Abstract
Fungi produce numerous low molecular weight molecules endowed with a multitude of biological activities. However, mining the full-genome sequences of fungi indicates that their potential to produce secondary metabolites is greatly underestimated. Because most of the biosynthesis gene clusters are silent under laboratory conditions, one of the major challenges is to understand the physiological conditions under which these genes are activated. Thus, we cocultivated the important model fungus Aspergillus nidulans with a collection of 58 soil-dwelling actinomycetes. By microarray analyses of both Aspergillus secondary metabolism and full-genome arrays and Northern blot and quantitative RT-PCR analyses, we demonstrate at the molecular level that a distinct fungal-bacterial interaction leads to the specific activation of fungal secondary metabolism genes. Most surprisingly, dialysis experiments and electron microscopy indicated that an intimate physical interaction of the bacterial and fungal mycelia is required to elicit the specific response. Gene knockout experiments provided evidence that one induced gene cluster codes for the long-sought after polyketide synthase (PKS) required for the biosynthesis of the archetypal polyketide orsellinic acid, the typical lichen metabolite lecanoric acid, and the cathepsin K inhibitors F-9775A and F-9775B. A phylogenetic analysis demonstrates that orthologs of this PKS are widespread in nature in all major fungal groups, including mycobionts of lichens. These results provide evidence of specific interaction among microorganisms belonging to different domains and support the hypothesis that not only diffusible signals but intimate physical interactions contribute to the communication among microorganisms and induction of otherwise silent biosynthesis genes.
Keywords: genome mining, lecanoric acid, orsellinic acid, Streptomyces
Microorganisms such as soil-dwelling bacteria and fungi produce a multitude of low molecular weight molecules that play an eminent role in drug discovery. It is conceivable that many of these compounds are produced as chemical signals or for defending the habitat; however, their true functions in their biological context are largely unknown (1, 2). Despite the large number of known bioactive compounds produced by fungi, the biosynthetic potential of these microorganisms is greatly underestimated. In fact, analyses of the increasing number of whole-genome sequences indicate that fungi encode the genetic information for the biosynthesis of a plethora of compounds that are not observed when cultured under standard laboratory conditions (3, 4). This applies in particular to the encoded thiotemplate assembly lines such as polyketide synthases (PKSs) (5) and nonribosomal peptide synthetases (NRPSs) (6). It appears that particular, in most cases unknown, triggers are required to activate such silent or cryptic biosynthesis pathways. To harness this untapped biosynthetic potential, various strategies have been developed over the past years, such as activation through gene expression in heterologous hosts (7), ectopic expression of pathway regulators (3), and epigenetic methods (8–10). Even so, the onset of secondary metabolite production in complex situations such as microbial communities or interspecies associations is particularly intriguing and yet underexplored. This is surprising, because it is generally believed that the interplay among organisms of the same or different species has resulted in vast natural product diversity (e.g., pheromones, predator-prey molecules, metabolites of symbiotic associations). The concept of interspecies crosstalk leading to chemical diversity has also been successfully applied to the laboratory to elicit the biosynthesis of previously undescribed metabolites (10–12). However, all these studies were entirely empirical at the chemical level.
Here, we report the first systematic microarray-based monitoring of induced expression of silent biosynthetic gene clusters in the model fungus Aspergillus nidulans through the interaction with a collection of actinomycetes sharing the same habitat. This integrative study led to the discovery of the long-sought after genetic locus coding for the biosynthesis of the archetypal polyketide orsellinic acid (OA; 1). In addition, we unveil the ability of A. nidulans to produce 1, the typical lichen metabolite lecanoric acid (2), and the cathepsin K inhibitors F-9775A (3) and F-9775B (4). In sum, we demonstrate that the fungus reacts on distinct interactions by the activation of specific secondary metabolism gene clusters, which contributes to understanding the crosstalk among different species of microorganisms.
Results and Discussion
Specific Induction of A. nidulans Secondary Metabolism Genes Through Cocultivation with Bacteria.
Bioinformatic analysis of the published A. nidulans genome sequence led to the identification of 28 putative polyketide and 24 putative nonribosomal peptide biosynthesis gene loci, which is in good agreement with the number reported by von Döhren (6). The abundance of putative biosynthesis gene clusters in A. nidulans clearly outnumbers the known secondary metabolites of this model organism. A reason for this observation might be that only a subset of biosynthesis pathway genes is expressed under standard laboratory culture conditions; therefore only a few potential chemical structures are produced. Apparently, these genes are only expressed on stimuli such as environmental cues, stress, or yet unknown biotic signals.
