and Hes7 (prehypertrophic chondrocytes), exhibit chondrogenic expression at E14.5 and E18.5 (Fig. S3 and data not shown). Taken together, these data demonstrate that endogenous NICD expression is activated in the prehypertrophic cartilage progenitors as they exit the proliferative zone, indicative of Notch pathway activation and function in normal chondrocyte differentiation. Furthermore, Col2α1Cre;RosaNotch mice express ectopic activated NICD earlier and at increased levels relative to NICD expression in cartilage precursors and differentiating cartilage of embryonic skeletal elements.
Fig. 2.
NICD activation in the chondrocyte lineage and premature activation in Col2α1Cre;RosaNotch mice. (A–C) Endogenous activated Notch signaling, NICD (red), is expressed in prehypertrophic and hypertrophic chondrocytes and osteoblasts in E18.5 humerus bone (arrows in B and C). (D–I) Transgenic and endogenous NICD expression (red) and transgenic GFP staining (green) of Col2α1Cre;RosaNotch mice is detected much earlier in proliferative chondrocytes and throughout the limb (arrows). Blue represents DAPI nuclei staining.
Increased Notch Pathway Activation Leads to Impaired Chondrocyte Differentiation and Decreased Ossification.
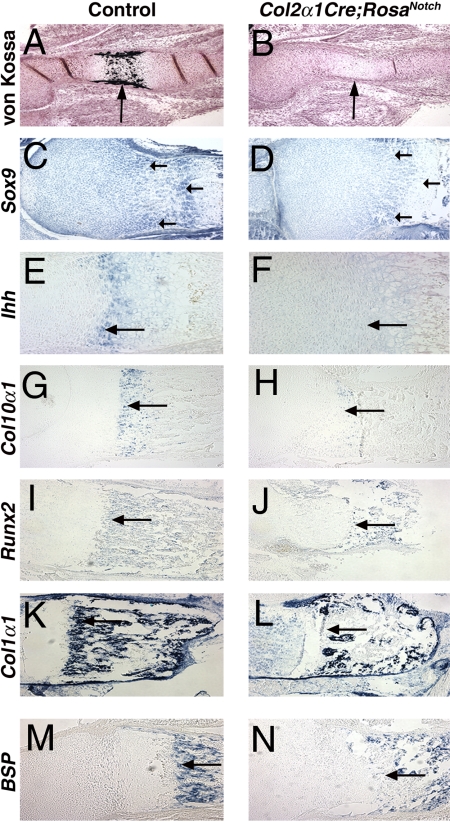
To investigate the role of Notch activation in chondrogenesis in more detail, we examined markers of specific stages of chondrocyte progenitor specification, differentiation, and endochondral bone formation. At E14.5, ossification is reduced in Col2α1Cre;RosaNotch mice, as demonstrated by decreased von Kossa staining (Fig. 3 A and B). However, by E18.5, positive von Kossa staining indicates that mineralization eventually occurs in Col2α1Cre;RosaNotch mice, although the ossified area is decreased (data not shown). To examine the molecular regulation of endochondral bone formation in Col2α1Cre;RosaNotch mice, we analyzed the expression of several genes that mark specific stages of chondrogenic differentiation. In situ hybridization analysis of E18.5 limb sections demonstrates deceased Sox9 expression in the early-stage cartilage progenitors of the proliferative zone in Col2α1Cre;RosaNotch embryos compared with controls (Fig. 3 C and D). Sox9 promotes cartilage progenitor proliferation and differentiation, and decreased expression of its target genes, Aggrecan and Cartilage link protein (Crtl1), also was observed in Col2α1Cre;RosaNotch mice (Fig. S4 and data not shown) (17, 18). In addition, hypertrophic chondrocyte differentiation, as measured by Indian hedgehog (Ihh), Col10α1, and matrix metalloproteinase 13 (Mmp13) expression, is nearly absent in E14.5 and E18.5 limb sections (Fig. 3E–H and data not shown). Although hypertrophic chondrocytes eventually form in Col2α1Cre;RosaNotch mice, the replacement of cartilage by bone also is markedly reduced, resulting in smaller appendicular skeletal elements. Runx2 is expressed by late hypertrophic chondrocytes as well as osteoblasts and promotes chondrocyte differentiation and bone development (19). Along with Col1α1 and bone sialoprotein (BSP), which mark osteoblasts exclusively, Runx2 expression is reduced in Col2α1Cre;RosaNotch mice (Fig. 3I–N). Osteoclast gene expression, as measured by matrix metalloproteinase 9 (Mmp9), is largely unchanged (data not shown). Together, these expression analyses indicate that the differentiation of chondrocyte progenitors and hypertrophic chondrocytes is inhibited with increased Notch signaling in Col2α1Cre;RosaNotch mice.
