Abstract
We present a droplet-based microfluidic technology that enables high-throughput screening of single mammalian cells. This integrated platform allows for the encapsulation of single cells and reagents in independent aqueous microdroplets (1 pL to 10 nL volumes) dispersed in an immiscible carrier oil and enables the digital manipulation of these reactors at a very high-throughput. Here, we validate a full droplet screening workflow by conducting a droplet-based cytotoxicity screen. To perform this screen, we first developed a droplet viability assay that permits the quantitative scoring of cell viability and growth within intact droplets. Next, we demonstrated the high viability of encapsulated human monocytic U937 cells over a period of 4 days. Finally, we developed an optically-coded droplet library enabling the identification of the droplets composition during the assay read-out. Using the integrated droplet technology, we screened a drug library for its cytotoxic effect against U937 cells. Taken together our droplet microfluidic platform is modular, robust, uses no moving parts, and has a wide range of potential applications including high-throughput single-cell analyses, combinatorial screening, and facilitating small sample analyses.
Keywords: cell encapsulation, viability assay, lab-on-a-chip, emulsion, optical coding
Droplet-based microfluidic approaches present a paradigm for screening, providing increased throughputs, reduced sample volumes and single-cell analysis capabilities. Droplet microfluidics uses a 2-phase system, in which each assay is compartmentalized in an aqueous microdroplet (1 pL to 10 nL) surrounded by an immiscible oil. The advantages of this droplet-based technique include the physical and chemical isolation of droplets eliminating the risk of cross-contamination, the fast and efficient mixing of the reagents that occurs inside droplets, the ability to digitally manipulate droplets at a very high-throughput, the ability to incubate stable droplets off-chip and reintroduce them into the microfluidic environment for further processing and analysis, and the absence of moving parts [such as chip-integrated valves (1) or pumps]. In addition, because a small number of cells can be analyzed in discrete droplets, this technology is particularly suitable for working with cells of limited availability, such as stem cells or primary cells from patients.
Previously, important steps have been made to develop specific components required to perform droplet-based manipulations, including cell encapsulation (2–8), droplet merging (9–13), droplet mixing (14, 15), on-chip incubation (16), droplet sorting (17), and introduction of multiple compounds (18). However, each of these droplet manipulations has been demonstrated as separate modules that have not been assembled into an integrated biological assay. In addition, direct quantitative analysis of mammalian cells inside intact droplets that would require on-chip staining and thus a more complex integration of droplet modules, has not been shown. Finally, there have been no reported demonstrations using stable premade optically encoded droplet libraries, which are required for performing very high-throughput screens in the droplet format.
Here, by combining an on-chip cell viability assay and the automated generation of coded droplet libraries, we were able to develop a fully integrated droplet-based workflow for conducting a mammalian cell cytotoxicity screen at a very high-throughput. The development of the on-chip viability assay served 2 purposes. First, it permitted analysis of the overall utility and biocompatibility of the droplet format, by allowing direct measurement of the viability of encapsulated cells. Second, it enabled cytotoxicity screening capabilities that are important in cancer research to either assess drug cytotoxicity or to screen cancer cells for therapeutic targets via synthetic lethality (19). This droplet workflow is generic and could be used with different types of libraries (e.g., DNA, siRNA, drug), fluorescent assay read-outs, or additional droplet manipulation modules.
Results
Droplet Screening Work-Flow.
A droplet screening workflow (Fig. 1) involves 4 steps: (i) the drug library is formatted using the library generation chip into a droplet emulsion with each member uniquely coded with an optical label; (ii) each droplet library member is combined with a cell-containing droplet on a merge chip; (iii) the combined emulsion is incubated for cell treatment; and (iv) the emulsion is reinjected into the assay chip, where each droplet's fluorescence is measured for both assay and drug coding readouts. As a prerequisite to using the droplet workflow to conduct a cytotoxicity screen on mammalian cells, we first developed an integrated viability assay and used it to demonstrate the survival of encapsulated cells over a period of several days.
Fig. 1.
Droplet screening workflow. The droplet screen has 4 steps. (A) A reformatting step to emulsify compound-code pairs and pool them into a droplet library. (B) Merging each library member with one of the cell-containing droplets that are continuously generated. Hence, each cell droplet has a specific composition defined by the compound droplet it has merged with. (C) Incubation. (D) Merged droplets are reinjected into an assay chip to identify each compound via their code and assess their specific effect on cells.