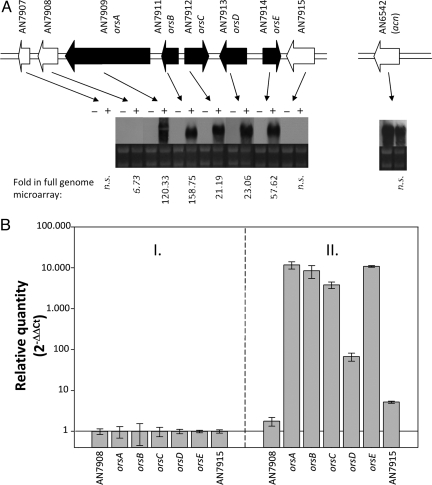
To monitor the expression of silent or cryptic loci systematically, we spotted probes representing each predicted biosynthetic pathway on a glass slide, yielding a specific A. nidulans secondary metabolism array (ASMA) [supporting information (SI) Fig. S1B]. The ASMA comprises genes encoding the central enzymes of each pathway (i.e., NRPSs and PKSs) as well as accessory and regulatory components. The expression profile monitored by the ASMA confirmed that the majority of gene clusters remain silent under laboratory conditions. To examine the impact of microbial interplay on fungal biosynthesis of gene expression, we cocultured the model fungus with a collection of soil-dwelling bacterial strains that share the same habitat. In total, 58 different actinomycetes that included different genera and species were probed (Table S1). On incubation, RNA was isolated, and after reverse transcription of A. nidulans mRNA, cDNA was hybridized with the ASMA. Surprisingly, only a single strain, designated Streptomyces hygroscopicus [American Type Culture Collection (ATCC) 29253], specifically induced fungal biosynthesis genes. According to the ASMA, 2 putative PKS (AN7909) and NRPS (AN7884) gene clusters were clearly up-regulated (Fig. 1A, Fig. S2, and Table S2). To confirm this observation and to analyze global changes in gene expression in the presence of the streptomycete, we carried out transcriptome analyses using full-genome microarrays (Fig. S1A). It should be noted that bacterial RNA does not interfere with hybridization, because we could not detect any 16S rRNA in the gels or in RNA quality control with the Agilent bioanalyzer; furthermore, cDNA that starts from the polyA tails of the eukaryotic (fungal) mRNA was used. During cocultivation of A. nidulans and S. hygroscopicus, a total of 395 genes were identified as being differentially expressed (248 up-regulated and 147 down-regulated) using the Bonferroni correction (13) for multiple testing based on the 0.05 P-value threshold. Among these were most of the genes spanning the region between AN7874 and AN7914, including the 2 biosynthesis gene clusters detected previously (Fig. S2) as well as the cryptic PKS gene AN7903. Northern blot analyses confirmed the induced expression of the whole region with the exception of genes AN7885–AN7893, AN7904, and AN7906–AN7908, which remained silent (Fig. 1A and Fig. S2). Quantitative (q) RT-PCR further confirmed the specific induction of the biosynthesis gene cluster from AN7909–AN7914 (Fig. 1B).
Fig. 1.
Impact of fungal-bacterial cocultivation on fungal gene expression pattern. (A) Northern blot analysis of ors genes. Total RNA from the A. nidulans WT (−) and the A. nidulans WT coincubated with S. hygroscopicus (+) was analyzed. Agarose gels as loading controls demonstrating the 18S and 28S rRNA are shown below the Northern blots. AN7909 and AN7911–AN7914 were designated as orsA–E, respectively. Numbers above the arrows indicate the annotated ORFs. Numbers below mark the “fold” detected by the full-genome DNA microarray of A. nidulans. An insignificant fold was marked as not significant (n.s.). ORF orsA represents the PKS gene of the ors locus. The acn gene of A. nidulans encoding β-actin was analyzed as a control for a gene not induced by S. hygroscopicus. (B) Relative quantity of the mRNA steady-state level determined by qRT-PCR of orsA–E (AN7909–AN7914). Relative quantity is given as the log2 of −ΔΔCt. Genes AN7908 and AN7915 flanking the ors gene cluster were analyzed as controls. They were not induced by cocultivation, as shown by microarray and Northern blot analyses. I. Cultivation of A. nidulans. II. Cocultivation of A. nidulans with S. hygroscopicus. Data obtained by cultivation of A. nidulans alone were set as 1.
Physical Interaction of S. hygroscopicus with A. nidulans Stimulates the Production of Aromatic Polyketides.
