Abstract
NOD1 and NOD2 are members of the NOD-like receptor (NLR) protein family that are involved in sensing the presence of pathogens and are a component of the innate immune system. Upon activation by specific bacterial peptides derived from peptidoglycans, NODs interact via a CARD-CARD interaction with the receptor-interacting protein kinase RIP2, an inducer of NF-κB activation. In this report, we show that NOD signaling is dependent on XIAP, a member of the inhibitor of apoptosis protein (IAP) family. Cells deficient in XIAP exhibit a marked reduction in NF-κB activation induced by microbial NOD ligands and by over-expression of NOD1 or NOD2. Moreover, we show that XIAP interacts with RIP2 via its BIR2 domain, which could be disrupted by XIAP antagonists SMAC and SMAC-mimicking compounds. Both NOD1 and NOD2 associated with XIAP in a RIP2-dependent manner, providing evidence that XIAP associates with the NOD signalosome. Taken together, our data suggest a role for XIAP in regulating innate immune responses by interacting with NOD1 and NOD2 through interaction with RIP2.
The NOD-like receptors (NLRs) constitute a family of innate immunity proteins involved in sensing the presence of intracellular pathogens and stimulating host defense responses (2). Two of these family members, NOD1 and NOD2, share structural and functional characteristics. Both, NOD1 and NOD2, contain C-terminal leucine-rich repeats (LRRs) thought to act as receptors for pathogen-derived molecules, a central nucleotide-binding oligomerization domain (NACHT) (3, 4), and N-terminal caspase recruitment domains (CARDs) that associate with down-stream signaling proteins (5, 6). NODs activation is stimulated by bacterial peptides derived from peptidoglycans, with diaminopimelic acid (DAP) stimulating NOD1 (7, 8), and muramyl dipeptide (MDP) activating NOD2 (9, 10). Upon recognition of these ligands, oligomerization of the NACHT domains initiates the recruitment of interacting proteins, binding the serine/threonine protein kinase RIP2/CARDIAK/RICK via CARD-CARD-interactions (5, 6). RIP2 is critical for NF-κB activation induced by NOD1 and NOD2 (11), although the molecular details of how the NOD/RIP2 complex stimulates NF-κB activation are only partly understood.
RIP2 not only binds to NOD1 and NOD2 via CARD-CARD interactions, but it also reportedly associates with other signaling proteins independently of the CARD, including members of the TNFR-associated factor (TRAF) family and members of the inhibitor of apoptosis protein (IAP) family, cIAP-1 and cIAP-2 (12, 13). IAP-family proteins play prominent roles in regulating programmed cell death by virtue of their ability to bind caspases (14–17), intracellular cysteine proteases responsible for apoptosis. A common structural feature of the IAPs is the presence of one or more baculoviral IAP-repeat (BIR) domains, which serve as scaffolds for protein interactions (18). One of the most extensively investigated members of the IAP-family is X-linked IAP (XIAP). XIAP contains three BIR domains (19), followed by a ubiquitin binding domain (UBA) (20) and a C-terminal RING that functions as E3-Ligase promoting ubiquitination and subsequent proteasomal degradation of distinct target proteins (21). In additional to its anti-apoptotic role as a caspase inhibitor, XIAP functions in certain signal transduction processes, which include activation of MAPKs (23) and NF-κB through interactions of TAB1/TAK1 with its BIR1 domain (25).
Hints that IAP-family members might be involved in innate immunity have come from studies demonstrating that flies depleted by shRNA of Drosophila IAP-2 (DIAP2) fail to activate NF-κB in response to bacterial challenge with Escherichia coli and show decreased survival rates when exposed to Enterobacter cloacae (26, 27). Recently, studies of xiap−/− mice have provided evidence of a contribution of XIAP to NF-κB and JNK activation induced by TLRs and NLRs during infection with Listeria monocytogenes, further supporting the hypothesis that IAPs may participate in innate immune responses (28). Here we show that XIAP is required at least in certain types of epithelial cells for NF-κB activation induced by NOD1 and NOD2, and demonstrate that XIAP binds RIP2 thereby associating with NOD1/NOD2 signaling complexes.
Results
XIAP Is Required for NOD Signaling.
Epithelial cells of the intestinal track are a first line of defense against many microorganisms. We took advantage of human tumor cell lines derived from colonic epithelium in which the XIAP gene had been ablated by homologous recombination to ask whether XIAP is required for cellular responses to synthetic NOD1 or NOD2 ligands. Accordingly, isogenic pairs of XIAP+/+ and XIAP−/− HCT116 and DLD-1 cells were stimulated for 24 h with NOD1 and NOD2 ligands, L-Ala-γ-D-Glu-mDAP (DAP) and muramyl dipeptide (MDP), respectively, then Interleukin-8 (IL-8) production was measured (Fig. 1 A and B). Both DAP and MDP induced increases in IL-8 production in the wild-type HCT116 and DLD-1 cells, with MDP more potent than DAP. In contrast, neither of these NOD ligands induced IL-8 production in cultures of XIAP-deficient HCT116 and DLD1. Whereas XIAP−/− cells failed to respond to NOD ligands, they remained responsive to TNF, which induced robust IL-8 production.