Development of the Integrated On-Chip Viability Assay Chip.
We successfully developed a fully integrated droplet viability assay to interrogate cells within intact droplets using a standard fluorescent assay (see Methods). The assay sequentially performs a series of tasks by integrating 5 different droplet modules into a single chip: (i) a generation and reinjection module, with 1 nozzle for encapsulating the viability fluorescent dyes into droplets and a second nozzle for encapsulating cells or reinjecting cell emulsions (Fig. 2A and Movie S1); (ii) a module to merge droplet pairs includes an expansion region and a pair of electrodes that delivers an electric field to control pairwise coalescence (Fig. 2B and Movie S2); (iii) a serpentine mixing module to rapidly trigger reactions (14, 15) (Fig. 2C and Movie S3); (iv) a delay line to allow on-chip enzymatic reactions to develop for 15 min (Fig. 2D and Movie S4); and (v) a detection module that elongates each droplet for consistent interrogation of every cell (Fig. 2E and Movie S5). Interrogation of each droplet's fluorescence was accomplished using laser line illumination and detection with photomultiplier tubes (PMTs) (Figs. 2E and 3A). Individual droplet signals decompose into a plateau that corresponds to the homogeneous droplet signal overlaid by a narrow peak that corresponds to a cell signal (Fig. 3B).
Fig. 2.
Development of an on-chip viability assay. The viability assay chip integrated a series of 5 modules that had been optimized for analysis of cell viability. These modules sequentially manipulated droplets to conduct a viability assay on a single chip. (A) A set of 2 nozzles encapsulated cells and live/dead fluorescent dyes respectively. The fork enabled the interdigitation of the streams resulting in cell-containing droplets alternating with dye-containing droplets (100 μm deep channels). (B) A fusion module that delivered an AC field permitted electrically-controlled merging of pairs of dye-containing droplets and cell-containing droplets (100 μm deep). (C) A mixing module facilitated rapid and thorough mixing of cells with dyes (100 μm deep). (D) A delay line optimized cell staining by enabling on-chip incubation of the droplet for 15 min (260 μm deep). (E) A detection module confined droplet laterally and vertically to collect the fluorescent signals excited with a laser slit (100 μm deep). Live cells and dead cells were scored with Calcein-AM and Sytox Orange, respectively.
Fig. 3.
On-chip viability assay characterization. Monocytic U937 cells were encapsulated and tested for their cell state with the integrated viability assay chip. (A) Typical 1 second raw signal trace collected by the photomultipliers. (B) A close-up of this raw trace showed typical droplet signals. The droplet fluorescence signature was characterized by an approximately rectangular cross-section. Peak signals corresponding to cell staining were characterized by higher maximum signal and narrower full-width at half-maximum. Each cell stain could be clearly distinguished on top of the homogeneous signal. In this trace, we could see 3 droplets, containing a live cell (520 nm), a dead cell (617 nm), and no cell, respectively. (C and D) Images of a live cell and of a dead cell inside droplets. (E) Assay performance characterization. Different mixtures of live and dead cells were scored, and the results were compared with control reactions performed in a microtiter assay. The results showed a strong correlation between the 2 methods.
For this study, we used a flow-focusing nozzle (20, 21) to encapsulate monocytic U937 cells into 700 pL (≈110 μm) droplets at a rate of 100 droplets/s. Droplets were stabilized with a fluorinated surfactant that was selected to optimize cell viability (see Methods). Cell encapsulation follows a Poisson distribution that defines the droplet occupancy statistics (2, 5, 6), and resulted here in approximately one-third of the droplets being empty and one-third containing a single-cell.
From a fluidics perspective, a number of design parameters constrain the integration of different modules into a single device, including the need to optimize the nozzle dimensions to achieve the appropriate droplet sizes and frequencies, and a requirement to minimize back-pressure across the entire chip. The impact of nozzle design on droplet parameters was experimentally established and optimized in a few iterations (21–23). On-chip incubation presented a more difficult challenge, as strategies to increase droplet residence time by lengthening the channels also proportionally increase the pressure drop (24). This issue was solved with the use of delay-lines that have large cross-section channels that allow longer droplet incubation times while limiting back-pressure (24). We used a 3-layer lithography process to allow for the generation and manipulation of droplets in shallow channels and droplet incubation in deeper channels (Fig. S1C).
