Abstract
Membrane traffic along the endocytic and exocytic pathways relies on the appropriate localization and activation of a series of different Rab GTPases. Rabs are activated by specific guanine nucleotide exchange factors (GEFs) and inactivated by GTPase-activating proteins (GAPs). GEF cascades, in which one Rab in its GTP-bound form recruits the GEF that activates the next Rab along the pathway, can account for the sequential activation of a series of Rabs, but it does not explain how the first Rab is inactivated after the next Rab has been activated. We present evidence for a counter-current GAP cascade that serves to restrict the spatial and temporal overlap of 2 Rabs, Ypt1p and Ypt32p, on the exocytic pathway in Saccharomyces cerevisiae. We show that Gyp1p, a GAP for Ypt1p, specifically interacts with Ypt32p, and that this interaction is important for the localization and stability of Gyp1p. Moreover, we demonstrate that, in WT cells, Ypt1p compartments are converted over time into Ypt32p compartments, whereas in gyp1Δ cells there is a significant increase in compartments containing both proteins that reflects a slower transition from Ypt1p to Ypt32p. GEF cascades working in concert with counter-current GAP cascades could generate a programmed series of Rab conversions responsible for regulating the choreography of membrane traffic.
Keywords: Golgi, membrane traffic, organelle
Rab proteins are guanine nucleotide-dependent molecular switches that are active when bound to GTP and inactive when bound to GDP. The cycle of nucleotide binding and hydrolysis is coupled to a cycle of membrane association and dissociation (1–3). Each Rab exhibits a distinctive pattern of subcellular localization (3, 4). By recruiting a unique set of effectors, they each regulate membrane traffic into or out of the compartments with which they are associated (1). Several examples of Rab guanine nucleotide exchange factor (GEF) cascades have been described in which one Rab, in its GTP-bound state, recruits the GEF that activates the next Rab along the pathway (5, 6). Although this mechanism could lead to the sequential activation of a series of Rab proteins along a membrane traffic pathway, we have proposed that Rab GTPase-activating protein (GAP) cascades work in a counter-current fashion with Rab GEF cascades to complete the Rab conversions (1) [supporting information (SI) Fig. S1 A and B]. The Rab GAP cascade model postulates that the activation of one Rab will serve to recruit the GAP that inactivates the preceding Rab on a membrane traffic pathway, thereby limiting the extent of overlap between adjacent Rab domains on a transport pathway.
The Rab GAP cascade model offers several testable predictions: first, the GAP for the upstream Rab should bind to the GTP-bound form of the downstream Rab, but not stimulate its GTPase activity. Second, the interaction with the downstream Rab should serve to recruit the GAP to the compartment marked by that Rab. Third, the GAP should act to limit the overlap between the 2 Rabs. To test these predictions, we chose to analyze the interaction of the GAP, Gyp1p, with the Rabs, Ypt32p and Ypt1p. Ypt32p is the closest yeast homologue of mammalian Rab11 (7, 8). It resides primarily on the very late compartments of the Golgi, where it regulates the export of secretory cargo and recycled plasma membrane constituents (7, 9). Ypt1p is associated with the Golgi, where it regulates endoplasmic reticulum (ER) to Golgi and intra-Golgi traffic, as well as traffic from the endosome to the Golgi (10, 11). Gyp1p is also associated with the Golgi, and phenotypic, genetic, and kinetic studies indicate that Gyp1p acts at that site to down-regulate Ypt1p by stimulating its GTPase activity (12–15). In contrast, Gyp1p shows no stimulation of the GTPase activity of Ypt32p (16, 17). The findings presented herein establish that Gyp1p acts in a GAP cascade that limits the overlap between Ypt1p and Ypt32p on the yeast secretory pathway.
Results
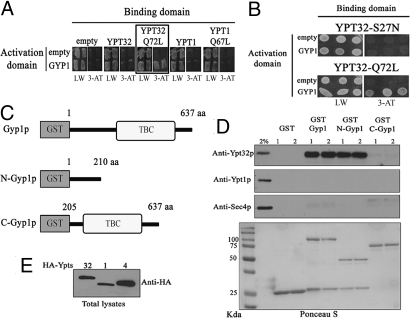
To test for an interaction between Gyp1p and Ypt32p we initially used the yeast 2-hybrid system. A positive interaction with GYP1 was observed in cells expressing the hydrolysis-deficient, YPT32-Q72L allele (YTP32-GTP; Fig. 1A, boxed panel). No interaction was observed in cells expressing YPT1 or the hydrolysis-deficient YPT1-Q67L allele (Fig. 1A) reflecting the low affinity (>200 μM) that this GAP has for its bona fide substrate (15). Furthermore, GYP1 showed an interaction with the YPT32-Q72L allele and not with YPT32-S27N, an allele that is thought to mimic the GDP-bound form (Fig. 1B), suggesting that Gyp1p preferentially binds to Ypt32-GTP bound. We also observed a positive interaction between GYP1 and the Q72L allele of YPT31, a yeast Rab that is 72% identical to, and functionally redundant with, YPT32 (Fig. S2A). To corroborate the yeast 2-hybrid results and to define the Ypt32p-binding site on Gyp1p, we performed pull-down experiments from yeast lysates using various Gyp1p constructs (Fig. 1C). The pull-down experiments confirmed that Gyp1p specifically interacts with Ypt32p and not with Ypt1p or Sec4p, and demonstrated that the interaction is confined to the amino-terminal portion of Gyp1p (Fig. 1D; GST-N-Gyp1 vs. GST-C-Gyp1). We observed the same specific interaction between Gyp1p and Ypt32p when we performed in vitro binding experiments using the GST-Gyp1 constructs and purified Ypt32p loaded with the non-hydrolyzable analogue GTPγS (Fig. S2B). These results demonstrate that Gyp1p specifically and directly interacts with active Ypt32p and that the interaction is through a region distinct from the catalytic TBC domain.
