Abstract
Regulation of ion-transport in the Na+/Ca2+ exchanger (NCX) occurs via its cytoplasmic Ca2+-binding domains, CBD1 and CBD2. Here, we present a mechanism for NCX activation and inactivation based on data obtained using NMR, isothermal titration calorimetry (ITC) and small-angle X-ray scattering (SAXS). We initially determined the structure of the Ca2+-free form of CBD2-AD and the structure of CBD2-BD that represent the two major splice variant classes in NCX1. Although the apo-form of CBD2-AD displays partially disordered Ca2+-binding sites, those of CBD2-BD are entirely unstructured even in an excess of Ca2+. Striking differences in the electrostatic potential between the Ca2+-bound and -free forms strongly suggest that Ca2+-binding sites in CBD1 and CBD2 form electrostatic switches analogous to C2-domains. SAXS analysis of a construct containing CBD1 and CBD2 reveals a conformational change mediated by Ca2+-binding to CBD1. We propose that the electrostatic switch in CBD1 and the associated conformational change are necessary for exchanger activation. The response of the CBD1 switch to intracellular Ca2+ is influenced by the closely located cassette exons. We further propose that Ca2+-binding to CBD2 induces a second electrostatic switch, required to alleviate Na+-dependent inactivation of Na+/Ca2+ exchange. In contrast to CBD1, the electrostatic switch in CBD2 is isoform- and splice variant-specific and allows for tailored exchange activities.
Keywords: alternative splicing, Ca2+-binding domain, homeostasis, Ca2+ sensor
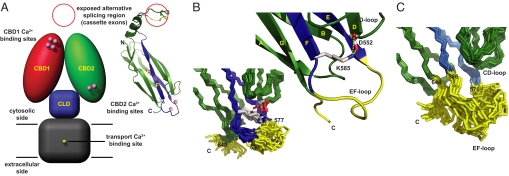
Driven by the Na+ gradient generated by the Na+/K+ ATPase, the Na+/Ca2+ exchanger (1) removes Ca2+ ions that entered the cytosol via L-type Ca2+ channels and/or Ca2+ release channels of the endoplasmic reticulum, predominantly expelling one Ca2+ ion for the uptake of three Na+ ions (2). Structurally, NCX appears to consist of nine transmembrane α-helices (3, 4) and a large, ≈500 residue-long cytosolic loop. Recently, we demonstrated the presence of two, rather than the previously assumed one, Ca2+-binding domain in the cytosolic exchanger loop (5). We also proposed a model (Fig. 1A) in which the Ca2+- binding sites of the first Ca2+-binding domain (CBD1) are ≈90 Å away from the transport Ca2+-binding site in the ion-conducting transmembrane domain. In contrast, Ca2+-binding sites of the second Ca2+-binding domain (CBD2) are close to a predicted third domain that shows homology with α-catenin and that we therefore designated as catenin-like domain (CLD). The NMR structures of Ca2+ bound CBD1 and CBD2 (5) revealed the architecture of a seven-stranded β-sandwich and suggested, in the absence of constraints to the metals, coordination of two Ca2+ ions in both domains. Recent X-ray structures of Ca2+-bound CBD1 (6) and CBD2 (7) not only provided a more detailed view of the Ca2+-binding sites but also showed unequivocally binding of four Ca2+ ions to CBD1.
Fig. 1.
Model of intact NCX and CBD2 NMR structures, representing the two major NCX1 splice variant classes. (A) Hypothetical NCX model consisting of four domains: Transmembrane domain (gray; residues 1–216 and 706–903), catenin-like domain (blue; residues 217–370 and 651–705), CBD1 (red; residues 371–500) and CBD2 (green; residues 501–650) with the numbering based on the canine NCX1 AD-splice variant (NCX1.4). At high intracellular Ca2+ concentrations four Ca2+ ions are bound to CBD1 whereas two Ca2+ ions are coordinated by CBD2-AD. The CBD2 ribbon diagram depicts the location of the regions encoding the mutually exclusive exon A or B in blue and the cassette exons in yellow. (B) Ensemble of NMR structures displaying the Ca2+-binding sites of CBD2-AD in the absence of Ca2+. Residues 578–581 and 648–657 are disordered and colored yellow. In the blow-up of the central chain, the crucial Lys-585 maintains a salt-bridge with Asp-552 and thereby stabilizes the CBD2-AD Ca2+-binding sites. (C) Ensemble of CBD2-BD NMR structures with an increased number of disordered residues (yellow) despite the presence of 10 mM CaCl2.