Fig. 3.
Chondrocyte differentiation and bone formation are delayed with decreased hypertrophic chondrocytes in Col2α1Cre;RosaNotch mice. (A and B) Von Kossa staining shows delayed ossification in the E14.5 limb of Col2α1Cre;RosaNotch mice (arrows). (C–N) E18.5 in situ hybridization of Col2α1Cre;RosaNotch limbs demonstrates reduced expression (arrows) of Sox9 (proliferative chondrocytes), Ihh and Col10α1 (hypertrophic chondrocytes), and Col1α1, Runx2, and BSP (osteoblasts) compared with controls.
NICD Expression in the Early Chondrocyte Cell Population Inhibits Axial Cartilage Development Through Impaired Sclerotome Differentiation in Col2α1Cre;RosaNotch Mice.
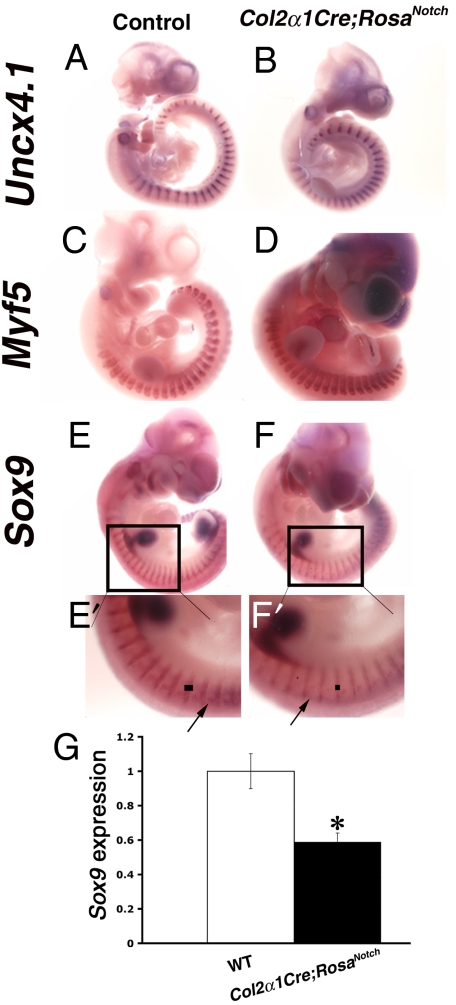
Notch pathway activation in the chondrocyte lineage results in nearly complete lack of cartilage elements and bone formation in the axial skeleton (Fig. 1B, F, G, and H). Because the vertebral column arises from sclerotome progenitors of the somites, we investigated this precursor cell population in further detail. Notch signaling is involved in the initial segmentation and compartmentalization of somites (20). We anticipated that this initial segmentation would be unaffected in Col2α1Cre;RosaNotch embryos, because the Col2α1Cre transgene is not expressed until the cartilage population has been specified (14). As expected, E10.5 whole-mount in situ hybridization demonstrates normal somite segmentation in Col2α1Cre;RosaNotch mice, as shown by segmentation and compartmentalization markers Uncx4.1 and Tbx18, as well as Notch pathway components Mesp2, Hes7, and Lunatic fringe (Lfng) (Figs. 4 A and B, Fig. S5 and data not shown). Thus, the somite segmentation clock and patterning pathways apparently are not affected in Col2α1Cre;RosaNotch mice.
Fig. 4.