Fig. 1.
Gyp1p specifically interacts with active Ypt32p. (A) A yeast 2-hybrid assay between GYP1 and YPT32, YPT1 or the hydrolysis-deficient YPT32-Q72L, and YPT1-Q67L alleles. Growth on solid medium lacking leucine/tryptophan (LW) indicates the presence of the plasmids, growth on solid medium lacking leucine/tryptophan/histidine + 10 mM 3-AT (3-AT) indicates a positive 2-hybrid interaction (boxed panel). (B) Yeast 2-hybrid analysis between GYP1 and YPT32 mutant alleles that mimic GDP- (YPT32-S27N) or GTP- (YPT32-Q72L) bound states. Growth on solid medium lacking leucine/tryptophan/histidine + 10 mM 3-AT medium indicates a positive 2-hybrid interaction. (C) Diagrams of the GST-Gyp1 construct that were used for the in vitro binding experiment in D. The GAP catalytic domain (TBC) of Gyp1p is shown. (D) Lysates of yeast strains over-expressing HA-Ypt32p, HA-Ypt1p, or HA-Sec4p were incubated with the various GST-Gyp1 fusion proteins shown in C. Lanes 1 and 2 represent duplicates of the binding reactions. Binding was detected by Western blot using antibodies specific to each GTPase. A 2% input of yeast total lysate was run for each GTPase. Ponceau S staining of one of the membranes shows the presence of the various GST-Gyps constructs. (E) Western blot against HA to detect the level of expression of each HA-Ypts on the lysates used for the binding experiments.
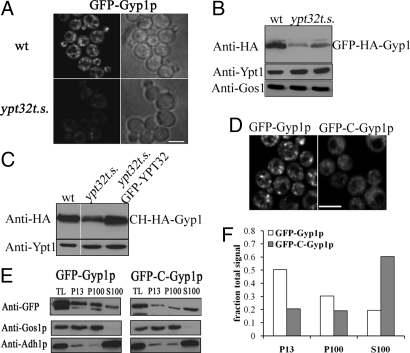
The GAP cascade model predicts that the interaction of Gyp1p and Ypt32p would be critical for the recruitment of Gyp1p to the appropriate membrane compartment. It has been previously reported that Gyp1p localizes to the Golgi apparatus (12), as does at least a portion of Ypt32p (7, 9), consistent with the proposed relationship. To determine the role of Ypt32p in the localization of Gyp1p, we expressed and analyzed a functional GFP-Gyp1p construct in a yeast strain that contains only a temperature-sensitive allele of ypt32 (ypt32ts) and lacks the functionally redundant YPT31 gene (9). We observed a decrease in fluorescent puncta, as predicted, but also an obvious reduction in total GFP-Gyp1p fluorescent signal in ypt32ts cells compared with WT cells, even at the permissive temperature (Fig. 2A). Quantification of the GFP-Gyp1p signal demonstrated that ypt32ts cells had 2.7-fold less total GFP-Gyp1p signal than WT cells [WT, 25.7 fluorescence arbitrary units (a.u.)/μm2 ± 12; ypt32ts, 9.4 fluorescence a.u./μm2 ± 3.3; n = 40 for each; P < 0.001; t test]. The difference in GFP-Gyp1p signal between WT and ypt32ts cells was even more dramatic (4.8-fold) following an incubation of the cells at 37 °C, reflecting an increase of the GFP-Gyp1p signal in WT cells, but not in ypt32ts cells (WT, 37.8 fluorescence a.u./μm2 ± 12 and ypt32ts, 7.9 fluorescence a.u./μm2 ± 3.3). Analysis of GFP-Gyp1 protein levels at the permissive temperature showed that ypt32ts cells had less GFP-Gyp1p than WT cells (Fig. 2B), demonstrating that the reduction in GFP-Gyp1p fluorescent signal was caused by a reduction in Gyp1p levels. The levels of 2 other Golgi-associated proteins, Ypt1p and Gos1p, were not reduced in ypt32ts cells (Fig. 2B). Moreover, Gyp1p was less stable after cycloheximide treatment in ypt32ts cells relative to WT cells (Fig. S3). The reduced level and stability of Gyp1p in ypt32ts cells were restored by expression of a WT copy of YPT32 (Fig. 2C, GFP-YPT32 lane; and Fig. S3), demonstrating that these changes are related to the partial loss of Ypt32p function. Interestingly, similar results have been reported in ypt32ts cells for Rcy1p, another interacting partner of Ypt32p (18).