Fig. 1.
XIAP is required for induction of cytokine production by NOD ligands. (A and B) HCT 116 XIAP +/+ (WT = Wild-Type) and XIAP −/− cells (KO = knock-out) (A) or DLD-1 XIAP+/+ or XIAP −/− cells (B) were stimulated with MDP (20 μg/mL), DAP (20 μg/mL), TNF-α (5 ng/mL), or left untreated for 24 h. Cell free supernatants were collected after centrifugation and analyzed for IL-8 secretion by ELISA. Data represent means ± SD of three independent experiments (pg/mL). (C) Reduced expression of NOD ligand-inducible genes in XIAP-deficient cells. HCT116 XIAP−/− (white bars) and XIAP+/+ (black bars) were stimulated for 1 h with various NF-κB inducers: 20 μg/mL γTri-DAP, 20 μg/mL MDP-LD, or 10 ng/mL TNF-α. RNA was isolated and relative levels of IκBα and IL-8 mRNAs were measured by Q-RT-PCR, normalized relative to 18S rRNA, expressed as relative levels compared with unstimulated cells (mean value = 1), and presented as mean + std dev of triplicate determinations performed in at least two independent experiments. (D) HCT116 XIAP−/− cells (KO) were transfected with FLAG-XIAP-encoding plasmid or empty FLAG-plasmid, then stimulated 24 h posttransfection with MDP (20 μg/mL), γTri-DAP (20 μg/mL), TNF-α (5 ng/mL), or left untreated. As a control, HCT116 XIAP+/+ (WT) were similarly stimulated. NF-κB reporter gene activity was measured after 24 h using the Dual Luciferase assay method. Normalized values represented mean ± SD (n = 3). (Inset) Lysates from the cells were prepared, normalized for total protein content, and analyzed by immunoblotting using anti-XIAP antibody. Reprobing blot with anti-beta-Actin antibody confirmed equal loading. (E) XIAP deficiency selective impacts NOD-mediated NF-κB activation. HEK293T cells containing a stably integrated NF-κB-luciferase reporter gene were infected with XIAP shRNA (KD = knock-down) (white bars) or scrambled control (CNTL) (black bar) lentiviruses. After 24 h, cells were stimulated with 10 μg/mL MDP-LD (MDP), 5 μg/mL γTri-DAP (DAP), 0.2 μg/mL doxorubicin (DOX), 10 ng/mL PMA/ionomycin (PMA), or 2 ng/mL TNF-α. NF-κB activity was measured 24 h later by luciferase activity, and data were expressed as fold-induction relative to control unstimulated values for each cell line (mean value = 1) and represent mean ± std dev of triplicates performed in at least two independent experiments. Inset shows immunoblot analysis of lysates from the cells (100 μg total protein) using anti-XIAP (Top) and anti-beta-actin antibodies (Bottom).
The observation that XIAP gene knock-out impairs NOD-signaling was further confirmed by quantitative RT-PCR analysis of the NF-κB target genes IκBα and IL-8, detecting decreased levels of IκBα and IL-8 mRNAs in XIAP-deficient HCT116 cells compared with wild-type HCT116 cells following stimulation with MDP or DAP (Fig. 1C). In contrast, TNF-α induced expression of these NF-κB target genes comparably in XIAP+/+ and XIAP−/− cells.
Similar observations were made using a NF-κB reporter gene to monitor responses to NOD ligands. In XIAP+/+ HCT116 cells, stimulation with MDP induced increases in NF-κB reporter gene activity (Fig. 1D). In contrast, MDP and DAP failed to stimulate NF-κB reporter gene activity in XIAP−/− HCT116 cells. Transfecting XIAP−/− HCT116 cells with a plasmid encoding XIAP (Fig. 1D) restored responsiveness to NOD ligands. Stimulating the same cells with suboptimal concentrations of TNF-α served as a control, showing XIAP-independent activation of NF-κB.
To explore the role of XIAP by an alternative approach, we used shRNA vectors to knock-down XIAP expression levels rather than gene ablation by homologous recombination. NF-κB activity was measured in control and XIAP knock-down (KD) HEK293T-cells stably expressing a NF-κB-driven luciferase reporter gene. Consistent with our observations in HCT116 and DLD-1 cells, MDP and DAP failed to activate NF-κB in cells deficient for XIAP when compared with the control vector-treated cells. In contrast, NF-κB activity was similarly induced in control vector- and XIAP shRNA-treated 293T cells after stimulation with other NF-κB inducers such as doxorubicin, PMA/ionomycin and TNF-α (Fig. 1E). In fact, PMA/ionomycin stimulated NF-κB reporter gene activity better in XIAP KD cells, for unknown reasons.
NOD1 and NOD2 Induced NF-κB Activation Depends on XIAP.