Col2α1Cre;RosaNotch mice exhibit somite segmentation but have reduced Sox9 expression and sclerotome differentiation. (A and B) Whole-mount in situ hybridization of E10.5 embryos shows normal somite segmentation of Col2α1Cre;RosaNotch embryos, as indicated by Uncx4.1 expression. (C and D) Appropriate myotome differentiation as shown by Myf5 expression. (E and F) Impaired sclerotome differentiation as indicated by reduced Sox9 expression in Col2α1Cre;RosaNotch somites compared with controls (arrows and lines in E ' and F'). (G) Significant reduction of somite Sox9 expression as determined by real-time RT-PCR analysis. * P ≤ 0.01
Somite sclerotome differentiation is impaired with increased Notch signaling in Col2α1Cre;RosaNotch embryos. This finding is consistent with Col2α1Cre activation at E9.5 in the differentiating somites (14). The sclerotome, which gives rise to cartilage and bone of the axial skeleton as marked by Sox9 and Scleraxis expression, is reduced in width and intensity, with a significant decrease of Sox9 mRNA expression (Fig. 4 E–G and data not shown). This correlates with the observed decrease in cartilage precursors of the developing vertebral column later during development (Fig. 1G). In contrast, the myotome, as marked by Myf5 expression, segments normally in Col2α1Cre;RosaNotch mice, demonstrating that muscle development is unperturbed with this genetic approach (Fig. 4 C and D). Taken together, whole-mount in situ hybridization studies demonstrate that NICD expression in cartilage precursors results in decreased sclerotome differentiation, leading to a severe reduction in cartilage progenitors of the vertebral column and near absence of the axial vertebrae.
Loss of Notch Function in the Chondrogenic Lineage Increases the Zone of Hypertrophic Chondrocytes.
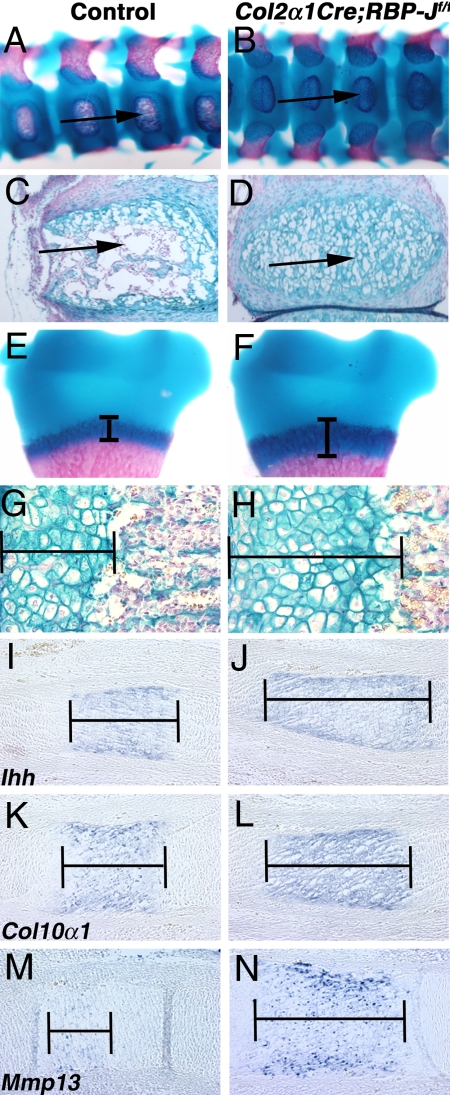
We have shown that Notch pathway activation in the chondrogenic lineage inhibits cartilage differentiation, resulting in an overall decrease in skeletal elements and consequent malformations. To determine the effects of decreased Notch pathway activity, the transcription factor RBP-J was targeted in the Col2α1Cre domain to generate mice with loss of Notch signaling in cartilage progenitors and differentiating chondrocytes. Col2α1Cre;RBP-Jf/f mice are perinatal lethal, with increased cartilage and decreased bone formation in the ossification centers of the vertebral column, as demonstrated by alcian blue– and alizarin red–stained skeletal preparations and histological sections (Fig. 5A–D). In addition, while Col2α1Cre;RBP-Jf/f mice have a similar long bone length as controls, they exhibit an overall increase in growth plate chondrocytes, accompanied by decreased ossified bone (Fig. 5 and Fig. S6). This increased cartilage with decreased bone coincides with an increased size of the hypertrophic chondrocyte zone, as shown in E18.5 forelimb growth plate sections (Fig. 5E–H and Fig. S6). Furthermore, expression of hypertrophic chondrocyte markers Ihh, Col10α1, and Mmp13 is increased and the zone of expression is expanded in Col2α1Cre;RBP-Jf/f E14.5 and E18.5 limb sections compared with controls (Fig. 5I–N and data not shown). In contrast to Col2α1Cre;RosaNotch mice, in Col2α1Cre;RBP-Jf/f mice, Sox9 expression was increased in limb sections at E14.5 and E18.5, further supporting a role for Notch regulation of Sox9 in chondrogenesis (Fig. S7 and data not shown). Taken together, these results suggest that loss of Notch signaling in the chondrocyte lineage causes increased, prolonged chondrogenesis and decreased osteogenesis. Coupled with the observed skeletal abnormalities with NICD activation in the chondrocyte lineage, these results indicate that early Notch signaling inhibits chondrogenesis by negatively regulating cartilage progenitor determination and expansion, as well as hypertrophic chondrocyte differentiation. As development proceeds, loss of Notch signaling leads to increased hypertrophic cartilage and decreased bone formation. Therefore, endogenous Notch signaling is required for proper timing and localization of chondrogenesis and endochondral ossification.