To test the ability of our technology to accurately discriminate different cell populations, we carried out a series of experiments to characterize the specificity, sensitivity, and reliability of the on-chip assay by scoring monocytic U937 cell viability. The assay showed a high degree of specificity, with less than 0.1% of the dead cells and 1% of the live cells showing double-staining with both live and dead cell stains. The assay sensitivity was determined by scoring cells prestained with an independent dye (Qdot 655- wheat germ agglutinin) showing that more than 99% of dead cells and 98% of live cells injected into the chip were detected. It is noteworthy that these rates are the result of the combined efficiencies of all of the modules used, demonstrating the overall robustness of the assay. Finally, we modeled the cytotoxic profiles expected from a screen with a series of known ratios of live and dead cells. The on-chip viability assay was able to properly score the different cytotoxic profiles as shown by the correlation between the results generated by the droplet and microplate assays (Fig. 3E). Altogether, these results showed the successful integration of the different droplet modules into a single on-chip viability assay, with a specificity and accuracy that is well suited for screening applications.
Encapsulated Cell Survival and Growth.
Next, we focused on optimizing encapsulated cell survival, a baseline requirement for screening applications. To assure robust and consistent cell encapsulation, we developed a magnetically stirred injection system that maintained cells in suspension and minimized shear-stress before on-chip injection (Fig. S2). Cells have been injected for more than 4 h using this system, with a constant cell flow and without a measurable negative effect on viability when assessed at the tube outlet.
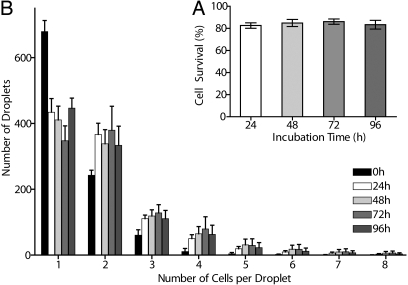
We used the quantitative integrated viability assay to optimize cell encapsulation methods. The cells were first encapsulated and collected into a syringe that was subsequently transferred to a conventional CO2 incubator. The emulsion was then reinjected into the quantitative viability assay for scoring. The high emulsion stability enables this off-chip incubation solution, which is more practical than a long on-chip incubation that would restrict the throughput and reduce the system flexibility. We found cell survival rates to be high and very stable, with the overall cell viability remaining >80% for up to 4 days (Fig. 4A). We also examined the cell parameters on a per droplet basis, to check the balance between cell growth and cell death during incubation. The data showed that only during the first 24 h did a small percentage of the cells divide (Fig. 4B). This limited cell growth could be explained by the fact that the effective density of 1 cell in each 700-pL droplet is equivalent to ≈1.4 million cells/mL, which is close to the plateau of the cell growth curve for U937 cells (Fig. S3). Possibly, only cells that were committed to cell division before encapsulation divided, whereas the rest of the cells experienced the small droplet volume as a high cell density environment that precluded growth, with the encapsulated environment capturing autocrine growth factors or metabolites. The overall limit of 80% viability at 24 h suggested an early loss of cell viability. Separate experiments (Fig. S4), showed that the encapsulation process itself induced a loss in cell viability (13% in these experiments) that was observable at 2 h, possibly because of shear stress or cell-surfactant interaction. Altogether, we can conclude that cells can be routinely encapsulated at a rate of 100 Hz with droplet experimental conditions that allow for excellent survival of U937 cells once encapsulated, and meeting the baseline viability requirements for high-throughput screening (HTS) experiments.
Fig. 4.
Quantitative in-droplet cell growth and survival. The integrated viability assay was used to optimize cell encapsulation and storage conditions to achieve maximum survival of encapsulated cells. Human monocytic U937 cells were encapsulated inside droplets, collected into a glass syringe, and incubated in a standard cell incubator. (A) After different incubation times, the encapsulated cells were assayed for viability using the integrated viability chip. Encapsulated cells maintained >80% survival up to 4 days post-encapsulation. (B) To assess the balance between cell growth and cell death, we analyzed the distribution of the total number of cells per droplet over time. During the first 24 h, the distribution of number of cells contained in each droplet showed a partial shift toward droplets containing more cells, the distribution remained very constant after this initial period. The results show that only limited cell growth occurred during the first 24 hours.
Mitomycin C Cytotoxicity Screen.