To further explore the role of XIAP in NOD signaling, we induced NF-κB activity by gene transfer mediated over-expression of NOD1 or NOD2, rather than using synthetic ligands to activate the endogenous proteins. HCT116 XIAP+/+ or XIAP−/− cells were transfected with increasing amounts of either myc-NOD1 or -NOD2 plasmids along with a NF-κB-driven luciferase reporter gene plasmid. Both NOD1 and NOD2 induced increases in NF-κB reporter gene activity in a dose-dependent manner, whereas no increase in NF-κB activity was observed in XIAP-deficient cells (Fig. 2 A and B). Reconstitution experiments in which XIAP−/− HCT116 cells were transfected with a plasmid encoding FLAG-XIAP showed restoration of NOD1 and NOD2-induced NF-κB activity (Fig. 2C).
Fig. 2.
NF-κB activity induced by over-expression of NOD1 or NOD2 requires XIAP. (A and B) HCT116 XIAP+/+ (WT) and XIAP−/− (KO) cells were seeded into 96-well plates at 2 × 104 cells per well. The next day cells were transfected with various amounts of plasmid DNA encoding Myc-NOD1 (A) or Myc-NOD2 (B), along with a fixed amount of NF-κB-Firefly luciferase and TK promoter-driven Renilla luciferase plasmids. NF-κB activity was measured 24 h posttransfection, normalizing Firefly relative to Renilla luciferase activity to determine relative levels of NF-κB activity (Firefly LUC/Renilla LUC) (mean ± SD; n = 3). (C) HCT116 XIAP−/− cells (KO) and XIAP+/+ cells (WT) were transfected in 96-well plates with 100 ng of Myc-NOD1 or -NOD2 per well along with 1 ng per well of either empty plasmid or FLAG-XIAP-encoding plasmid. NF-κB activity was measured 24 h after transfection by the Dual luciferase assay (mean ± std dev; n = 3). (D) HEK293T cells stably over-expressing NOD1 or NOD2 with stably integrated NF-κB-luciferase reporter gene were transduced with control scrambled or XIAP shRNA lentiviruses (multiplicity of infection, MOI >100). Luciferase activity was measurement 12–14 h later, expressing data as mean ± std dev of greater than or equal to three replicate determinations performed in at least two independent experiments. (E and F) HEK293T cells were seeded and transfected with plasmids encoding pcDNA Myc-epitope tagged NOD1 (E), NOD2 (F), XIAP shRNA, and/or a control vector together with NF-κB-luciferase reporter gene and Renilla luciferase plasmid for normalization of data. NF-κB activity was measured 24 h posttransfection, and expressed as fold induction relative to cells transfected with control plasmid (mean ± SD; n = 3) and are representative of three independent experiments. (G and H) HEK293T cells stably expressing an XIAP shRNA were seeded and transfected with plasmids encoding pcDNA Myc-epitope tagged NOD1 (G), or NOD2 (H), or a control vector together with a NF-κB-luciferase report gene. NF-κB activity was measured 24 h later, reporting data as fold activity induction (mean ± std dev; n = 3) (G, Right) Immunoblot analysis was performed on HEK293T stable transfectants for XIAP expression. Lysates were normalized for protein content (20 μg) and blots were probed with antibodies recognizing XIAP and β-actin.
To confirm these observations by an alternative method in another cell line, we used HEK293T cells stably over-expressing NOD1 or NOD2 and containing a NF-κB-responsive luciferase reporter gene and infected these cells acutely with XIAP shRNA lentivirus to achieve reductions in XIAP protein. NF-κB reporter gene activity driven by stable NOD1 or NOD2 over-expression was significantly reduced in these cells treated with XIAP shRNA virus compared with control virus (Fig. 2D), thus corroborating the results obtained with XIAP knock-out cell lines. Similar results were obtained in experiments where NOD1 and NOD2 were over-expressed by transient transfection (Fig. 2 E and F) or where XIAP was stably knocked down using shRNA (Fig. 2 G and H).
XIAP Directly Interacts with RIP2.
The protein kinase and adapter protein RIP2 is a known contributor to NOD signaling, which interacts with NOD1 and NOD2 via CARD-CARD interactions (11). RIP2 has also been reported to associate with c-IAP1 and c-IAP2 (29). To investigate if RIP2 similarly interacts with XIAP, we performed co-immunoprecipitation (co-IP) assays using HEK293T cells expressing FLAG-XIAP and GFP-RIP2 by transfection. In addition, we compared interactions of XIAP with full-length RIP2 and mutant versions of RIP2 lacking either the N-terminal CARD domain or the C-terminal kinase domain (KD) of RIP2. XIAP demonstrated binding to both full-length RIP2 and the RIP2ΔCARD but not to RIP2ΔKD (Fig. 3A). The interaction of endogenous XIAP with endogenous RIP2 was also demonstrated by co-IP using lysates of THP-1 monocytes, and anti-RIP2 antibodies for immunoprecipitation, showing that XIAP protein is recovered in immunoprecipitates generated using anti-RIP2 but not control antibody (Fig. 3B).
Fig. 3.