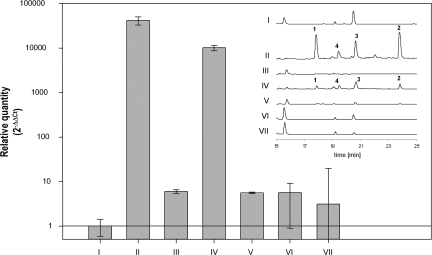
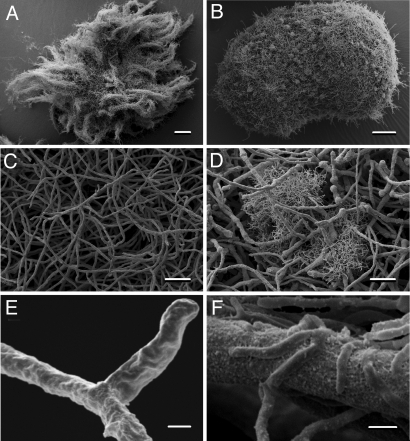
The specific response of the fungus could be caused by bacterial metabolites that are released into the environment. To test whether diffusible low molecular weight signaling molecules are involved in triggering fungal gene expression, we treated the fungal culture with the supernatant of the bacterial culture, with (co)culture extracts as well as heat-inactivated bacteria. Furthermore, we also carried out a cocultivation experiment in which bacteria and fungi were separated using a dialysis tube. To exclude the involvement of signal molecules that cannot diffuse through the membrane or the scenario in which a signal molecule is only produced in coculture, we also tested the influence of the supernatant of a coculture of bacterium and fungus lacking the PKS gene on fungal gene expression and metabolite production. Surprisingly, in no case was the fungal response observed, as demonstrated by qRT-PCR analyses, which show dramatic changes (5 orders in magnitude) in PKS gene expression on bacterial-fungal contact (Fig. 2). Obviously, the induction of fungal gene expression depends on the direct contact between the fungus and the bacterium. This assumption was unambiguously confirmed by scanning electron microscopy of biomass obtained from cocultivation. The micrographs impressively demonstrate that S. hygroscopicus is nested within the fungal mycelium and that the bacterial mycelium is, in fact, partially anchored to the fungal hyphae (Fig. 3). This intimate trans domain interaction very likely simulates a scenario occurring in the field. Our finding is also supported by the recent observation of Siemieniewicz and Schrempf (14) that viable networks of hyphae of the highly chitinolytic Streptomyces olivaceoviridis extend massively at the expense of Aspergillus proliferans hyphae. However, the tight association, as in S. hygroscopicus and A. nidulans, with a concomitant specific metabolic response is, to the best of our knowledge, unprecedented.
Fig. 2.
Relative quantity of the mRNA steady-state level determined by qRT-PCR of the polyketide gene orsA (AN7909) and the corresponding HPLC profiles of supernatants of cultures. Relative quantity is given as the log2 of −ΔΔCt. Constitutive expression of the β-actin gene (AN6542) was used as an internal control. I: Control, A. nidulans. II: Cocultivation of A. nidulans and S. hygroscopicus, inoculation with 1/20 volume of S. hygroscopicus culture. III: A. nidulans culture, inoculation with 1/20 volume of filtered S. hygroscopicus culture supernatant. IV: Cocultivation of A. nidulans and S. hygroscopicus, inoculation with 1/100 volume of S. hygroscopicus culture. V: Cocultivation of A. nidulans and S. hygroscopicus (1/100 volume of S. hygroscopicus culture) separated by a dialysis bag. VI: Cultivation of A. nidulans, inoculation with 1/20 volume culture supernatant of a coculture of A. nidulans WT and S. hygroscopicus. VII: Cultivation of A. nidulans with 1/20 volume culture supernatant of a coculture of A. nidulans ΔorsA (ΔAN7909) strain and S. hygroscopicus.
Fig. 3.
Physical interaction of A. nidulans with S. hygroscopicus. (A) Scanning electron micrograph of A. nidulans. (Scale bar: 200 μm.) (B) Scanning electron micrograph of A. nidulans coincubated with S. hygroscopicus. (Scale bar: 200 μm.) (C) Magnification of A. (Scale bar: 20 μm.) (D) Magnification of B. (Scale bar: 20 μm.) (E) Further magnification of C. (Scale bar: 1 μm.) (F) Further magnification of D showing the close contact between the filamentous bacteria and fungal mycelia. (Scale bar: 1 μm.)