In pioneering work in 1992, Hilgemann and coworkers dissected the regulatory properties of cardiac NCX into Ca2+-dependent activation and Na+-dependent inactivation using the giant patch method (8, 9). Ion-transport of NCX is activated by binding of Ca2+ ions to its Ca2+-binding domains whereas Na+ concentrations rising >15 mM can lead to Na+-dependent inactivation. In NCX1, Na+-dependent inactivation is influenced by alternative splicing as variants containing exon A can alleviate inactivation at elevated intracellular Ca2+ concentrations whereas those variants containing the mutually exclusive exon B (10, 11) cannot. Intriguingly, exchanger variants containing exon A seem to be almost exclusively expressed in excitable cells, where high Ca2+ fluxes are needed whereas exon B-containing variants are generally restricted to nonexcitable cells (12). In the CBD2 structure, exons A and B encode strands E and F as well as the first acidic segment of the Ca2+-binding sites (Fig. 1A). In proximity to the CBD1 Ca2+-binding sites, cassette exons C–F, whose functions are largely unknown, form a region of variability. In addition to NCX1, there are two other genes that encode homologous Na+/Ca2+ exchangers, NCX2 (13) and NCX3 (14). Showing ≈70% sequence identity to one another, the three isoforms are reported to share very similar regulatory behavior despite their tissue-specific expression (15).
Although the structures of the individual Ca2+-binding domains, CBD1 and the splice variant CBD2-AD (containing exons A and D), are well established in their Ca2+-bound forms (5–7), the molecular mechanisms that link Ca2+ binding and release to altered transport function in the intact exchanger remain elusive. By the same token, it is unclear what role tissue-specific alternative splicing in CBD2 plays in NCX regulation. Here, we report solution structures of the two major splice variant classes in NCX1, the Ca2+-free form of CBD2-AD and the structure of CBD2-BD in the presence of Ca2+. They complement the current structural information on the Ca2+-binding domains and provide fuel for a mechanism of Ca2+ regulation in NCX. Based on striking differences in the electrostatic potential between their Ca2+-free and -bound forms, we propose that Ca2+-binding sites of CBD1 and CBD2 form electrostatic switches analogous to C2-domains (16). SAXS analysis of constructs containing both, CBD1 and CBD2, reveals a substantial conformational change, mediated by Ca2+ binding to the CBD1 Ca2+-binding sites. The combination of cassette exons in CBD2 may influence the affinities of the CBD1 Ca2+-binding sites and thereby may modulate the response of the switch in CBD1 to varying intracellular Ca2+ concentrations. Furthermore, we provide insight into Na+-dependent exchanger inactivation by demonstrating the inability of exon B-containing NCX1 splice variants to bind Ca2+. We also show by sequence analysis and calorimetric studies that CBD2 from NCX1, NCX2, and NCX3 have different numbers of Ca2+-binding sites with distinct affinities. Correlation of these specific Ca2+-binding properties with functional studies, hints at isoform- and splice variant-specific exchanger inactivation that likely results in tailored exchange activities and may be one reason for tissue-specific expression of NCX.
Results
CBD2-AD Apo-Structure.
To obtain insight into the Ca2+-binding mechanism and the function of CBD2-AD, we determined the structure of its apo-form by multidimensional heteronuclear NMR spectroscopy at neutral pH (for refinement statistics see Table S1). The NMR structure displays the architecture of a seven-stranded β-sandwich that provides a stable scaffold for the Ca2+-binding sites formed by the AB-, CD-, and EF-loops as well as residues succeeding the G-strand (Fig. 1 A and B). Superposition of the apo- with the Ca2+ bound form (PDB code: 2QVM) reveals an rmsd of 1.14 Å on 102 Cα positions with the largest differences lying in the EF-loop. In contrast to the Ca2+-bound form, only very few resonances could be observed for residues 578–581 (EF-loop) and 649–657 (C terminus), thus reflecting an increased flexibility of these regions in the apo-form (Fig. 1B). Nevertheless, the apo-form of CBD2-AD maintains its structural integrity in the absence of Ca2+ as the crucial Lys-585 points into Ca2+-binding site II where it forms a salt-bridge with Asp-552. This is in contrast to a recent X-ray structure of CBD2-AD (7), obtained from crystals grown at pH 4.9 at which NCX is reportedly inactive (15). In the X-ray structure, Lys-585 rigidifies the Ca2+-binding sites by formation of salt bridges to both, Asp-552 and Glu-648 as well as a hydrogen bond to the backbone carbonyl oxygen of Glu-580. However, in the NMR data, acquired at pH 7, we do not find any NOEs to Glu-580 and Glu-648 that would support these interactions nor do we observe any structurally relevant cross-peaks for residues 649–657 that in the X-ray structure appear to be even better defined than in the corresponding Ca2+-bound form.