XIAP binds RIP2. (A) HEK293T cells were co-transfected with plasmids encoding FLAG-XIAP, GFP-RIP2WT, GFP-RIP2ΔCARD, GFP-RIP2Δkinase domain (KD) or empty pEGFP-C2, as indicated. After 24 h, cell lysates were prepared, normalized for protein content, and GFP-tagged proteins were immunoprecipitated using anti-GFP antibody. Immunoprecipitates were analyzed by immunoblotting using antibodies specific for FLAG epitope (Top) or GFP (Middle). Alternatively, cell lysates were analyzed directly by SDS/PAGE/immunoblotting (Bottom). Molecular weight (MW) markers are indicated in kilo-Daltons (kDa). (*HC and *LC indicate Ig heavy and light chains). (B) Lysates of THP-1 cells were immunoprecipitated with control IgG or rat anti-RIP2 antibody. The resulting immunoprecipitates were analyzed by immunoblotting using mouse monoclonal anti-XIAP antibody (Top). The cell lysate (50 μg protein) was also analyzed by SDS/PAGE/immunoblotting using mouse-monoclonal anti-XIAP or rat monoclonal anti-RIP2 (Bottom). (C) Lysates of transfected HEK293T cells expressing FLAG-RIP2 were incubated with recombinant GST-XIAP, various GST-XIAP fragments, or GST-Survivin immobilized on glutathione Sepharose and bound proteins were analyzed by SDS/PAGE/immunoblotting using mouse monoclonal anti-FLAG (Top) and anti-GST (Bottom) antibodies. Asterisks denote nonspecific bands.
To further elucidate which domain of XIAP mediates binding to RIP2, we performed in vitro protein binding studies by the GST-pull-down method, using a panel of GST-fusion proteins containing a variety of fragments of XIAP and incubating with lysates from HEK293T cells transfected with FLAG-RIP2. FLAG-RIP2 bound to fragments of XIAP containing the BIR2 domain, including a fragment comprised only of the BIR2 domain, whereas all fragments lacking the BIR2 domain failed to bind (Fig. 3C). In contrast, none of GST fusion proteins displayed interactions with a control protein, FLAG-SIP (Fig. S1). Thus, the BIR2 domain of XIAP is both necessary and sufficient for RIP2 binding.
XIAP Binds RIP2 via the SMAC-Binding Site of BIR2.
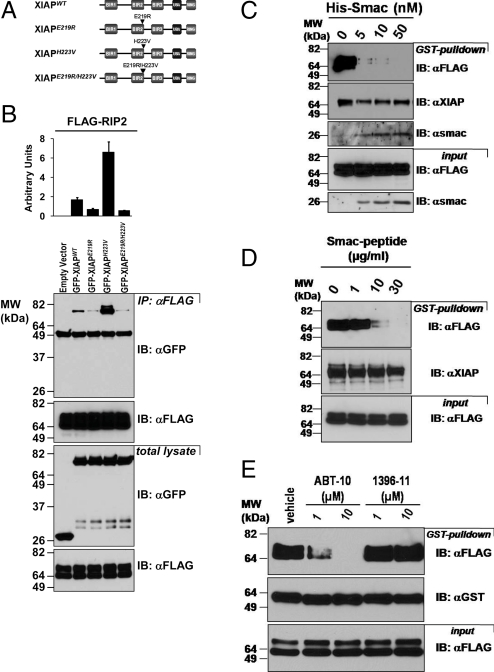
Because XIAP also binds SMAC/DIABLO via its BIR2 domain, we performed binding assays using XIAP constructs mutated at the SMAC-binding site of BIR2 (Fig. 4A). Previously, E219R and H223V mutations were shown to ablate SMAC binding to this domain, affecting critical residues for binding the Ala-Val-Ile-Pro tetrapeptide motif through which SMAC associates with a crevice on BIR2 (30). We therefore transfected HEK293T cells with FLAG-RIP2 and plasmids encoding GFP-fusion proteins containing wild-type versus mutant XIAP and performed co-IP experiments. Interestingly, RIP2 showed decreased binding to the E219R XIAP mutant, whereas the H223V mutant showed increased binding to RIP2 compared with wild-type XIAP (Fig. 4B). In contrast, the XIAP mutants showed the expected SMAC binding properties, with the E219R and H223V mutations ablating SMAC protein binding to BIR2 but having no impact on SMAC binding via BIR3 (Fig. S2). Thus, mutation of residues in the same crevice on BIR2 that is involved in SMAC binding modulate binding to RIP2. Consistent with these observations, recombinant SMAC protein (but not control SseL protein) competed for XIAP binding to RIP2 in vitro, showing concentration-dependent inhibition of XIAP/RIP2 interaction at nanomolar concentrations (Fig. 4C and Fig. S3). A synthetic peptide corresponding to the N terminus of SMAC, which binds the aforementioned BIR2 crevice, also inhibited XIAP/RIP2 interaction in a concentration-dependent manner in vitro, although requiring micromolar concentrations (Fig. 4D). Note that SMAC protein is dimeric and binds both the BIR2 and BIR3 domains of XIAP, resulting in high affinity association via simultaneous two-side binding, whereas the peptide is monomeric (31). Similarly, ABT-10, a small molecule compound that targets the SMAC-binding crevice on BIR domains, inhibited XIAP/RIP2 association in vitro (32), whereas compound TPI-1396–11 that binds a nonSMAC site near BIR2 did not interfere with RIP2/XIAP association (33) (Fig. 4E). When applied to cells, the ABT-10 compound also demonstrated inhibition of XIAP/RIP2 interaction, as assessed by co-IP experiments using lysates derived from the treated cells (Fig. S4).