Fig. 2.
Gyp1p levels and localization depend on its interaction with functional Ypt32p. (A) GFP-Gyp1p localization in WT (NY2772) and ypt32ts (NY2773) cells at 25 °C. (B) GFP-HA-Gyp1p levels in total protein lysates from WT or ypt32ts (2 independent samples) detected by immunoblotting for the HA epitope. A Ypt1p immunoblot is shown as loading control. (C) Immunoblotting for HA to analyze the levels of mCherry-HA-Gyp1p (CH-HA-Gyp1) in WT (NY2775) cells, ypt32ts (NY2776) cells, or ypt32ts cells expressing functional GFP-YPT32 (NY2777). (D) A Gyp1p construct lacking the Ypt32p-interaction region (GFP-C-Gyp1p; NY2774) is mis-localized in comparison to the localization of full-length Gyp1p (GFP-Gyp1p) in WT cells. (Scale bar, 5 μm in A and D.) (E) Subcellular fractionation of GFP-Gyp1p and GFP-C-Gyp1p. TL lane represents 2% of the total protein lysate that was fractionated. P13 and P100 represent the membrane-bound fractions obtained after 10,000 × g and 100,000 × g centrifugations, respectively. S100 represents the soluble fraction obtained after the 100,000 × g centrifugation. An equal amount of sample was loaded for each fraction. Immunoblotting for GFP was used to detect GFP-Gyp1p or GFP-C-Gyp1p. Immunoblots for Gos1p (membrane) and Adh1p (cytosolic) are shown as controls. (F) Quantification of the data shown in E. Image acquisition data are provided in SI Methods.
To further explore the importance of the Ypt32p interaction for Gyp1p recruitment, we analyzed the localization of GFP-Gyp1p and GFP-C-Gyp1p, a truncation that lacks the amino-terminal region required for interaction with Ypt32p, but retains the catalytic TBC domain. GFP-C-Gyp1 did not exhibit the punctate localization observed in cells expressing GFP-Gyp1p (Fig. 2D). To verify that the changes in GFP-C-Gyp1p localization were associated with changes in the membrane association of the protein, we analyzed the sub-cellular fractionation pattern of GFP-Gyp1p and GFP-C-Gyp1p by differential centrifugation (Fig. 2E). The ratio of soluble to membrane-bound GFP-C-Gyp1 was 1.5, compared with 0.25 for GFP-Gyp1p (Fig. 2F), demonstrating that GFP-C-Gyp1p preferentially accumulates in the cytosol. Analysis of control proteins Gos1p (membrane protein) or Adh1p (cytosolic protein) confirmed that the separation of membranes and cytosol was effective in this experiment (Fig. 2E). This change in the subcellular fractionation of GFP-C-Gyp1p corroborated the change we observed in its localization, and together demonstrate the importance of the Ypt32p interaction for the recruitment of Gyp1p.