We used the droplet screening workflow to test a fluorescently encoded drug library (containing different concentrations of mitomycin C) for its cytotoxic effects on U937 cells. A number of challenges associated with producing a stable droplet library were addressed in this work. First, a library reformatting system was designed to robotically interface between a microtiter plate (macroscale) format and a library generation chip (microscale), allowing automated preparation of the droplet library by sipping each member from the well of a microtiter plate and injecting it into the library generation chip for encapsulation at a rate of 350 Hz. Droplets were collected into a glass vial for storage and subsequent manipulation. The library emulsions generated using this system showed a high degree of stability during storage, incubation, and when assessed during reinjection. Finally, each drug concentration was fluorescently labeled with a specific concentration of a soluble red dye (Alexa Fluor 680 R-phycoerythrin) (Fig. 5A). Each drug-label combination was encapsulated into 200 pL (73 μm) droplets using the library generation chip (Fig. 1, Step 1, and Fig. S1A).
Fig. 5.
Drug library coding and decoding. We used the integrated viability assay to screen an optically encoded mitomycin C drug library for its cytotoxic effect against human monocytic U937 cells. (A) Each drug concentration was coded with a specific concentration of a dye fluorescent at 710 nm (Alexa Fluor 680 R-phycoerythrin). The resulting 8-member library contained the mitomycin C drug diluted 6-fold stepwise (4 μM for the highest concentration) and the coding dye diluted 2-fold stepwise (1.8 mg/mL for the highest concentration). (B) The coding signal is collected in the third detection channel (710 nm), in addition to the signal traces of the viability stained cells in the first 2 detection channels (520 nm; 617 nm) (compare to Fig. 3B). (C) The histogram clearly shows that the 8 members of the library are optically resolved during the read-out after the 2 merges during the experiment. Each condition tested roughly equal numbers of cells (≈900 cells corresponding to ≈900 droplets per condition). The signal of the highest concentration member was clipped because of PMT saturation.
The screening process began with the drug library being reintroduced and combined with cell-containing droplets (700 pL, ≈110 μm) on a merge chip at a rate of 100 Hz (Fig. 1, Step 2, and Fig. S1B). After the combination step, the emulsions were collected into a glass syringe and incubated in a conventional cell incubator for 24 h (Fig. 1, Step 3). The emulsions were subsequently injected into the integrated assay chip for quantitative fluorescent-based cytotoxicity and drug coding read-out at 100 Hz (Fig. 1, Step 4).
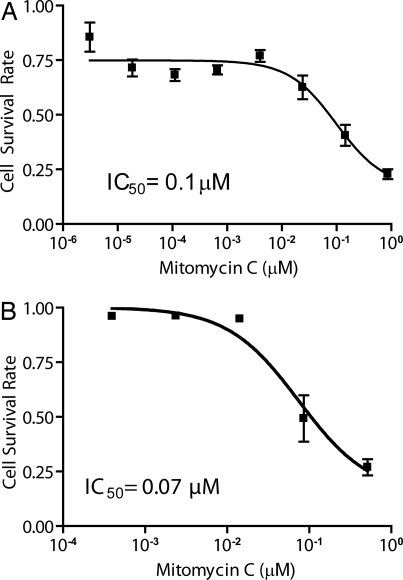
Upon assay read-out (Fig. 5B), the 8 library members (representing the 8 concentrations of the drug) could be easily resolved (Fig. 5C) despite the 2 successive merges that broaden each member's distribution, thus demonstrating the tight control over droplet volumes during generation and manipulation. Each drug treatment was represented by a roughly equal number of droplets (≈900), showing sufficient mixing of the library before use. Finally, we assessed the specific effect of each drug concentration on cell viability and generated a dose-response curve by binning droplets based on their optical code (Fig. 6A). Both the shape of the dose-response curve and the IC50 measured in the droplet assay were similar to those obtained in the control experiment performed in a microtiter plate (compare to Fig. 6B). The 80% cell viability limit in the droplet assay is explained by the 10%–20% loss of cell viability during cell encapsulation seen in previous experiments. It is important to note that each data point represents 900 equivalent tests, making the droplet assay very robust. In addition, the low standard deviation from 3 replicate experiments underlines the high reproducibility of the droplet method. Finally, the results also highlight the fact that cells in droplets are not only viable, but respond to the drug treatment in a manner similar to that seen when the cells are in solution in a microtiter plate.
Fig. 6.