Fig. 4.
SMAC binding site of BIR2 domain of XIAP is required for RIP2 binding. (A) Schematic representation of GFP-XIAP mutants. (B) Transfected HEK293T cells expressing FLAG-RIP2 together with GFP-XIAPWT, GFP-XIAPE219R, GFP-XIAPH223V, GFP-XIAPE219R/H223V or GFP-control were lysed and subjected to immunoprecipitation using anti-FLAG antibody. Immunoprecipitates were analyzed by SDS/PAGE/immunoblotting using anti-FLAG and anti-GFP antibodies. Protein binding was quantified by densitometry analysis, measuring the integrated density value expressed as arbitrary units of the GFP-XIAP bands. Values are expressed as mean ± SD of three independent experiments. (C–E) Lysates (1 mg) of transfected HEK293T cells expressing FLAG-RIP2 were incubated with 2 μg of recombinant GST-XIAP immobilized on glutathione-Sepharose along with various amounts of His-6-SMAC protein C, SMAC peptide (D), or SMAC-mimicking compounds ABT-10, nonSMAC-mimicking compound TPI-1396–11, or vehicle control (E). Beads were analyzed by immunoblotting using anti-FLAG-HRP, anti-XIAP/anti-GST or anti-SMAC antibodies as indicated. An aliquot of lysates was also directly analyzed by immunoblotting (“input”).
RIP2 Mediates Associates of XIAP with NOD1 and NOD2.
RIP2 binds to NOD1 and NOD2 via a CARD-CARD interaction, whereas our data indicate that XIAP binds to RIP2 independent of its CARD. Consequently, we surmised that RIP2 could molecularly bridge XIAP to the NOD1 and NOD2 complexes, by binding these proteins through different domains (kinase domain [KD] versus CARD). To test this hypothesis, we used recombinant GST-XIAP for assays where we attempted to pull down myc-NOD1 or myc-NOD2 produced by gene transfection in HEK293T cells, comparing lysates in which RIP2 full-length protein or fragments of RIP2 were co-expressed (Fig. 5 A and B). Expressing RIP2 resulted in clear pull-down of myc-NOD1 and myc-NOD2 with GST-XIAP. In contrast, neither RIP2ΔCARD nor RIP2ΔKD supported pull-down of myc-NOD1 or myc-NOD2 with GST-XIAP.
Fig. 5.
XIAP protein associates with the NOD/RIP2 complex. Myc-NOD1 (A) or Myc-NOD2 (B) were expressed in HEK293T cells along with GFP-RIP2 (wild-type [WT]), GFP-RIP2ΔCARD or GFP-RIP2Δkinase domain (KD). Protein lysates (1 mg) were incubated with GST-XIAP immobilized on glutathione-Sepharose and adsorbed proteins were analyzed by immunoblotting using anti-Myc and anti-GFP antibodies. An aliquot of lysates (input) was analyzed directly by immunoblotting.
Finally, because the CARDs of NOD1 and NOD2 are required for binding RIP2 (5, 6), we compared full-length NOD1/NOD2 and CARD deletion mutants of NOD1/2 with respect to their ability to be pulled down by GST-XIAP. Whereas both full-length myc-NOD1 and myc-NOD2 were recovered from RIP2-containing lysates by GST-XIAP pull-down, the NOD1/NOD2 mutants with deletion of CARDs failed to associate with GST-XIAP (Fig. S5). Altogether, these data are consistent with the hypothesis that RIP2 serves as a bridge between XIAP and NOD1/NOD2.
Discussion
In this study, we present evidence that XIAP participates in NLR signaling by interacting with RIP2. The requirement for XIAP for NOD1 and NOD2-mediated activation of NF-κB was shown by studies of both homozygous XIAP gene knock-out cells and by using shRNA to knock-down XIAP expression. Furthermore, XIAP was found to be required when NF-κB induction was stimulated with either synthetic ligands that activate endogenous NOD1 and NOD2 or by gene transfer mediated over-expression of NOD1 and NOD2. In contrast, XIAP deficiency did not impair the ability of other NF-κB inducers such as doxorubicin, PMA/ionomycin, and TNF-α to stimulate NF-κB activity. Thus, XIAP appears to participate selectively in the NF-κB pathway induced by NLR family members such as NOD1 and NOD2.