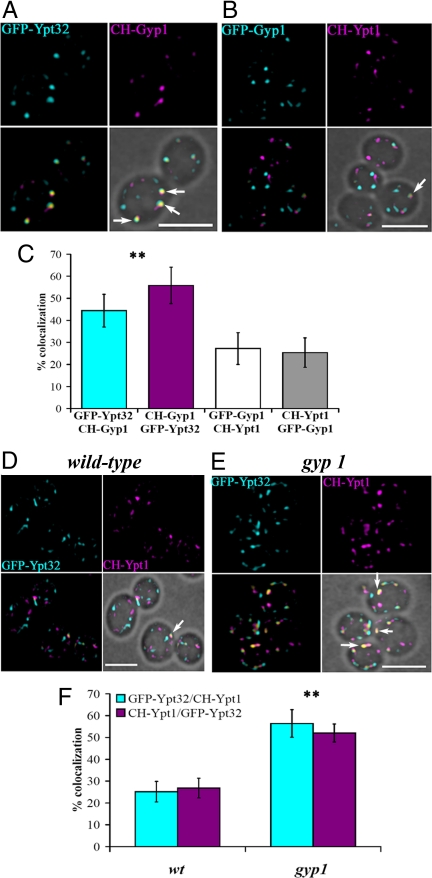
Gyp1p localization to the Golgi apparatus had been previously demonstrated by co-localization with a Golgi marker (12). Ypt1p and Ypt32p had also been localized to the Golgi, where they play important roles in traffic into and out of this structure (7–10, 19). Nonetheless, the yeast Golgi is a complex and dynamic structure with individual cisterna constantly maturing through the loss of early markers and the acquisition of late Golgi markers (20, 21), and therefore more detailed analysis was needed to establish the spatial and temporal relationships of these proteins. Our prediction, based on the interaction of Ypt32p and Gyp1p, was that Gyp1p should co-localize more extensively with Ypt32p than with Ypt1p. To evaluate the degree of co-localization of Gyp1p with Ypt32p or Ypt1p in live cells, we tagged them at their N-termini with GFP or mCherry (CH) fluorescent proteins. We established that these fluorescently tagged Rab proteins were functional by rescuing the corresponding temperature-sensitive strains (Fig. S4A). Both CH-Ypt1p and GFP-Ypt32p were over-expressed relative to the endogenous proteins to acquire images with the shortest possible exposure times (Fig. S4B). As predicted, we observed a higher degree of co-localization between GFP-Ypt32p and CH-Gyp1p than between GFP-Gyp1p and CH-Ypt1p (Fig. 3 A and B, yellow spots labeled with arrows). Analysis of the percentage of co-localization between the proteins showed that Gyp1p co-localized twice as well with Ypt32p compared with Ypt1p (Fig. 3C), demonstrating that Gyp1p preferentially resides in compartments containing Ypt32p.
Fig. 3.
Co-localization of Gyp1p with Ypt32p and the role of Gyp1p in limiting co-localization of Ypt1p and Ypt32p. (A) Fluorescent images of gyp1 cells expressing GFP-Ytp32p and CH-Gyp1p (NY2778). Merged fluorescent images were superimposed with the bright-field image (Lower Right) to show the cell contour. Arrows indicate spots where GFP-Ypt32p and CH-Gyp1p co-localized (yellow spots). (B) Fluorescent images from gyp1 cells expressing GFP-Gyp1p and CH-Ypt1p (NY2779). Arrow indicates a spot with co-localized GFP-Gyp1p and CH-Ypt1p. (C) Bar graph shows the percentage of co-localization of the following proteins: GFP-Ypt32p spots containing CH-Gyp1p signal (cyan bar), CH-Gyp1p spots containing GFP-Ypt32p signal (magenta bar), GFP-Gyp1p spots containing CH-Ypt1p (white bar), and CH-Ypt1p spots containing GFP-Gyp1p (gray bar; ≈100 cells, 2–5 spots/cell, error bar indicates SD); **P < 0.001 (t test) between Gyp1p/Ypt32p and Gyp1p/Ypt1p. (D) Fluorescent images of WT cells expressing GFP-Ypt32p and CH-Ypt1p (NY2780). Arrow indicates a spot with both GFP-Ypt32p and CH-Ypt1p. (E) Fluorescent images of gyp1 cells expressing GFP-Ytp32p and CH-Ypt1p (NY2781). Arrows indicate spots in which GFP-Ypt32p and CH-Gyp1p co-localize. (F) Bar graph shows the percentage of compartments in which GFP-Ypt32p and CH-Ypt1p (cyan bars) co-localize, or compartments in which CH-Ypt1p and GFP-Ypt32p (magenta bars) co-localize in WT and gyp1 cells (≈100 cells, 2–5 spots/cell, error bar indicates SD); **P < 0.001 (t test) between WT and gyp1. (Scale bar, 5 μm.) Image acquisition data are provided in SI Methods.
The Rab GAP cascade model predicts that Ypt32p and Ypt1p would show a low degree of co-localization because recruitment of Gyp1p to a membrane compartment by Ypt32p would lead to inactivation and loss of Ypt1p from that compartment. Although Ypt1p and Ypt32p exhibit a superficially similar pattern of punctate localization (7–10), there has been no reported analysis of co-localization. We analyzed the co-localization of Ypt1p and Ypt32p in WT cells, and more importantly, in gyp1 cells, where we expect a higher degree of co-localization as a result of a lack of Ypt1p inactivation. We observed that approximately 25% of CH-Ypt1p- and GFP-Ypt32p-containing compartments showed co-localization in WT cells (Fig. 3 D and F). However, in gyp1 cells, we observed 55% co-localization (Fig. 3 E and F). Furthermore, we observed a similar twofold increase in the degree of co-localization between the Ypt1p-effector Cog3p (22) and the trans-Golgi network marker Sec7p in gyp1 cells (Fig. S5). The increase in the overlap between CH-Ypt1p and GFP-Ypt32p observed in gyp1 cells demonstrates the role of Gyp1p in defining a boundary between Ypt1p and Ypt32p at the late Golgi. We also analyzed cells lacking GYP8, as Gyp8p has also been shown to have GAP activity with Ypt1p in vitro (23); however, we did not observe any change in the percentage of co-localization of CH-Ypt1p and GFP-Ypt32p in gyp8 cells compared with WT. Furthermore, gyp1gyp8 double mutant cells did not show any increase in co-localization compared with gyp1 cells (Fig. S6), demonstrating that the increase in co-localization is solely a result of the loss of Gyp1p. Gyp8p may act to down-regulate another Rab in vivo or it may define a boundary between Ypt1p and a Rab other than Ypt32p (see Discussion).