Mitomycin C cytotoxicity characterization. As a screening proof-of-principle, we generated a dose-response curve of the effect of mitomycin C on U937 in cells. (A) By binning each library member based on its coding signal, we could assess the specific effect of each compound concentration. (B) Control dose-response curve in a microtiter plate assay. The dose-response curve generated on-chip and the IC50 inferred were similar to microtiter plate controls. Both the shape and the IC50 were similar to the on-chip screen results. The main difference resided in the ∼15% loss of cell viability during cell encapsulation, which resulted in a slightly compressed assay dynamic range when this initial cell death was not subtracted.
Discussion
We have successfully developed a droplet-based microfluidic technology to perform high-throughput single-cell screening. This work relied on a series of key innovative features: the development of an integrated on-chip assay, the quantitative demonstration that mammalian cells could survive up to 4 days inside droplets, and the ability to manufacture and optically encode droplet-formatted drug libraries.
The on-chip integration of the viability assay enabled a previously unattainable quantitative view of the viability and growth of single encapsulated cells. The design of droplet microfluidic chips is modular, and new functionalities can be added as long as the pressure drop across the chip remains within the working conditions of the pumps and PDMS-glass bonding. Hence, the breadth of applications could be further expanded by the use of postscreening droplet manipulation, like sorting (9) or genetic analysis (25) or by using a wide range of different fluorescent assays (26). Importantly, the detected molecules and assay fluorophores do not have to be retained inside cells or be bound to their surface, as they remain encapsulated inside the droplets (27). Finally, in addition to small-molecule drugs, the library content can be expanded to include molecules such as siRNA, DNA, or proteins.
Cell viability critically depends on the cell-droplet interface interaction and is thus highly dependent on the surfactant formulation used (2). This interaction can be tailored by the use of aqueous additives, such as Pluronic F68 in this study (Fig. S5), to provide both excellent biocompatibility and emulsion stability. The use of additives is an effective route for finding biocompatible formulations and should represent a parallel effort to the synthesis of new surfactants (2, 6). In addition, encapsulation in 700-pL droplets resulted in limited cell growth, and assays that require cellular proliferation could benefit from increasing the droplet volume (Fig. S6) (2). Finally, we foresee the use of this system for screening anchorage-dependent cells by either preseeding cells on beads coated with the required anchor proteins or by encapsulating cells into biocompatible hydrogels (28).
The throughput (100 Hz) of the droplet-based assay shows significant gains over conventional HTS techniques, which can process up to 1 compound per second (2). For a given flow rate, the throughput is inversely proportional to the droplet volume, which is fixed by the biological assay. Further engineering can improve the throughput, and we were able to reach a throughput of 500 Hz for the merge chip by stabilizing the flows with a feedback loop control. In addition, because the microfluidic devices are stamped, the total throughput can be enhanced by simply fabricating parallel circuits. For single-cell applications, efficient throughput depends on the fraction of droplets containing only 1 cell, which could be improved by using nonstochastic single-cell encapsulation methods that maximize the number of droplets carrying a single cell (3, 29).
We have demonstrated the practical use of a stable droplet library, addressing a number of significant challenges. First, we automated the library generation with a robotic process to ensure the proper reformatting of the library from a standard microplate format into a droplet format. Second, the emulsion stability provided by the surfactant formulation enabled the storage and subsequent manipulations of the library, including reintroduction to a microfluidic chip. Finally, we designed a fluorescent coding scheme that permitted the identification of each member of the drug library during screening. Additional effort will be required to extend the number of library members that can be coded for screening. The optical coding scheme demonstrated here can be expanded by using fluorescence polarization as an additional parameter. For example, 10 intensity levels combined with 10 fluorescence polarization levels would generate 100 different codes for each coding color. Multiplexing this coding scheme for 2 different colors will bring the range of accessible codes to 10,000 optical codes. A number of alternative coding strategies can be envisioned, including fluorescent techniques based on microbeads (30, 31) or coded particles (32) that would permit both real-time deconvolution and postprocessing data analysis, positional encoding (18), or the use of DNA (33, 34) or peptide molecules for coding, which would alleviate the challenges of spectral availability but would require a sorting step and postprocessing data analysis.
Altogether the system we have developed is versatile and modular in 3 major ways: (i) many different types of operations or manipulations can be performed on-chip; (ii) many different types of fluorescence read-out can be used; and (iii) a variety of library types can be interrogated.
Precise cell encapsulation, high-throughput droplet generation and manipulation, and the possibility of combinatorial screening (by combining 2 libraries for instance) suggest that droplet technology can perform as an ideal platform for a range of “-omics” approaches. The droplet technology we have developed is a generic, fast, and robust tool that can be used not only in the context of any large-scale screen, but is also well-suited for analysis of small samples of medical relevance.