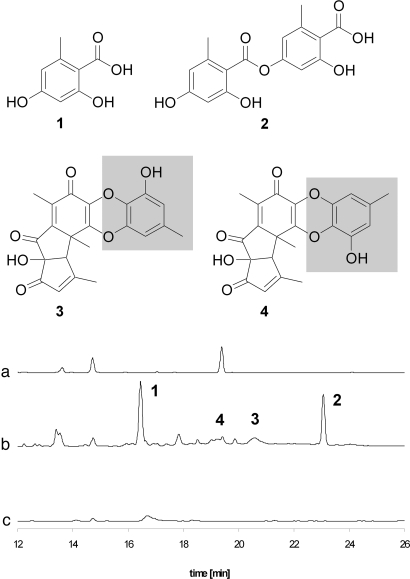
The induced expression of biosynthesis genes prompted us to monitor the metabolome of the coculture for new metabolites. Comparison of HPLC profiles of the extracts from individual strains and from the coculture revealed new metabolites produced by the induced fungus (Fig. 4). These new metabolites were only present when the fungus and the bacterium could physically interact (Fig. 2). The new metabolites were isolated from the extract of an upscaled culture (14 L), and their structures were elucidated by 1D and 2D NMR and MS measurements. Compound 1 has a molecular formula of C8H8O4, as established by high-resolution electrospray ionization mass spectrometry (HRESI-MS) analysis, and on the basis of 1D and 2D NMR data (Fig. S3), the structure was established as OA (Fig. 4). The molecular formula of compound 2 was deduced from HRESI-MS measurements as C16H14O7, and 13C NMR data suggested the presence of a dimer of compound 1. The chemical shift of C-7 (δ 167.1 ppm) indicates an ester bond between both aromatic compounds, and the heteronuclear multiple bond correlation (HMBC) couplings (Fig. S3) revealed the structure of compound 2 as lecanoric acid (Fig. 4). The NMR data were in accordance with published data (15). Interestingly, lecanoric acid is a typical lichen metabolite (16). Yet, there is only a single report on filamentous fungi producing this ATP synthesis and electron transfer inhibitor (17). In addition to 1 and 2, 2 yellow-orange pigments are produced as minor compounds (3 and 4). Electrospray ionization mass spectrometry (ESI-MS) measurements revealed a molecular mass of 396 for both compounds, and from HRESI-MS, a molecular formula of C21H16O8 was deduced. Compound database searches and comparison of the NMR data of 3 and 4 with literature data identified the compounds as the known polyketides F-9775A (3) and F-9775B (4). Both compounds were originally isolated from Paecilomyces carneus and are inhibitors of cathepsin K, thus damping osteoporosis (18). The structures of 3 and 4 suggest that these compounds are partially derived from OA (Fig. 4).
Fig. 4.
HPLC profiling of microbial cultures. Chromatographic profiles of extracts from A. nidulans WT (a), A. nidulans WT coincubated with S. hygroscopicus (b), and orsA deletion strain cocultured with S. hygroscopicus (c). Structures of OA (1), OA-derived lecanoric acid (2), F-9775A (3), and F-9775B (4).
The Cryptic Gene Locus Codes for Archetypal Polyketide Biosynthesis.
OA is the simplest acetate-derived aromatic compound that requires no reduction during its biosynthesis, and thus represents the archetypal phenolic polyketide. Despite its widespread occurrence in fungi and the growing body of knowledge on fungal polyketide biosynthesis, the molecular basis of its biosynthesis has remained unknown to date. The activity of orsellinic acid synthase (OAS) from Penicillium madriti was demonstrated in a crude cell extract as early as 1968 (19). However, neither the gene nor the protein amino acid sequence has been elucidated to date, and nothing is known yet about the catalytic domains or their organization (5). To correlate the biosynthesis of the phenolic compounds with the interaction with S. hygroscopicus, we performed transcription analyses and targeted gene inactivation. Northern blot analyses indicated that the production of 1-4 strictly correlates with the expression of the PKS gene. Furthermore, to prove unequivocally that the identified gene encodes the required enzyme for the biosynthesis of OA, we deleted the PKS gene (orsA) in the genome of A. nidulans (Fig. 1A and Fig. S4). HPLC monitoring of the cocultivation of the resulting mutant with S. hygroscopicus indicated that the production of OA (1), lecanoric acid (2), and F-9775A/B (3/4) was fully abolished (Fig. 4). Taken together, these data clearly prove that the identified PKS gene is required for the biosynthesis of the epitome polyketide 1 and suggest that 1 serves as a biosynthetic building block for 2-4.
Functional and Phylogenetic Relation of the OAS.
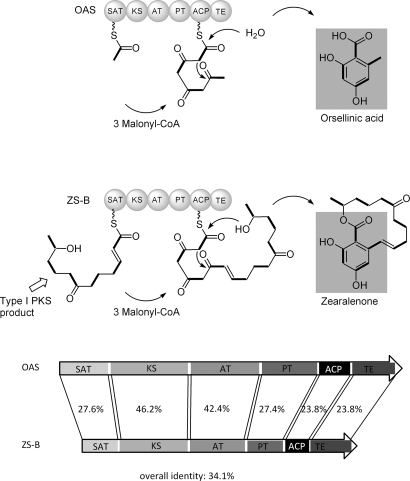
The discovery and functional proof of the OAS from A. nidulans now allow a first glance into the model pathway. The deduced amino-acid sequence of orsA shows conserved motifs for starter acyl transferase (SAT), ketosynthase (KS), acyltransferase (AT), product template (PT), acyl carrier protein (ACP), and thioesterase (TE) domains, which are hallmarks for nonreducing PKSs (Fig. 5). To identify the potential orthologs of OAS (OrsA) and to understand its relation to the other fungal PKSs, we performed National Center for Biotechnology Information (NCBI) protein database searches and phylogenetic analyses (SI Text). The resulting phylogenetic tree (Fig. S5) corresponds well to the previously published phylogenies of fungal PKSs (20, 21). The OA producers form a clade that has a basal position relatively to the other nonreducing PKSs, which fits well with the postulated early evolutionary rise of these proteins. The whole branch is characterized by the loss of the methyltransferase domain and gain of the TE domain. Proteins with the highest similarity all feature the same domain architecture and represent a wide spectrum of species across the fungal domain, including lichens. Some of the more distant PKSs are known as melanin synthases, but approximately 20 close relatives to OrsA (Fig. S5) have not yet been functionally characterized. These proteins are good candidates for OAS-like enzymes.