We considered 2 possible mechanisms to explain the increase in Ypt1p-Ypt32p co-localization in gyp1 cells: either, in the absence of Gyp1p, both Rabs accumulate in static, abnormal membrane compartments; or the increase in co-localization is related to a delay in the removal of Ypt1p after Ypt32p has been recruited to the compartment. To distinguish between these mechanisms, we performed 3D time-lapse microscopy. This type of analysis has been recently used to establish that, in budding yeast, individual Golgi cisternae mature by losing early markers and acquiring late markers (20, 21). Based on the roles that Ypt1p and Ypt32p play in membrane traffic through the Golgi, the maturation of individual Golgi cisternae in yeast, and the low level of co-localization we observed between Ypt1p and Ypt32p in WT cells, we anticipated that Ypt1p compartments would be converted to Ypt32p compartments in a time-dependent manner.
Fast sequential acquisition of images in the z axis for fluorescent proteins and 3D visualization with Imaris software (Bitplane) allowed us to spatially resolve the fluorescent signal of individual compartments over time. Quantification demonstrated that WT and gyp1 cells had a similar number of Ypt1p and Ypt32p structures, and the principle difference between these cells resides in the fraction of compartments showing co-localization (white bars, Fig. S7 A and B).
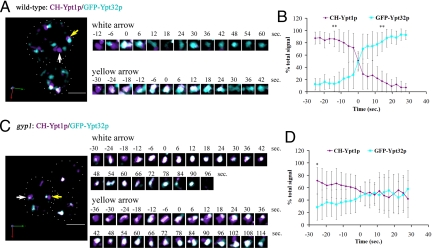
In WT cells, analysis of changes in the fluorescent signals of individual compartments demonstrated that CH-Ypt1p compartments are converted to GFP-Ypt32p compartments in a time-dependent manner (Fig. 4A, arrows; and Movies S1 and S2). To better illustrate this transition, we cropped the images to show specific compartments during a representative time series (Fig. 4A and Fig. S8A). From a total of 26 CH-Ypt1p compartments observed on the first frame of the 3 movies captured for WT, we analyzed the changes in total fluorescent signal of 18 compartments (69%). Of these 18 compartments, we observed that 16 (89%) completed the transition from CH-Ypt1p to GFP-Ypt32p. Approximately 30% of the compartments were not included in the analysis because they appeared to undergo either fission or fusion during the time course or were too close to others to resolve as individual compartments.
Fig. 4.
Three-dimensional time-lapse fluorescence microscopy demonstrates the conversion of a Ypt1p compartment to a Ypt32p compartment. (A) Images from Movie S1 demonstrate conversion of a CH-Ypt1p compartment to a GFP-Ypt32p compartment (arrows) in WT cells (NY2780). Numbers indicate the time in seconds relative to the frame where the percent of total signal was 50% (i.e., time 0). White spots represent the co-localization between CH-Ypt1p and GFP-Ypt32p. (B) Averaged trace of CH-Ypt1p and GFP-Ypt32p signals in WT cells (n = 16, error bars indicate SD); **P < 0.001 (t test) between Ypt1p and Ypt32p signals. (C) Images from Movie S3 demonstrate conversion of a CH-Ypt1p compartment to a GFP-Ypt32p compartment (arrows) in gyp1 cells (NY2781). (D) Averaged trace of CH-Ypt1p and GFP-Ypt32p signals in gyp1 cells (n = 18, error bars indicate SD); *P < 0.007 (t test) between Ypt1p and Ypt32p signals. (Scale bar, 5 μm.) Image acquisition data are provided in SI Methods.
Analysis of changes in the fluorescent signals in gyp1 cells revealed an extended period of concurrent labeling of individual compartments by both CH-Ypt1p and GFP-Ypt32p (Fig. 4C, arrows; Movies S3 and S4; and Fig. S8B) relative to WT cells. From a total of 40 CH-Ypt1p compartments observed on the first frame of the 3 movies captured for gyp1 cells, we analyzed the changes in total fluorescent signal of 24 compartments (60%). However, from the 24 compartments analyzed in gyp1 cells, we observed only 8 (33%) that completed the transition from CH-Ypt1p to GFP-Ypt32p. The remainder of the compartments showed a prolonged presence of both fluorescent signals throughout the analysis.