Materials and Methods
Microfabrication and Fluidics Set-Up.
Fluidic chips were fabricated in PDMS (Dow Corning) using standard soft lithography (24) and were irreversibly bonded to a tin oxide-coated microscope slide (Delta Technologies). The electrodes were incorporated by injecting low-melting solder (McMaster-Carr) in dedicated channels (35). The electric field (≈300 V.cm−1, 100 kHz) was supplied by a high-voltage module (JKL Components). The channels were coated with Aquapel (PPG Industries). The flow rates were controlled by syringe pumps (Harvard Apparatus).
Cell Encapsulation.
Cells were encapsulated while being in the logarithmic phase growth. They were washed twice in Dulbecco-PBS (Invitrogen) and resuspended in fresh medium (RPMI, 10% fetal bovine serum; Invitrogen) with 1% Pluronic F-68 (MP Biochemicals) and 0.1% (wt) BSA (Sigma-Aldrich) at a density of 1.4 × 106 cells/mL.
The cells were transferred into the injection vial (see Fig. S2) and injected into the chip via a FEP tubing (Upchurch Scientific). The cells were displaced from the injection vial by pure fluorinated oil at 250 μL/h and were pinched by the surfactant-oil phase at 1200 μL/h at the nozzle (80 μm wide, 100 μm deep) resulting in the cell encapsulation into 700 pL (≈110 μm) droplets. The cells were encapsulated using 1 nozzle of the drug-cell combination chip (described below). The fluorocarbon phase consisted in a fluorinated oil (RainDance Technologies) that contained an ammonium salt of carboxy-PFPE surfactant (36) to stabilize the droplets against coalescence while ensuring good biocompatibility of the interface when used with Pluronic F68.
After collection into 5-mL glass syringes, most of the oil was removed, and a 0.2-μm sterile Puradisc filter (Whatman) was inserted. The emulsions were stored in a standard incubator (37 °C, 5% CO2), with the filter tip in contact with Dulbecco-PBS for buffering purpose. For cell growth experiments, images were taken during encapsulation and were used to count cell occupancy for the 0 h time points (ImageJ).
Viability and Cytotoxicity Experiments.
Live and dead cells were scored with Calcein-AM and Sytox Orange (Invitrogen), while fluorescein (Invitrogen) was added at 0.1 μM to outline the droplets (Fig. 3B). The live-dead assay was conducted either with the encapsulation oil formulation (RainDance Technologies) with a fluorosurfactant (Zonyl-FSO, 1.3% wt; Dupont) added to the dye solution, or with a fluorinated PEG surfactant (37) in F oil (RainDance Technologies), with no notable difference.
For specificity and sensitivity experiments, live cells and dead cells were scored separately. Dead cells were prepared by incubation in 70% ethanol at 4 °C overnight. For sensitivity experiments, cells were prestained before on-chip scoring with Qdot 655 wheat germ agglutinin (Invitrogen). For the mixture experiments, the ratios of live and dead cells were measured at the end of the experiment by scoring cells going out of the injection vial. The viability chip operation was described in the main text.
The chip dimensions were: Nozzle for reinjecting the cell emulsion 80 μm wide × 100 μm deep, nozzle for the dye solution encapsulation 100 μm wide × 100 μm deep; the merge module was 340 μm wide, 320 μm long, and 100 μm deep; the mixing module consists of 9 U-turns that were 52.5 μm wide and 177 μm long with an angle of 60 °; the incubation line was 600 μm wide × 260 μm deep and 1.5 m long; finally the detection module was 30 μm wide, 50 μm long, and 100 μm deep.
Data Acquisition and Analysis.
A 488-nm laser (Picarro) and a 565-nm laser (Coherent) were focused into the channels through achromat and cylindrical lenses (Edmund Optics). Fluorescence emissions were collected with a set of filters (535–40 nm, 617–73 nm, 710–40 nm; Semrock) and photomultiplier tubes (Hamamatsu). Fluorescence detection was driven at 100 kHz by a custom data-acquisition system (Labview; National Instruments) that also allowed signal processing and statistical analysis. Three thresholds were used to identify droplets, live cells, and dead cells. A collection of parameters was measured, and each cell peak was linked to droplet parameters (number of cells, size, and code).