Fig. 5.
Model for OA and zearalenone biosynthesis and domain architectures of the involved polyketide synthases: ACP, AT, KS, OAS, PT domain, SAT, TE, and ZS-B.
Notably, among the closely related and functionally characterized fungal iterative PKSs, zearalenone synthase B (ZS-B) from Gibberella zea (anamorph Fusarium graminearum) showed the highest overall identity (34.1%) with OAS (Fig. 5). Zearalenone is a mycotoxin with chronic estrogenic effects on mammals (22). Its biosynthesis involves 2 PKSs: a reducing PKS (ZS-A) produces a hexaketide chain, which serves as starter unit for a nonreducing PKS (ZS-B) (23). Both OAS (OrsA) and ZS-B are tetraketide synthases that elongate the starter with 3 nonreduced malonate-derived C2 units. Regions of lowest conservation were determined by protein sequence alignment and comparison with the reported domain deconstruction by Townsend and coworkers (24). The separated domains of OAS and ZS-B show highest conservation in the KS and AT domains with 46.2% and 42.2% identity, respectively, reflecting the identical polyketide backbone synthesis. However, the core PKSs clearly deviate in their ACP domains (23.8% identity), which might be attributable to an additional phosphopantetheinyl-binding motif in ZS-B, as in the A. nidulans WA synthase (25). Not unexpectedly, SAT domains of both enzymes share rather low overall sequence similarity (27.6%), because the starter units differ significantly (i.e., acetate for OAS and a hexaketide for ZS-B). The PT domains, which support cyclization and aromatization of the enzyme-bound polyketide chain (26), are conserved with an identity of 27.4%. Lowest similarity was found for TE/cyclase domains of both enzymes with 23.8% identity. This is not surprising, because OAS TE apparently catalyzes the release of a carboxylic acid by hydrolysis, whereas the N-terminal TE domain of ZS-B acts as a cyclase and releases zearalenone with the formation of a macrocycle by lactonization. These data highlight the functional similarity and deviation of these related iterative fungal PKSs.
Significance and Conclusions.
In recent years, various highly diverse interactions between bacteria and fungi have been investigated (27). Such microbial interplay may have a dramatic effect on the survival, colonization, and pathogenesis of the interacting partners. However, considering the multitude of possible microbial interactions, the intimacy and impact of fungal-bacterial associations are often underestimated. In addition to the wellknown lichen-forming fungal-cyanobacterial symbiosis, there are even instances in which (myco)toxins are actually produced by bacteria residing within the fungal cytosol (28) and in which the fungal sporulation strictly depends on the presence of endobacteria (29).
In this study, we used previously undescribed array technology to monitor the selective induction of silent fungal biosynthesis genes through bacterial-fungal interactions. Through individual cocultivation of the model fungus A. nidulans with a collection of 58 actinomycetes, we identified a bacterium that selectively triggers the expression of silent biosynthesis genes that are not expressed under standard laboratory conditions. Most surprisingly, the fungal gene expression does not rely on diffusible chemical signals secreted by the bacterium. In fact, dialysis experiments and electron microscopy unveiled that an intimate physical interaction of the bacterial and fungal mycelia is required to elicit the specific response. Chemical analyses, Northern blot analysis, qRT-PCR analysis, and gene inactivation revealed that the cryptic PKS gene codes for the long-sought after OAS. This previously undescribed synthase is mechanistically related to zearalenone synthase, as deduced from detailed domain analyses. Furthermore, a phylogenetic study demonstrated that orthologs of this synthase are widespread among the fungal domain, including lichen mycobionts. Notably, apart from this archetypal polyketide, A. nidulans also produces the OA derivative lecanoric acid (2), which is a typical lichen metabolite. It is intriguing that this compound is usually found in a fungal/bacterial mutualism, and thus likely plays a role in microbial communication. On the other hand, because lecanoric acid (2) inhibits ATP synthesis and electron transfer, it is also conceivable that the bacterium has elicited a fungal defense strategy affecting organisms that are susceptible to lecanoric acid-mediated energy breakdown. Because the inducing bacterium is not affected, one may even speculate about a symbiotic fungal-bacterial relation. From a pharmacological point of view, the induced production of an ATP synthesis inhibitor (2) and the antiosteoporosis agents 3 and 4 in A. nidulans indicates that dormant genes may code for the biosynthesis of valuable biologically active compounds and potential therapeutics that are not produced in the absence of particular triggers.