Our data are consistent with RIP2 serving as the link between XIAP and the NODs, where the CARD domain of RIP2 binds the CARDs of NOD1/NOD2 and the nonCARD regions (presumably the kinase domain) of RIP2 binds XIAP. Whereas further studies of the endogenous NOD1/NOD2 protein complexes will be required to gain a complete understanding, including eventually in vitro reconstitution from purified recombinant proteins, we speculate that XIAP could provide a platform on which to assemble components of an IKK-activating complex, in as much as the BIR1 domain of XIAP binds the TAB/TAK complex, a known upstream activator of IKKs (25). In this regard, RIP2 has been reported to bind the noncatalytic IKKgamma (NEMO) subunit of the IKK complex (34). Thus, with BIR2 of XIAP binding RIP2 (which binds IKKgamma) and BIR1 binding TAB/TAK (which phosphorylates IKKs), XIAP theoretically could bring the necessary components into close apposition for successful activation of IKKs and thus NF-κB.
The role of ubiquitination mediated by the RING domain of XIAP in this context remains to be defined. In the case of RIP1, association with c-IAP1 or c-IAP2 (typically together with TRAFs) results in K63-linked ubiquitinylation of RIP1, a posttranslational modification that is believed to recruit TAB/TAK and a modification that also occurs on IKKgamma in the context of some pathways leading to NF-κB activation (35, 36). Analogously, XIAP may interact with ubiquitin conjugating enzymes (e.g., UBC13) responsible for K63-linked phosphorylation when incorporated into NOD signalosomes, using its E3 ligase activity in facilitate IKK activation.
The participation of XIAP in NOD1/NOD2 signaling is reminiscent of the role of DIAP2 in innate immunity responses in Drosophila. In the fly, RNA interference screens have identified DIAP2 as an essential player in Drosophila innate immune signaling (26, 27, 37). DIAP2 operates downstream of the PGRP-Lc receptor, in a signaling cascade involving IMD (fly ortholog of RIP1/RIP2), dTAB2, and dTAK1 that activates Rel (NF-κB) and JNK-dependent target gene expression (26).
Our mutagenesis and competition experiments suggest that the SMAC binding crevice on the surface of BIR2 mediates interactions between XIAP and RIP2. In this regard, protein interactions involving this site include the proteolytically processed N terminus of SMAC and HtrA2/OMI and the processed N terminus of the small catalytic subunits of caspase-3 and -7 (16, 38, 39), in each case representing an N terminus created by proteolysis. We have no reason to suspect that RIP2's binding to this site on BIR2 requires proteolysis, but cannot entirely exclude it. Also, our mutagenesis data suggest that the residues lining the SMAC-binding crevice that contribute to RIP2 binding are at least partly different from those involved in SMAC and HtrA2/OMI binding, in as much as the H223V mutation inhibited SMAC but enhanced RIP2 binding. Future structural studies of the RIP2/BIR2 complex will be insightful in terms of confirming directly whether the SMAC-binding crevice is responsible for RIP2 binding and, if so, elucidating how a single protein interaction site can accommodate different protein ligands. In this regard, we cannot exclude the possibility that RIP2 interacts with an alternative site on BIR2, with the SMAC-binding pocket allosterically regulating binding.
The observation that a chemical SMAC mimic inhibited XIAP/RIP2 association reveals another unanticipated function of these compounds. Recently, it was reported that SMAC-mimicking compounds binding c-IAP1 and c-IAP2 stimulate their E3 ligase activity, causing the destruction of these proteins and impacting the NF-κB signaling mechanism by causing the accumulation of NIK and altering regulation of RIP1 (40). Our data predict that such compounds would inhibit NF-κB activity induced via the NOD-XIAP pathway, whereas simultaneously stimulating NF-κB via the aforementioned mechanisms involving NIK and possibly RIP1. Attempts to explore this possibility have been difficult to interpret because of the multiple simultaneous cellular activities of these SMAC-mimicking compounds, which stimulate some NF-κB pathways (40, 41), presumably inhibit other NF-κB pathways (e.g., NOD/XIAP), and which induce apoptosis by dislodging caspases from BIR domains of IAP family proteins.
Our analysis of the role of XIAP in NOD1 and NOD2 signaling is limited thus far to epithelial cell lines. Thus, it remains to be determined what relevance XIAP is to NOD1/NOD2 signaling in other cell lineages. While this article was in preparation, Bertrand et al. reported that NOD1 and NOD2 associate with c-IAP1 and c-IAP2, collaborating in innate immunity signaling (29). Thus, c-IAP1 and c-IAP2 may substitute for XIAP, and vice versa, in some cell lineages. Future investigations of the interactions of IAP-family proteins with NOD signaling complexes will help to reveal the biochemical mechanisms by which NLR family members such as NOD1 and NOD2 activate IKKs and other classes of kinases involved in host defense responses.
Materials and Methods
Additional methodological details are provided as SI Text.
Reagents.
Muramyl dipeptide (MDP) and L-Ala-γ-D-Glu-mDAP (γTri-DAP) were purchased from Invivogen. SMAC peptide and XIAP chemical inhibitors ABT-10 and TPI-1396–11 have been previously described (31–33).