Drug Library.
Mitomycin C (Sigma) was mixed with the coding dye Alexa Fluor 680 R-phycoerythrin (Invitrogen) and diluted in RPMI before encapsulation. A custom-built library reformatting system was used to prepare each library. Each member was systematically encapsulated from a 96-well microtiter plate by being dispensed into a library generation chip. The members were collected into a vial, and the library was mixed before use. The layout of the fluidics chip consisted in a simple nozzle (80 μm wide × 50 μm deep) and 2 outlets, of which 1 was used for collection (see Fig. S1A). The drug concentrations were encapsulated into 200 pL (73 μm) droplets by using 250 μL/h flow-rate for the aqueous phase and 1250 μL/h flow-rate for the oil phase.
Drug Library and Cells Combination.
The combination chip layout comprised a set of 2 nozzles (Fig. S1B). One nozzle (60 μm wide and 100 μm deep) was used to space the library droplets, the other nozzle (80 μm wide and 100 μm deep) was used to encapsulate cells into 700 pL (≈110 μm) droplets. Both streams were collated before entering a merging module where they were combined as pairs. A short incubation-line (600 μm × 0.24 m) assured optimal droplet stability before collection.
Supplementary Material
Acknowledgments.
We thank Phenix-Lan Quan, Yves Charles, Jeffrey Branciforte, Yue Suo, Haakan Joensson, Michael Weiner, Wolfgang Hinz, Bernard Mathey-Prevot, Chris Bakal, and RainDance Technologies for technical support and useful discussions. This work was supported by Small Business Innovation Research, National Institutes of Health (SBIR-NIH) Grant 5R43HG003925–02. N.P. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903542106/DCSupplemental.
References
- 1.Unger MA, et al. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 2.Clausell-Tormos J, et al. Droplet-based microfluidic platforms for the encapsulation and screening of Mammalian cells and multicellular organisms. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Edd JF, et al. Controlled encapsulation of single-cells into monodisperse picolitre drops. Lab Chip. 2008;8:1262–1264. doi: 10.1039/b805456h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He M, et al. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal Chem. 2005;77:1539–1544. doi: 10.1021/ac0480850. [DOI] [PubMed] [Google Scholar]
- 5.Huebner A, et al. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem Commun. 2007;12:1218–1220. doi: 10.1039/b618570c. [DOI] [PubMed] [Google Scholar]
- 6.Koster S, et al. Drop-based microfluidic devices for encapsulation of single cells. Lab Chip. 2008;8:1110–1115. doi: 10.1039/b802941e. [DOI] [PubMed] [Google Scholar]
- 7.Luo C, et al. Picoliter-volume aqueous droplets in oil: Electrochemical detection and yeast cell electroporation. Electrophoresis. 2006;27:1977–1983. doi: 10.1002/elps.200500665. [DOI] [PubMed] [Google Scholar]
- 8.Sgro AE, Allen PB, Chiu DT. Thermoelectric manipulation of aqueous droplets in microfluidic devices. Anal Chem. 2007;79:4845–4851. doi: 10.1021/ac062458a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn K, et al. Electrocoalescence of drops synchronized by size-dependent flow in microfluidic channels. Appl Phys Lett. 2006;88:264105. [Google Scholar]
- 10.Chabert M, Dorfman KD, Viovy JL. Droplet fusion by alternating current (AC) field electrocoalescence in microchannels. Electrophoresis. 2005;26:3706–3715. doi: 10.1002/elps.200500109. [DOI] [PubMed] [Google Scholar]
- 11.Link DR, et al. Electric control of droplets in microfluidic devices. Angew Chem Int Ed Engl. 2006;45:2556–2560. doi: 10.1002/anie.200503540. [DOI] [PubMed] [Google Scholar]
- 12.Niu X, Gulati S, Edel JB, deMello AJ. Pillar-induced droplet merging in microfluidic circuits. Lab Chip. 2008;8:1837–1841. doi: 10.1039/b813325e. [DOI] [PubMed] [Google Scholar]
- 13.Priest C, Herminghaus S, Seemann R. Controlled electrocoalescence in microfluidics: Targeting a single lamella. Appl Phys Lett. 2006;89:134101. [Google Scholar]
- 14.Sarrazin F, et al. Mixing characterization inside microdroplets engineered on a microcoalescer. Chem Eng Sci. 2007;62:1042–1048. [Google Scholar]