To better illustrate and quantify the difference in the Ypt1p-to-Ypt32p transition between WT and gyp1 compartments, we aligned the time course of each compartment trace based on the time at which the CH-Ytp1p and GFP-Ytp32p signals became equal (50% total signal, time 0 on Fig. 4 B and D). After the transition point was established for each trace, the percentages of total signal before and after the transition point were grouped and averaged to generate the averaged traces shown on Fig. 4 B and D (n = 16 spots for each graph). The averaged trace for the WT compartments (Fig. 4B) clearly demonstrated that, 25 seconds before the transition point, the compartments were predominantly labeled with CH-Ypt1p, and 14 seconds after the transition point, the compartments became predominantly labeled with GFP-Ypt32p. Conversely, for gyp1 compartments, the averaged trace (Fig. 4D) shows that, 25 seconds before the transition point, the compartments were predominantly labeled with CH-Ypt1p, but 30 seconds after the transition point, the compartments still showed similar levels of labeling for both protein signals. The gyp1 compartments became predominantly labeled with GFP-Ypt32p only after 64 seconds, which is 4.5-fold longer than the WT compartments (Fig. S8C, n = 13 for each graph). This analysis demonstrates that the increase in the co-localization of Ypt1p and Ypt32p observed in gyp1 cells is a result of the increase in time needed for the Ypt1p to Ypt32p conversion. This, in turn, is caused by the failure to inactivate and extract Ypt1p.
Discussion
In principle, a Rab GAP cascade, working in a counter-current fashion with a Rab GEF cascade, could form a self-organizing system that generates a programmed switch in Rab association over time. By recruiting a new set of effectors, this would lead to a change in the functionality of the membrane. Our data establish that Gyp1p is the central component of a Rab GAP cascade mechanism that is needed to maintain the spatial and temporal boundary between Ypt1p and Ypt32p domains on Golgi compartments in yeast. Loss of Gyp1p leads to a blurring of the Ypt1p and Ypt32p domains and hence a mixing of functions that are normally separated in space and time.
Our live cell imaging studies are generally consistent with the cisternal maturation (20, 21) model of Golgi function in as much as individual compartments that were initially labeled with CH-Ypt1p typically transitioned over time to become labeled with GFP-Ypt32p. However, close examination suggests that other processes may contribute as well. Golgi compartments were seen to be dynamic, undergoing a certain amount of fission and fusion. In some cases (30%), a Ypt32p compartment appeared to fuse to a Ypt1p compartment to yield a mixed compartment or a mixed compartment appeared to undergo segregation and fission to yield separate Ypt1p and Ypt32p compartments. Nonetheless, our data clearly indicate that Gyp1p is needed to restrict the overlap between these 2 Rabs, be it a temporal or spatial restriction.
The loss of Gyp1p function results in several trafficking defects, including a partial defect in the sorting of carboxypeptidase Y (CPY) from the Golgi to the vacuole (12) and defects in the recycling of the exocytic vSNARE Snc1p and the lipophilic dye FM4–64 (14). These defects appear to be the result of an excessive level of activated Ypt1p, as overexpression of Ypt1p exaggerates each of the trafficking defects of a gyp1Δ strain, leading to lethality. Conversely, a partial loss of function mutation in ypt1 suppresses the growth and trafficking defects of a gyp1Δ strain. Furthermore, the gyp1Δ trafficking defects can be largely mimicked by a ypt1 allele deficient in GTP hydrolysis (12, 14). The effects of the loss of Gyp1p on trafficking may relate to a role for Ypt1p and one of its effectors, the conserved oligomeric Golgi (COG) complex, in transport from the endosome to the Golgi (22). We show here that, in gyp1Δ cells, the distribution of Cog3p is shifted toward that of Sec7p, marking the trans-Golgi network. This shift in COG localization may affect the recycling to the cell surface. The CPY mis-sorting defect is most likely an indirect effect of a block in recycling of the CPY receptor Vps10p from the endosome back to the Golgi (14). Synthetic lethality has been observed between gyp1Δ and a large number of other mutations, including many that affect components of the Golgi apparatus (13, 14). Thus, the disorganization of the Golgi that results from the loss of Gyp1p antagonizes mutants already deficient in some other aspect of Golgi function.
Although we have shown that a Rab GAP cascade helps define the boundary between 2 different Rab domains midway through the exocytic pathway, and one could potentially extrapolate this mechanism to the boundaries between other Rabs, it is interesting to contemplate how Rabs are inactivated at the extreme ends of the pathway. Sec4p, the final Rab on the yeast secretory pathway, is inactivated by either of 2 closely related Rab GAPs, Msb3p and Msb4p (24). They are recruited to the plasma membrane, not by binding to a downstream Rab as there is no Rab downstream of Sec4p, but to Cdc42p, a GTPase of the Rho family that serves to establish polarity at the cell cortex (25). At the other end of the secretory pathway, yet another mechanism may be used to recruit a Rab GAP. Gyp8p and its human homologue, TBC1D20, have been implicated in the down regulation of Ypt1p and its homologue, Rab1, respectively (23, 26). However, unlike Gyp1p, TBC1D20 is predominantly associated with the ER, rather than the Golgi (26). This is consistent with our observation that the loss of Gyp8p had no effect on the extent of overlap between Ypt1p and Ypt32p within the Golgi. The localization of TBC1D20 is mediated, not by binding another Rab, but through its interaction with a reticulon, a multi-spanning membrane protein found only in the reticulated regions of the ER (26). Thus, TBC1D20 may serve to restrict Rab1 from certain sub-domains of the ER at the very start of the secretory pathway. Understanding the choreography of membrane traffic by Rab proteins will require the identification of all the relevant GEFs and GAPs and clarification of the various mechanisms by which they are spatially and temporally regulated.