Fig. 5.
Loss of Notch pathway function in the chondrocyte lineage results in skeletal defects associated with increased cartilage formation. (A–D) Col2α1Cre;RBP-Jf/f mice presenting with decreased ossification centers (arrows) in the E18.5 vertebral column as shown by skeletal preparations (A and B) and alcian blue staining of sections (C and D). (E–H) Expansion of the Col2α1Cre;RBP-Jf/f hypertrophic chondrocyte zone in the E18.5 humerus growth plate (brackets). (I–N) Increased Ihh, Col10α1, and Mmp13 expression in E14.5 Col2α1Cre;RBP-Jf/f limb sections compared with controls (brackets).
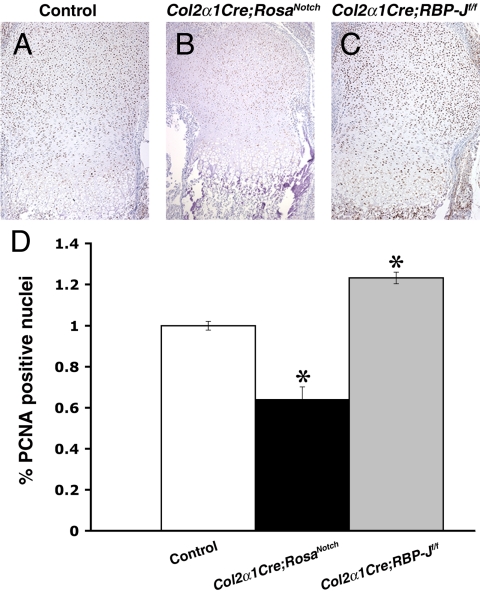
The reduced size of cartilage and bone elements in Col2α1Cre;RosaNotch mice suggests that increased Notch signaling inhibits chondrocyte proliferation and/or survival. Compared with controls, Col2α1Cre;RosaNotch limb sections exhibit significantly reduced proliferation throughout the growth plate, specifically in the prehypertrophic chondrocyte zone, as shown by proliferating cell nuclear antigen (PCNA) analysis (Fig. 6A, B, and D). Furthermore, Col2α1Cre;RBP-Jf/f limb sections exhibit significantly increased proliferation throughout the growth plate compared with controls (Fig. 6A, C, and D). However, there was no apparent difference in apoptosis in endochondral bones in controls versus Col2α1Cre;RosaNotch or Col2α1Cre;RBP-Jf/f mice, as indicated by caspase 3 and TUNEL assays (data not shown). These findings suggest that abnormal proliferation, but not a difference in apoptosis, contributes to abnormal chondrogenesis with increased or decreased Notch signaling levels. Taken together, these results demonstrate that the Notch signaling pathway is involved in the regulation of chondrocyte proliferation and differentiation in appendicular and axial skeleton development.
Fig. 6.
Notch signaling regulates proliferation in chondrogenesis. PCNA staining of E18.5 limb sections reveals significantly decreased proliferation in Col2α1Cre;RosaNotch mice and significantly increased proliferation in Col2α1Cre;RBP-Jf/f mice compared with controls. *P ≤ .001.
Discussion
In this study, we have demonstrated an essential role for Notch signaling in chondrogenic proliferation, differentiation, and skeletal development. Our data suggest that regulation of Notch signaling is required for a proper balance of chondrogenic proliferation and differentiation at initial stages of somite compartmentalization and long bone development. During normal chondrogenic differentiation in endochondral bone development, NICD is not expressed in the proliferative zone, but it is activated in prehypertrophic and hypertrophic cartilage. Increased NICD inhibits proliferation and prehypertrophic and hypertrophic chondrocyte differentiation, which ultimately results in decreased bone formation. Loss of Notch signaling in the chondrocyte lineage causes increased proliferation and an expanded hypertrophic chondrocyte zone, with an accompanying decrease in bone formation. Therefore, the level of Notch pathway expression must be tightly regulated to allow for proper chondrogenic cell proliferation, differentiation, and ossification during endochondral bone formation.