Cell Culture and Transfection.
HEK293T and THP-1 cells were maintained in DMEM and RPMI medium 1640 (Irvine Scientific), respectively, supplemented with 10% heat inactivated FBS, 1 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. XIAP+/+ (wild-type) and XIAP−/− colonic carcinoma cell lines HCT116 and DLD-1, gifts from B. Vogelstein (John Hopkins University), were maintained in McCoy′s 5A (Irvine Scientific) with the same supplements (43). All cell lines were cultured at 37°C in 5% CO2. Subconfluent HEK293T or HCT116 cells were transfected using Lipofectamine 2000 (Invitrogen). Medium was changed 4–6 h after transfection.
Expression Plasmids.
Plasmids encoding human FLAG-XIAP, FLAG-SIP, GFP-RIP2WT, GFP-RIP2ΔCARD, GFP-RIP2Δkinase domain, human Myc-NOD1, Myc-NOD1ΔCARD, Myc-NOD2, Myc-NOD2ΔCARD1 and Myc-NOD2ΔCARDs have been recently described (1, 25, 44). XIAP-targeting shRNA vector was created by desiging a 83-mer oligonucleotide containing an XbaI site at the 5′ end and sense and antisense shRNA strands separated by a short spacer, plus a partial sequence of the H1-RNA promoter at the 3′ end. Standard PCR procedures (Advantage 2 PCR kit, Clontech) were performed by using specific shRNA oligonucleotides and T3 primer plus pSuper-like plasmid (22) as a template to provide H1-mediated shRNA cassettes with an additional XbaI site at the 3′ end. The following shRNA oligonucleotides were used: 5′- CTGTCTAGACAAAAAGTGGTAGTCCTGTTTCAGCTCTCTTGAAGCTGAAACAGGACTACCACGGGGATCTGTGGTCTCATACA -3′ for XIAP, and 5′- CTGTCTAGACAAAAAGCTTCTGCTCGCCAATAAATCTCTTGAATTTATTGGCGAGCAGAAGCGGGGATCTGTGGTCTCATACA -3′ as scrambled control. PCR products were purified (Qiagen), digested with XbaI, and cloned into the 3′ LTR NheI site of a CMV-GFP lentiviral vector as described (22). For additional plasmid constructions, see SI Text.
Lentivirus Production.
Vesicular stomatitis virus G envelope protein-pseudotyped lentiviruses were produced in HEK293T cells and purified as described (22, 24, 42).
Luciferase Gene Reporter Assay.
Wild-type and XIAP deficient HCT116 cells were seeded at a density of 2 × 104 cells per well in 96-well plates. The next day, cells were transfected with 50 ng pNF-κB-LUC (Clontech) and 5 ng Renilla luciferase gene driven by a constitutive TK promoter (pRL-TK; Promega) along with indicated plasmids. After 24 h of transfection in some experiments, cells were stimulated with various agents for 24 h or directly lysed and luciferase activities were assayed using the Dual Luciferase kit (Promega). The results for firefly luciferase activity were normalized to renilla luciferase activity.
In experiments with wild-type or transduced HEK 293T cells with stably integrated 5× κB-mediated luciferase reporter gene, cells were seeded into 96-well plates at 104 to 105 cells per well, and treated with respective inducers for 16 to 24 h. Luciferase activity was measured as suggested by manufacturer's protocol (BriteLite reagent, Perkin-Elmer). The mean results were obtained from triplicates.
Immunoprecipitation and Immunoblotting.
For immunoprecipitation (IP) cells were lysed in IP buffer [20 mM Tris pH 7.5, 135 mM NaCl, 1 mM EDTA or 1 mM EGTA (for binding assays involving NODs), 0.5% Nonidet P-40, 10% glycerol, 10 mM NaF, 1 mM DTT, 2 mM Na3VO4, 20 μM leupeptin, 1 mM PMSF, 20 mM N-ethylmaleimide, 0.5 μM iodoacetic acid, 1× protease inhibitor mix (Roche Applied Science)]. Clarified protein lysates (1–2 mg) were incubated with 2 μg monoclonal anti-FLAG antibody (Sigma Aldirch), 2 μg monoclonal anti-GFP antibody (Santa Cruz Biotechnology), or 8 μg monoclonal anti-RIP2 antibody (Alexis Biochemicals) prelinked to 25–50 μg recombinant protein G Sepharose (Invitrogen) at 4°C. For GST pulldown experiments, recombinant GST-fusion proteins were preincubated with 25 μg glutathione-Sepharose 4B (GE Healthcare) at 4°C and mild rotation for 1 h. Beads were centrifuged at 3,400 rpm for 5 min, supernatants removed and incubated with 1 mg cell lysates in IP buffer at 4°C with rotation. After incubation overnight bound immune complexes were washed four times in IP buffer, boiled in 2× Laemmli buffer and analyzed by SDS/PAGE and immunobloted using various antibodies as specifically indicated. Lysates (50 μg) were also directly analyzed by immunoblotting after normalization for total protein content.