Methods
Yeast strains and plasmids used for this work are described in SI Methods and Tables S1 and S2.
Ypt32p Pull-Down Assay from Yeast Lysates with Bacterially Purified GST-Gyp1.
Yeast strains expressing HA-YPT1, HA-YPT32, or HA-SEC4 from the inducible GAL1/10 promoter (NY1706, NY1708, and NY1710) (27) were used. The strains were grown overnight in YP + 2% raffinose at 25 °C. After the cultures reached an absorbance of approximately 1 to 1.5 (Abs600 nm), 2% galactose was added and the cultures were incubated for 4 h at 25 °C. Twenty-five Abs units from each culture were withdrawn, centrifuged, washed with 1.2 M sorbitol + 10 mM NaN3, and stored at -80 °C. The cell pellets were resuspended with 750 μL of ice-cold lysis buffer (1× PBS solution, 0.1% Triton X-100, and 1 mM DTT) containing a mixture of protease inhibitors (no EDTA; Roche) and PMSF. Zirconia beads were added to three fourths of the volume and the cells were broken using a bead beater for 3 min at 4 °C. The lysates were centrifuged at 13,000 × g for 10 min at 4 °C. Equal volumes of the lysates were used for a Western blot with an HA antibody to detect the level of expression of each HA-tagged protein after the induction (Fig. 1E). A total of 25 μL from each lysate were used in 200 μL binding reactions. Approximately 200 nM of the purified GST constructs bound to GSH beads (between 2 and 5 μL of beads from each construct + 5 to 8 μL of GSH empty beads) were used per reaction. The binding buffer contained: 1× PBS solution, 1 mg/mL BSA, 1 mM DTT, 1 mM MgCl2, and 0.1% Triton X-100. The binding reactions were done at room temperature for 1 h on a rotation plate. Each reaction was washed 5 times with binding buffer (without BSA), and the beads were resuspended in 15 μL of wash buffer + 15 μL of 2× loading buffer. The proteins were eluted by boiling the samples for 5 min. Half of each reaction was run on a 12% SDS/PAGE, transferred, and blotted using polyclonal antibodies specific for each of the GTPases.
In Vivo Fluorescence Microscopy and Co-Localization Analysis.
Fluorescence microscopy images were acquired on a Yokagawa spinning disc confocal microscopy system (PerkinElmer). The system was mounted onto an inverted microscope (IX71; Olympus) equipped with a 1 kb × 1 kb electron multiplying CCD camera (Hamamatsu Photonics), which was controlled by Ultraview ERS software (PerkinElmer). Cells were imaged with a 100 × 1.4 NA oil phase objective, yielding a pixel size of 87 nm. Excitation of GFP or mCherry was achieved using 488-nm argon and 568-nm argon/krypton lasers, respectively (Melles Griot). Cells were grown in synthetic defined (SD) media to an absorbance of 0.3 to 0.6 (Abs600 nm), washed and concentrated with fresh SD media, and incubated on ice. To avoid cell movement and to have cells in a similar plane during microscopy, the cells were mounted on microscope slides containing a dried 1% low melt agarose pad dissolved in SD medium. For each sample, a z-stack of 14 to 16 slices was generated, with optical sections spaced 400 nm apart. Before analysis, each optical section was digitally enhanced as described in SI Methods. For presentation and co-localization analysis, we used the maximum intensity projection of 4-middle slices for each stack. The percentage of co-localization between GFP and mCherry signal was calculated on a pixel-by-pixel basis using the co-localization threshold plug-in from MacBiophotonics ImageJ, which generates a scatter-plot of the pixel intensities to calculate the threshold for each channel (28). The scatter-plot is then used to calculate the number of co-localized pixels and their intensities. To calculate the percentage of co-localization, we used the sum of intensities greater than the threshold that showed co-localization divided by the sum of intensities greater than the threshold of the respective channel that did not co-localize.
Four-Dimensional Videomicroscopy.