The unprecedented intimate trans domain interaction of Aspergillus and Streptomyces very likely simulates a scenario occurring in the field and raises important questions with regard to the molecular basis of such microbial interactions and fungal signal transduction. Future studies of this and related systems may not only provide fundamental insights into microbial communication but have the potential to lead to the discovery of otherwise overlooked bioactive compounds.
Materials and Methods
Strains.
A. nidulans strain RMS011 (pabaA1, yA2; ΔargB::trpCΔB, trpC801, veA1) (30) was used in this study. Plasmids were propagated in Escherichia coli DH5α. The A. nidulans orsA deletion strain was obtained by transformation of A. nidulans (31) using a linear DNA fragment that encoded approximately 1,500-bp sequences homologous to the regions upstream and downstream of orsA, the codon-humanized coding sequence of the Gaussia luciferase gene (Prolume), and the argB gene of A. nidulans as a selectable marker gene (Fig. S4).
Media and Cultivation of Strains.
E. coli was grown at 37 °C in LB supplemented with ampicillin (50 μg mL−1). Actinomycetes were cultivated in M79 medium (32) at 28 °C for 2–3 days with shaking. A. nidulans strains were grown in Aspergillus minimal medium (AMM) (33). Required supplements were added as follows: arginine (final concentration of 50 μM), uracil or uridine (2.2 mg mL−1 and 1 mg mL−1, respectively), and p-aminobenzoic acid (3 μg mL−1). As a preculture, a 100-mL AMM overnight culture inoculated with 108 conidia mL−1 was used. The biomass was separated from the medium using Miracloth (Calbiochem) and inoculated into 100 mL of fresh AMM. Five milliliters of a freshly prepared actinomycete culture was added, and the Aspergillus-actinomycetes coculture was further incubated at 37 °C with shaking. After 3 h, samples were taken for transcriptome analysis. After 24 h, 50 mL of the culture containing both hyphae and medium was harvested for HPLC analysis. Cocultivation, heat inactivation, and dialysis cultures were performed as described in SI Text.
ASMA.
Custom spotted (BF-Biolabs) CodeLink glass slides (GE Healthcare) were used. Seven to 10 different 25-mers corresponding to the 3′ half of each presented gene were spotted in hexaplicates (Fig. 1B).
Analysis of Global Gene Expression.
Custom full-genome microarrays of A. nidulans were generated (febit biomed). They were hybridized with cDNA synthesized from RNA, which was isolated from cocultures of A. nidulans with S. hygroscopicus ATCC29253.
Preparation of RNA, RT, Labeling, Array Hybridization, and Data Processing.
Total RNA from A. nidulans was isolated, labeled, and hybridized as described in SI Text. Scans were analyzed with GenePix Pro 6.1 software (Molecular Devices). A transcript was represented as a triplicate of probesets. Each probeset contained 10 probes (different 25-mer oligonucleotides) per gene. The signal intensity of a probeset was calculated using Tukey's biweight robust estimation procedures (34). Investigation of reproducible differences among samples was performed using the Linear Models for Microarray Data (LIMMA) packages (35) of Bioconductor (36). Data were processed using quantile normalization. Background correction was performed using the method “minimum.” To obtain the genes with the most evidence of differential expression, a linear model fit was calculated for each gene using the LIMMA package.
Northern Blot Analyses.
Northern blot probes were labeled with digoxigenin-11-dUTP (Roche). TriSURE reagent (Bioline)-isolated total RNA samples from A. nidulans were separated on formaldehyde-containing agarose gels and blotted on Hybond-N+ positive-charged nylon membranes (GE Healthcare). Hybridization was carried out in DIG-easy Hyb buffer, followed by binding of antidigoxigenin-alkaline phosphate Fab fragments and application of CDP-Star ready-to-use solution (Roche). Thereby, fluorescence signals were generated and visualized on Super RX x-ray films (Fuji).
qRT-PCR.