The severe chondrodysplasia and dwarfism seen in the Col2α1Cre;RosaNotch mutant mice are characterized by an almost complete absence of cartilage and bone in the axial skeleton and reduced cartilage and bone throughout the embryo. Among the genes affected by altered Notch signaling in axial and appendicular skeleton development is Sox9, which promotes cartilage progenitor proliferation and differentiation and marks the progenitor population of chondrogenic cells (17, 18). Decreased Sox9 expression could result in loss of the vertebral column as well as reduced hypertrophic cartilage in Col2α1Cre;RosaNotch mice. The Col2α1Cre;RosaNotch skeletal phenotype is similar to that of haploinsufficient Sox9 mice, as well as mice lacking Sox9 in the chondrogenic lineage, which also have a lack of proper chondrocyte differentiation, resulting in skeletal defects (18, 21). In cultured chondrocytes, overexpression of Notch1 inhibits chondrocyte differentiation and proliferation, as evidenced by decreased Sox9, Col2α1, Aggrecan, and Scleraxis gene expression, whereas loss of Notch signaling results in increased Sox9 expression (12, 13). These in vitro data are consistent with our in vivo data showing that Sox9 and its target gene Crtl1 are subject to regulation by Notch signaling (Fig. S4 and Fig. S7). Therefore, these data provide support for a mechanism whereby Notch signaling modulates Sox9 gene expression in chondrogenic cells to regulate the timing and localization of chondrogenic progenitor proliferation and differentiation.
The role of the Notch pathway in the appendicular skeleton has been previously investigated in osteoblastic lineages, but the role of Notch signaling in chondrogenic axial and appendicular skeletal development has not been reported (8–10). Chondrogenic NICD activation results in a reduced sclerotome and, consequently, a nearly absent vertebral column, demonstrating that increased Notch signaling inhibits chondrogenic differentiation. In contrast, Notch pathway loss of function results in increased hypertrophic cartilage and decreased bone mineralization in the vertebral column. These data point to a previously unidentified role for Notch signaling in axial chondrogenic differentiation and development. Furthermore, Notch pathway gain and loss of function in the chondrocyte lineage result in inappropriate appendicular skeleton formation as a consequence of an abnormal hypertrophic chondrocyte zone. The hypertrophic zone is truncated in Col2α1Cre;RosaNotch mice and is increased in Col2α1Cre;RBP-Jf/f mice, with corresponding skeletal malformations. Increased Notch signaling inhibits Sox9 expression, while decreased Notch signaling leads to increased Sox9, consistent with the reported functions of Sox9 in chondrogenic proliferation and differentiation (Fig. S7). Expression of Hes7, a direct downstream target of Notch signaling in somitic cells, also is affected in appendicular cartilage, whereby increased NICD leads to ectopic Hes7 expression in proliferative chondrocytes, while Notch pathway loss of function results in undetectable expression in the chondrocyte lineage (Fig. S7) (22). The relationship of Hes7 and Sox9 in the chondrogenic lineage has not yet been determined.
Previous studies have examined Notch signaling in bone development and disease. Notch1 directly or indirectly inhibits Runx2 expression in osteogenic cells, and Runx2 deficiency leads to skeletal malformations (8, 9). This agrees with the observation that Runx2 expression is decreased with NICD gain of function in the limb. The in vivo activation and inactivation of Notch signaling in the osteoblastic lineage results in skeletal defects; however, these mice are viable (8). The difference in viability with manipulations of Notch signaling in cartilage versus bone can be attributed to abnormalities in the earlier cartilage lineage that affect chondrocyte proliferation and differentiation and ultimately result in more severe skeletal defects, including lethality. Likewise, deletion of Notch1 and 2 or Presenilin-1 and -2 in the early limb bud mesenchyme with Prx1-Cre is similar to that in Col2α1Cre;RBP-Jf/f mice, with both demonstrating disrupted chondrocyte differentiation and increased hypertrophic chondrocytes (9). This increase in hypertrophic chondrocytes postnatally corresponds to increased trabecular bone in Prx1Cre;Psenf/fPsen2−/− mice, which could not be investigated in the present study because of the lethality of Col2α1Cre;RBP-Jf/f at birth. Although delayed and reduced, bone formation does occur in Col2α1Cre;RosaNotch and Col2α1Cre;RBP-Jf/f mice, suggesting that Notch signaling does not completely prevent osteogenesis. It is likely that bone loss with increased Notch signaling is due to reduced cartilage primordia and defective chondrocyte differentiation, whereas the relative decrease of bone with Notch pathway loss of function results from an increase in hypertrophic cartilage at the expense of bone.