For 4-dimensional imaging (i.e., 3D plus time), we used the same microscope and conditions as described earlier. Movies of 450 seconds duration were made for each strain expressing GFP-Ypt32p and mCherry-Ypt1p, with time points acquired every 2.8 sec (i.e., 160 time points). We alternated acquisition of the red and green fluorescent signals. For each time point, a z-stack consisting of 9 to 10 optical sections was generated, with optical sections spaced 350 nm apart. Each optical section was digitally enhanced as described in SI Methods before fluorescence intensity analysis. The digitally enhanced images were up-loaded into Imaris software (Bitplane) for 3D visualization, co-localization analysis, and 3D surface generation, and to track the changes in signal intensity (i.e., sum intensity) for each channel during the time course. The 3D surface generation for each channel is described in SI Methods. The movies were made with Volocity software.
Supplementary Material
Acknowledgments.
We thank members of the Novick and Ferro-Novick labs for useful discussions and comments; R. Tsien and S. Ferro-Novick for plasmids and antibodies used; D. Toomre, J. Goss, and R. M. Perera for training and usage of the spinning disc microscope in the Cinema Lab facility. This work was supported by grants from the National Institute of Health (P.J.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 14185.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906536106/DCSupplemental.
References
- 1.Grosshans B, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pfeffer S. Rab GTPases: specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer S, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. doi: 10.1038/nrm1500. [DOI] [PubMed] [Google Scholar]
- 4.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz D, Medkova M, Walch-Solimena C, Novick P. Ypt32 recruits the Sec4p guanine nucleotide exchange factor, Sec2p, to secretory vesicles; evidence for a Rab cascade in yeast. J Cell Biol. 2002;157:1005–1015. doi: 10.1083/jcb.200201003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 7.Benli M, Döring F, Robinson D, Yang X, Gallwitz D. Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 1996;15:6460–6475. [PMC free article] [PubMed] [Google Scholar]
- 8.Buvelot Frei S, et al. Bioinformatic and comparative localization of Rab proteins reveals functional insights into the uncharacterized GTPases Ypt10p and Ypt11p. Mol Cell Biol. 2006;26:7299–7317. doi: 10.1128/MCB.02405-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jedd G, Richardson C, Litt R, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bacon R, Salminen A, Ruohola H, Novick P, Ferro-Novick S. The GTP-binding protein Ypt1 is required for transport in vitro: the Golgi apparatus is defective in ypt1 mutants. J Cell Biol. 1989;109:1015–1022. doi: 10.1083/jcb.109.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du L, Novick P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol Biol Cell. 2001;12:1215–1226. doi: 10.1091/mbc.12.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du L, Novick P. Pag1p, a novel protein associated with protein kinase Cbk1p, is required for cell morphogenesis and proliferation in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:503–514. doi: 10.1091/mbc.01-07-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafourcade C, Galan J, Gloor Y, Haguenauer-Tsapis R, Peter M. The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p. Mol Cell Biol. 2004;24:3815–3826. doi: 10.1128/MCB.24.9.3815-3826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan X, Eathiraj S, Munson M, Lambright D. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- 16.Du L, Collins R, Novick P. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J Biol Chem. 1998;273:3253–3256. doi: 10.1074/jbc.273.6.3253. [DOI] [PubMed] [Google Scholar]
- 17.Albert S, Will E, Gallwitz D. Identification of the catalytic domains and their functionally critical arginine residues of two yeast GTPase-activating proteins specific for Ypt/Rab transport GTPases. EMBO J. 1999;18:5216–5225. doi: 10.1093/emboj/18.19.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S, et al. Ypt31/32 GTPases and their novel F-box effector protein Rcy1 regulate protein recycling. Mol Biol Cell. 2005;16:178–192. doi: 10.1091/mbc.E04-03-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calero M, et al. Dual prenylation is required for Rab protein localization and function. Mol Biol Cell. 2003;14:1852–1867. doi: 10.1091/mbc.E02-11-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Losev E, et al. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- 22.Suvorova E, Duden R, Lupashin V. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Antoni A, Schmitzová J, Trepte H, Gallwitz D, Albert S. Significance of GTP hydrolysis in Ypt1p-regulated endoplasmic reticulum to Golgi transport revealed by the analysis of two novel Ypt1-GAPs. J Biol Chem. 2002;277:41023–41031. doi: 10.1074/jbc.M205783200. [DOI] [PubMed] [Google Scholar]
- 24.Albert S, Gallwitz D. Msb4p, a protein involved in Cdc42p-dependent organization of the actin cytoskeleton, is a Ypt/Rab-specific GAP. Biol Chem. 2000;381:453–456. doi: 10.1515/BC.2000.059. [DOI] [PubMed] [Google Scholar]
- 25.Gao X, et al. The GAP activity of Msb3p and Msb4p for the Rab GTPase Sec4p is required for efficient exocytosis and actin organization. J Cell Biol. 2003;162:635–646. doi: 10.1083/jcb.200302038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas A, et al. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 27.Grote E, Novick P. Promiscuity in Rab-SNARE interactions. Mol Biol Cell. 1999;10:4149–4161. doi: 10.1091/mbc.10.12.4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costes S, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.