To quantify transcript levels by qRT-PCR, total RNA was purified and its quality was controlled as described previously. Ten micrograms of DNase I-treated RNA was utilized as a template for cDNA synthesis for 3 h at 48 °C using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed on an Applied Biosystems StepOne Real-Time PCR system in triplicate for each sample. The A. nidulans β-actin gene AN6542 was used as an internal standard for calculation of expression levels. The cDNA samples were diluted 10 times for amplification to obtain EvaGreen (Biotium)-labeled PCR fragments (GeneAmp Fast PCR Master Mix; Applied Biosystems) by using gene-specific primers for the genes AN7908–AN7915 (Table S3). The expression of β-actin was monitored using forward primer 5′-CACCCTTGTTCTTGTTTTGCTC-3′ and reverse primer 5′-AAGTTCGCTTTGGCAACGC-3′. The sizes of the PCR amplicons of AN7908–AN7915 and AN6542 were 81, 92, 94, 80, 107, 107, 86, and 105 bp, respectively. The cycling parameters included an initial DNA denaturation step at 95 °C for 2 min, followed by 45 cycles with DNA denaturation at 95 °C for 5 s and primer annealing and extension at 62 °C for 15 s. Controls with no added template were included for each primer pair to exclude primer dimers from interfering with amplification detection. qRT-PCR results were analyzed using StepOne software (version 2.0; Applied Biosystems). The cycle number at which the fluorescence passed the cycle threshold (Ct) was determined by StepOne software (version 2.0). It was used for quantitation of the expression level. Relative expression levels for each cDNA sample were obtained by the ΔΔCt method via normalization to β-actin using the formula 2−(Ct AN79xx − Ct AN6542) for all samples derived from experimental cultures as well as from the calibration culture (A. nidulans cultivated in AMM under identical growth conditions).
Preparation of Chromosomal DNA and Southern Blot Analysis.
Genomic DNA from A. nidulans mycelia was isolated using the MasterPure Yeast DNA purification kit (Epicentre Biotechnologies) according to a modified isolation protocol (37). Cells were disrupted in a Precellys24 homogenisator (peqlab) for 3 cycles (30 s grinding/30 s pause) at a rotation speed of 6,500 rpm in an Innuspeed Lysis Tube A (Analytik Jena). Southern blot analysis was carried out by using a nonradioactive labeled DNA probe as described for Northern blot analysis.
Extraction, Isolation, and Structure Elucidation of 1-4.
See SI Text.
Scanning Electron Microscopy.
A. nidulans and S. hygroscopicus were cocultivated as described. Fungal pellets were washed twice with 0.1 M cacodylate buffer (pH 7.2), fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M cacodylate buffer, postfixed with osmium tetroxide, dehydrated in a graded series of ethanol solutions, critical-point dried from liquid CO2, mounted on a stumb, and coated with gold. The specimens were examined using a LEO 1450 VP Scanning Electron Microscope (Leo Electron Microscopy).
Supplementary Material
Acknowledgments.
We thank C. Täumer, M.G. Schwinger, and K. Perlet for technical assistance in cultivation of microorganisms and Southern and Northern blot analyses and A. Perner and F. Rhein for MS and NMR measurements, respectively. We are grateful to the Electron Microscopy Center of the University Hospital Jena for providing electron microscopic photographs. This research was supported by the excellence graduate schools Jena School for Microbial Communication, International Leibniz Research School for Microbial and Biomolecular Interactions, and Hans Knöll Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0901870106/DCSupplemental.
References
- 1.Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc London B. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goh EB, et al. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann S, et al. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 4.Chiang YM, et al. Molecular genetic mining of the Aspergillus secondary metabolome: Discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox RJ. Polyketides, proteins and genes in fungi: Programmed nano-machines begin to reveal their secrets. Org Biomol Chem. 2007;5:2010–2026. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]
- 6.von Döhren H. A survey of nonribosomal peptide synthetase (NRPS) genes in Aspergillus nidulans. Fungal Genet Biol. 2008;46:S45–S52. doi: 10.1016/j.fgb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Schümann J, Hertweck C. Advances in cloning, functional analysis and heterologous expression of fungal polyketide synthase genes. J Biotechnol. 2006;124:690–703. doi: 10.1016/j.jbiotec.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 8.Williams RB, et al. Epigenetic remodeling of the fungal secondary metabolome. Org Biomol Chem. 2008;6:1895–1897. doi: 10.1039/b804701d. [DOI] [PubMed] [Google Scholar]
- 9.Henrikson JC, Hoover AR, Joyner PM, Cichewicz RH. A chemical epigenetics approach for engineering the in situ biosynthesis of a cryptic natural product from Aspergillus niger. Org Biomol Chem. 2009;7:435–438. doi: 10.1039/b819208a. [DOI] [PubMed] [Google Scholar]