Supplementary Material
Acknowledgments.
We thank T. Siegfried and M. Hanaii for manuscript preparation and acknowledge the generous support of the National Institutes of Health (AI-56324) and the Deutsche Forschungsgemeinschaft (KR 3496/1–1). We also thank Drs. R. Houghten, B. Vogelstein, J. Tschopp, Y.J. Kang, J. Han, S. Krajewski, and X. Wang for sharing reagents.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907131106/DCSupplemental.
References
- 1.Matsuzawa SI, Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell. 2001;7:915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 2.Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Bell JK, et al. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 4.Opitz B, et al. Nucleotide-binding oligomerization domain proteins are innate immune receptors for internalized Streptococcus pneumoniae. J Biol Chem. 2004;279:36426–36432. doi: 10.1074/jbc.M403861200. [DOI] [PubMed] [Google Scholar]
- 5.Inohara N, et al. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 6.Ogura Y, et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 7.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 8.Girardin SE, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 9.Girardin SE, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 10.Inohara N, et al. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn's disease. J Biol Chem. 2003;278:5509–5512. doi: 10.1074/jbc.C200673200. [DOI] [PubMed] [Google Scholar]
- 11.Kobayashi K, et al. RICK/Rip2/CARDIAK mediates signaling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy JV, Ni J, Dixit VM. RIP2 is a novel NF-kappaB-activating and cell death-inducing kinase. J Biol Chem. 1998;273:16968–16975. doi: 10.1074/jbc.273.27.16968. [DOI] [PubMed] [Google Scholar]
- 13.Thome M, et al. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol. 1998;8:885–888. doi: 10.1016/s0960-9822(07)00352-1. [DOI] [PubMed] [Google Scholar]
- 14.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: Why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveraux QL, et al. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17:2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;388:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 17.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun C, et al. NMR structure and mutagenesis of the inhibitor-of-apoptosis protein XIAP. Nature. 1999;401:818–822. doi: 10.1038/44617. [DOI] [PubMed] [Google Scholar]
- 19.Duckett CS, et al. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15:2685–2694. [PMC free article] [PubMed] [Google Scholar]
- 20.Gyrd-Hansen M, et al. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol. 2008;10:1309–1317. doi: 10.1038/ncb1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 22.Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanna MG, Duckett CS, Richter BW, Thompson CB, Ulevitch RJ. Selective activation of JNK1 is necessary for the anti-apoptotic activity of hILP. Proc Natl Acad Sci USA. 1998;95:6015–6020. doi: 10.1073/pnas.95.11.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu M, et al. XIAP induces NF-kappaB activation via the BIR1/TAB1 interaction and BIR1 dimerization. Mol Cell. 2007;26:689–702. doi: 10.1016/j.molcel.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gesellchen V, Kuttenkeuler D, Steckel M, Pelte N, Boutros M. An RNA interference screen identifies Inhibitor of Apoptosis Protein 2 as a regulator of innate immune signalling in Drosophila. EMBO Rep. 2005;6:979–984. doi: 10.1038/sj.embor.7400530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh JR, et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- 28.Bauler LD, Duckett CS, O'Riordan MX. XIAP regulates cytosol-specific innate immunity to Listeria infection. PLoS Pathog. 2008;4:e1000142. doi: 10.1371/journal.ppat.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertrand MJ, et al. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009 doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 30.Scott FL, et al. XIAP inhibits caspase-3 and -7 using two binding sites: Evolutionarily conserved mechanism of IAPs. EMBO J. 2005;24:645–655. doi: 10.1038/sj.emboj.7600544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, et al. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature. 2000;408:1004–1008. doi: 10.1038/35050006. [DOI] [PubMed] [Google Scholar]
- 32.Oost TK, et al. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J Med Chem. 2004;47:4417–4426. doi: 10.1021/jm040037k. [DOI] [PubMed] [Google Scholar]
- 33.Schimmer AD, et al. Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell. 2004;5:25–35. doi: 10.1016/s1535-6108(03)00332-5. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa M, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- 37.Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist Gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki Y, et al. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 40.Varfolomeev E, et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 41.Vince JE, et al. IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell. 2007;131:682–693. doi: 10.1016/j.cell.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 42.Pfeifer A, Verma IM. Gene therapy: Promises and problems. Annu Rev Genomics Hum Genet. 2001;2:177–211. doi: 10.1146/annurev.genom.2.1.177. [DOI] [PubMed] [Google Scholar]
- 43.Cummins JM, et al. X-linked inhibitor of apoptosis protein (XIAP) is a nonredundant modulator of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis in human cancer cells. Cancer Res. 2004;64:3006–3008. doi: 10.1158/0008-5472.can-04-0046. [DOI] [PubMed] [Google Scholar]
- 44.Krieg A, Le Negrate G, Reed JC. RIP2-beta: A novel alternative mRNA splice variant of the receptor interacting protein kinase RIP2. Mol Immunol. 2009;46:1163–1170. doi: 10.1016/j.molimm.2008.11.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.