- 15.Song H, Chen DL, Ismagilov RF. Reactions in droplets in microfluidic channels. Angew Chem Int Ed Engl. 2006;45:7336–7356. doi: 10.1002/anie.200601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song H, Tice JD, Ismagilov RF. A microfluidic system for controlling reaction networks in time. Angew Chem Int Ed Engl. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 17.Ahn K, et al. Dielectrophoretic manipulation of drops for high-speed microfluidic sorting devices. Appl Phys Lett. 2006;88 024104. [Google Scholar]
- 18.Boedicker JQ, Li L, Kline TR, Ismagilov RF. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip. 2008;8:1265–1272. doi: 10.1039/b804911d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus KJ, Barrett IJ, Nouhi Y, Hieter P. Specific synthetic lethal killing of RAD54B-deficient human colorectal cancer cells by FEN1 silencing. Proc Natl Acad Sci USA. 2009;106:3276–3281. doi: 10.1073/pnas.0813414106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loscertales IG, et al. Micro/nano encapsulation via electrified coaxial liquid jets. Science. 2002;295:1695–1698. doi: 10.1126/science.1067595. [DOI] [PubMed] [Google Scholar]
- 21.Anna SL, Bontoux N, Stone HA. Formation of dispersions using “flow focusing” in microchannels. Appl Phys Lett. 2003;82:364–366. [Google Scholar]
- 22.Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 23.Ward T, Faivre M, Abkarian M, Stone HA. Microfluidic flow focusing: Drop size and scaling in pressure versus flow-rate-driven pumping. Electrophoresis. 2005;26:3716–3724. doi: 10.1002/elps.200500173. [DOI] [PubMed] [Google Scholar]
- 24.Frenz L, Blank K, Brouzes E, Griffiths AD. Reliable microfluidic on-chip incubation of droplets in delay-lines. Lab Chip. 2009;9:1344–1348. doi: 10.1039/b816049j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiss MM, et al. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal Chem. 2008;80:8975–8981. doi: 10.1021/ac801276c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez JE, Negulescu PA. Intracellular detection assays for high-throughput screening. Curr Opin Biotechnol. 1998;9:624–631. doi: 10.1016/s0958-1669(98)80141-9. [DOI] [PubMed] [Google Scholar]
- 27.Joensson HN, et al. Detection and analysis of low-abundance cell-surface biomarkers using enzymatic amplification in microfluidic droplets. Angew Chem Int Ed Engl. 2009;48:2518–2521. doi: 10.1002/anie.200804326. [DOI] [PubMed] [Google Scholar]
- 28.Cushing MC, Anseth KS. Materials science. Hydrogel cell cultures. Science. 2007;316:1133–1134. doi: 10.1126/science.1140171. [DOI] [PubMed] [Google Scholar]
- 29.Chabert M, Viovy JL. Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells. Proc Natl Acad Sci USA. 2008;105:3191–3196. doi: 10.1073/pnas.0708321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fournier-Bidoz S, et al. Facile and rapid one-step mass preparation of quantum-dot barcodes. Angew Chem Int Ed Engl. 2008;47:5577–5581. doi: 10.1002/anie.200800409. [DOI] [PubMed] [Google Scholar]
- 31.Han M, Gao X, Su JZ, Nie S. Quantum-dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 32.Pregibon DC, Toner M, Doyle PS. Multifunctional encoded particles for high-throughput biomolecule analysis. Science. 2007;315:1393–1396. doi: 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- 33.Melkko S, et al. Isolation of high-affinity trypsin inhibitors from a DNA-encoded chemical library. Angew Chem Int Ed Engl. 2007;46:4671–4674. doi: 10.1002/anie.200700654. [DOI] [PubMed] [Google Scholar]
- 34.Portney NG, et al. Length-based encoding of binary data in DNA. Langmuir. 2008;24:1613–1616. doi: 10.1021/la703235y. [DOI] [PubMed] [Google Scholar]
- 35.Siegel AC, et al. Cofabrication of electromagnets and microfluidic systems in poly(dimethylsiloxane) Angew Chem Int Ed Engl. 2006;45:6877–6882. doi: 10.1002/anie.200602273. [DOI] [PubMed] [Google Scholar]
- 36.Johnston KP, et al. Water-in-carbon dioxide microemulsions: An environment for hydrophiles including proteins. Science. 1996;271:624–626. [Google Scholar]
- 37.Holtze C, et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip. 2008;8:1632–1639. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.