- 10.Scherlach K, Hertweck C. Triggering cryptic natural product biosynthesis in microorganisms. Org Biomol Chem. 2009;7:1753–1760. doi: 10.1039/b821578b. [DOI] [PubMed] [Google Scholar]
- 11.Angell S, Bench BJ, Williams H, Watanabe CM. Pyocyanin isolated from a marine microbial population: Synergistic production between two distinct bacterial species and mode of action. Chem Biol. 2006;13:1349–1359. doi: 10.1016/j.chembiol.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Oh DC, Kauffman CA, Jensen PR, Fenical W. Induced production of emericellamides A and B from the marine-derived fungus Emericella sp. in competing co-culture. J Nat Prod. 2007;70:515–520. doi: 10.1021/np060381f. [DOI] [PubMed] [Google Scholar]
- 13.Benjamini Y, Hochberg Y. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 14.Siemieniewicz KW, Schrempf H. Concerted responses between the chitin-binding protein secreting Streptomyces olivaceoviridis and Aspergillus proliferans. Microbiology. 2007;153:593–600. doi: 10.1099/mic.0.2006/001073-0. [DOI] [PubMed] [Google Scholar]
- 15.Narui T, et al. NMR assignments of depsides and tridepsides of the lichen family Umbilicariaceae. Phytochemistry. 1998;48:815–822. [Google Scholar]
- 16.Stocker-Worgotter E. Metabolic diversity of lichen-forming ascomycetous fungi: Culturing, polyketide and shikimate metabolite production, and PKS genes. Nat Prod Rep. 2008;25:188–200. doi: 10.1039/b606983p. [DOI] [PubMed] [Google Scholar]
- 17.Umezawa H, et al. Isolation of lecanoric acid, an inhibitor of histidine decarboxylase from a fungus. J Antibiot. 1974;27:587–596. doi: 10.7164/antibiotics.27.587. [DOI] [PubMed] [Google Scholar]
- 18.Satou A, Morishita T, Hosoya T, Ishikawa Y. 1999 Japan Patent 11–001480. [Google Scholar]
- 19.Gaucher GM, Shepherd MG. Isolation of orsellinic acid synthase. Biochem Biophys Res Commun. 1968;32:664–671. doi: 10.1016/0006-291x(68)90290-8. [DOI] [PubMed] [Google Scholar]
- 20.Grube M, Blaha J. On the phylogeny of some polyketide synthase genes in the lichenized genus Lecanora. Mycol Res. 2003;107:1419–1426. doi: 10.1017/s0953756203008724. [DOI] [PubMed] [Google Scholar]
- 21.Kroken S, et al. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc Natl Acad Sci USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuiper-Goodman T, Scott PM, Watanabe H. Risk assessment of the mycotoxin zearalenone. Regul Toxicol Pharmacol. 1987;7:253–306. doi: 10.1016/0273-2300(87)90037-7. [DOI] [PubMed] [Google Scholar]
- 23.Gaffoor I, Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol. 2006;72:1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Udwary DW, Merski M, Townsend CA. A method for prediction of the locations of linker regions within large multifunctional proteins, and application to a type I polyketide synthase. J Mol Biol. 2002;323:585–598. doi: 10.1016/s0022-2836(02)00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii I, Watanabe A, Sankawa U, Ebizuka Y. Identification of Claisen cyclase domain in fungal polyketide synthase WA, a naphthopyrone synthase of Aspergillus nidulans. Chem Biol. 2001;8:189–197. doi: 10.1016/s1074-5521(00)90068-1. [DOI] [PubMed] [Google Scholar]
- 26.Crawford JM, et al. Deconstruction of iterative multidomain polyketide synthase function. Science. 2008;320:243–246. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wargo MJ, Hogan DA. Fungal-bacterial interactions: A mixed bag of mingling microbes. Curr Opin Microbiol. 2006;9:359–364. doi: 10.1016/j.mib.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Partida-Martinez LP, Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 29.Partida-Martinez LP, Monajembashi S, Greulich KO, Hertweck C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr Biol. 2007;17:773–777. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 30.Stringer MA, Dean RA, Sewall TC, Timberlake WE. Rodletless, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 1991;5:1161–1171. doi: 10.1101/gad.5.7.1161. [DOI] [PubMed] [Google Scholar]
- 31.Ballance DJ, Turner G. Development of a high-frequency transforming vector for Aspergillus nidulans. Gene. 1985;36:321–331. doi: 10.1016/0378-1119(85)90187-8. [DOI] [PubMed] [Google Scholar]
- 32.Prauser H, Falta R. Phage sensitivity, cell wall composition and taxonomy of actinomycetes (Translated from German) Z Allg Mikrobiol. 1968;8:39–46. doi: 10.1002/jobm.3630080106. [DOI] [PubMed] [Google Scholar]
- 33.Hortschansky P, et al. Interaction of HapX with the CCAAT-binding complex—A novel mechanism of gene regulation by iron. EMBO J. 2007;26:3157–3168. doi: 10.1038/sj.emboj.7601752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mosteller F, Tukey J. Exploratory Data Analysis and Regression. Reading, MA: Addison-Wesley; 1977. [Google Scholar]
- 35.Smyth GK. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 36.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80.1–R80.16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wickes B. DNA isolation from a filamentous fungus using the MasterPure Yeast DNA purification kit. Epicentre Forum. 2004;11:7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.