More than 150 human congenital defects include vertebral anomalies, and Notch pathway components have been implicated in several of these (7). Dll3, Lfng, Mesp2, and Hes7 have been associated with spondylocostal dysostosis, but to date no Notch receptor has been implicated in this condition (7, 20, 23). Clinical characteristics of spondylocostal dysostosis include short-trunk dwarfism, malformed vertebrae, rib abnormalities, and protruding abdomen, all of which are apparent in the Col2α1Cre;RosaNotch mutant mice (7). The decreased expression of Sox9 with activated NICD in chondroprogenitors is intriguing. A Sox9-associated congenital disease, campomelic dysplasia, is characterized by skeletal malformations similar to those seen in the Col2α1Cre;RosaNotch mice (18, 21). In addition, Notch1 and other Notch pathway components are known to be up-regulated in osteoarthritis and have been linked to osteopenia, osteoporosis, and osteosclerosis, demonstrating that Notch1 has homeostatic functions in the adult skeleton (8, 9, 24). In summary, there is increasing evidence for Notch signaling pathway regulation of chondrocyte proliferation, differentiation, and ossification in the appendicular and axial skeleton.
Materials and Methods
Additional information is provided in SI Materials and Methods.
Mice.
Col2α1Cre (Jackson Labs), RosaNotch, and RBP-Jf/f mice have been described previously (14–16).
Skeletal Preparation and Histology.
Whole-mount skeletal preparations stained with alcian blue 8GX (Sigma Aldrich) and alizarin red S (Sigma Aldrich) were prepared as described previously (25). For histological sections, embryos were fixed in 4% paraformaldehyde/PBS at 4 °C overnight, processed, and embedded in paraffin. Sections were cut to 5 μm or 14 μm thickness and subjected to H&E staining, alcian blue staining, immunostaining, and in situ hybridization. See SI Materials and Methods for details.
Immunohistochemistry.
Paraffin sections were incubated at 4 °C overnight with primary antibodies, and then incubated with the appropriate secondary antibody at room temperature, processed further with an ultrasensitive ABC IgG Staining Kit (Pierce), and visualized with diaminobenzidine (DAB) staining or immunofluorescence. Anti-Notch1 antibody staining was performed as described previously (26).
In Situ Hybridization.
For whole-mount in situ hybridization, embryos were harvested at E10.5 and processed as described previously (27). In situ hybridization on paraffin sections was conducted as described previously (28). Digoxygenin-labeled probes were amplified from mouse limb cDNA. The Col1a1 probe has been described previously (29). The in situ probes and PCR primers used are listed in Table S1.
Supplementary Material
Acknowledgments.
We thank Doug Melton (RosaNotch) and Raphael Kopan (RBP-Jf/f) for the generous gift of the mice used in this study; Vincent Christoffels (Tbx18), Jeff Molkentin (Myf5), and Eric Olson (Scleraxis) for their generous gift of RNA probes; Nadean Brown for the anti-Hes1 antibody; Kristin Melton for helpful discussions; and Michelle Combs for editorial assistance. This work was supported by a National Institutes of Health/National Heart, Lung and Blood Institute SCCOR grant P50HL074728 (to K.E.Y.) and Teratology training grant T32 ES07051 (to T.J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902306106/DCSupplemental.
References
- 1.Nakashima K, de Crombrugghe B. Transcriptional mechanisms in osteoblast differentiation and bone formation. Trends Genet. 2003;19:458–466. doi: 10.1016/S0168-9525(03)00176-8. [DOI] [PubMed] [Google Scholar]
- 2.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 3.Gridley T. Notch signaling and inherited disease syndromes. Hum Mol Genet. 2003;12(Special Issue 1):R9–R13. doi: 10.1093/hmg/ddg052. [DOI] [PubMed] [Google Scholar]
- 4.Kovall RA. More complicated than it looks: Assembly of Notch pathway transcription complexes. Oncogene. 2008;27:5099–5109. doi: 10.1038/onc.2008.223. [DOI] [PubMed] [Google Scholar]
- 5.Nye JS, Kopan R. Developmental signaling: Vertebrate ligands for Notch. Curr Biol. 1995;5:966–969. doi: 10.1016/s0960-9822(95)00189-8. [DOI] [PubMed] [Google Scholar]
- 6.Boni A, et al. Notch1 regulates the fate of cardiac progenitor cells. Proc Natl Acad Sci USA. 2008;105:15529–15534. doi: 10.1073/pnas.0808357105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Shifley ET, Cole SE. The vertebrate segmentation clock and its role in skeletal birth defects. Birth Defects Res C Embryo Today. 2007;81:121–133. doi: 10.1002/bdrc.20090. [DOI] [PubMed] [Google Scholar]
- 8.Engin F, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilton MJ, et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanotti S, et al. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149:3890–3899. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowe R, Zikherman J, Niswander L. Delta-1 negatively regulates the transition from prehypertrophic to hypertrophic chondrocytes during cartilage formation. Development. 1999;126:987–998. doi: 10.1242/dev.126.5.987. [DOI] [PubMed] [Google Scholar]
- 12.Watanabe N, et al. Suppression of differentiation and proliferation of early chondrogenic cells by Notch. J Bone Miner Metab. 2003;21:344–352. doi: 10.1007/s00774-003-0428-4. [DOI] [PubMed] [Google Scholar]
- 13.Fujimaki R, Toyama Y, Hozumi N, Tezuka K. Involvement of Notch signaling in initiation of prechondrogenic condensation and nodule formation in limb bud micromass cultures. J Bone Miner Metab. 2006;24:191–198. doi: 10.1007/s00774-005-0671-y. [DOI] [PubMed] [Google Scholar]
- 14.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–146. [PubMed] [Google Scholar]
- 15.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 17.Lincoln J, Kist R, Scherer G, Yutzey KE. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol. 2007;305:120–132. doi: 10.1016/j.ydbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stricker S, Fundele R, Vortkamp A, Mundlos S. Role of Runx genes in chondrocyte differentiation. Dev Biol. 2002;245:95–108. doi: 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- 20.Gridley T. The long and short of it: Somite formation in mice. Dev Dyn. 2006;235:2330–2336. doi: 10.1002/dvdy.20850. [DOI] [PubMed] [Google Scholar]
- 21.Bi W, et al. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci USA. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niwa Y, et al. The initiation and propagation of Hes7 oscillation are cooperatively regulated by Fgf and notch signaling in the somite segmentation clock. Dev Cell. 2007;13:298–304. doi: 10.1016/j.devcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 23.Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL. Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet. 2008;17:3761–3766. doi: 10.1093/hmg/ddn272. [DOI] [PubMed] [Google Scholar]
- 24.Karlsson C, Brantsing C, Egell S, Lindahl A. Notch1, Jagged1, and HES5 are abundantly expressed in osteoarthritis. Cells Tissues Organs. 2008;188:287–298. doi: 10.1159/000121610. [DOI] [PubMed] [Google Scholar]
- 25.Kuczuk MH, Scott WJ., Jr Potentiation of acetazolamide-induced ectrodactyly in SWV and C57BL/6J mice by cadmium sulfate. Teratology. 1984;29:427–435. doi: 10.1002/tera.1420290314. [DOI] [PubMed] [Google Scholar]
- 26.Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev Dyn. 2007;236:2594–2614. doi: 10.1002/dvdy.21246. [DOI] [PubMed] [Google Scholar]
- 27.Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn. 2004;230:239–250. doi: 10.1002/dvdy.20051. [DOI] [PubMed] [Google Scholar]
- 28.Shelton EL, Yutzey KE. Tbx20 regulation of endocardial cushion cell proliferation and extracellular matrix gene expression. Dev Biol. 2007;302:376–388. doi: 10.1016/j.ydbio.2006.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chakraborty S, Cheek J, Sakthivel B, Aronow BJ, Yutzey KE. Shared gene expression profiles in developing heart valves and osteoblast progenitor cells. Physiol Genomics. 2008;35:75–85. doi: 10.1152/physiolgenomics.90212.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.