CBD2-BD Structure.
Among the numerous NCX1 splice variants only exon A-containing exchangers are capable of overcoming Na+-dependent inactivation at high intracellular Ca2+ concentrations. To offer a structural explanation for this functional observation, we determined the structure of CBD2-BD in the presence of 10 mM CaCl2 (Fig. 1C). Strikingly, no resonances could be detected in any spectra for residues 577–585 that contribute half of the Ca2+-coordinating residues in CBD2-AD and very few resonances for the remaining potential Ca2+-coordinating residues, Glu-516, Asp-552, and Glu-648. Hence, Ca2+-binding sites in the NCX1 CBD2-BD NMR structure are disordered.
Ca2+-Binding Determinants of CBD1 and CBD2.
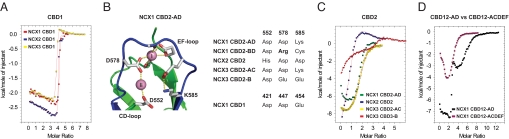
Sequence comparison of all available Na+/Ca2+ exchanger sequences (Figs.S1 and S2 ) reveals strict conservation of the Ca2+-coordinating residues in CBD1. To confirm experimentally that these residues also exhibit comparable Ca2+-binding properties, we titrated CBD1 of canine NCX1, mouse NCX2, and NCX3 under identical conditions with CaCl2 (Fig. 2A and Fig. S3 A–C). Indeed, the ITC patterns look very similar and are consistent with binding of four Ca2+ ions, in agreement with the Ca2+-bound NCX1 CBD1 X-ray structure (6). However, due to the steepness of their binding isotherms, caused by three high-affinity Ca2+-binding sites, it was not possible to confidently determine the individual macroscopic binding constants (for approximate affinity ranges see Fig. S3 A–C).
Fig. 2.
Thermodynamical analyses of CBD1 and CBD2 Ca2+-binding sites using ITC. (A) ITC curves of NCX1, NCX2, and NCX3 CBD1, obtained at pH 7 in 30 mM NaCl and 130 mM KCl, confirm binding of four Ca2+ ions with likely comparable affinities. (B) Isoform- and splice variant-specific key-residues 552, 578, and 585 that form the CBD2 Ca2+-binding sites between the CD- and EF-loops. (C) Comparison of the NCX1 CBD2-AD binding isotherm (5) with those of NCX2 CBD2, NCX3 CBD2-AC, and NCX3 CBD2-B suggests binding of one, two, and three Ca2+ ions, respectively. (D) Comparison of NCX1 CBD12-AD and CBD12-ACDEF Ca2+-binding isotherms.
In strong contrast in CBD2, residue types at positions 552, 578, and 585, located around Ca2+-binding site II, can vary depending on the isoform and splice variant (Fig. 2B and Fig. S2) whereas Ca2+-coordinating residues in site I are conserved with the exception of position 578 in exon B-containing splice variants. To characterize the impact of the different residue types, we compared Ca2+ binding of NCX1 CBD2-AD (macroscopic binding constants of 1 and 9 μM) (5) to WT forms of NCX1 CBD2-BD, NCX2 CBD2, NCX3 CBD2-AC, and NCX3 CBD2-B (Fig. 2C and Fig. S3 D–F) as well as their double mutants (Fig. S3 G and H), designed to restore the Ca2+-coordination scheme of NCX1 CBD2-AD (Fig. 2B).
Unlike in NCX1 CBD2-AD, where Asp-578 is crucial to the coordination of Ca2+ ions at site I and II (5, 7) and Lys-585 prevents binding of a second Ca2+ ion on the side of the CD-loop as in CBD1, NCX1 CBD2-BD possesses at positions 578 and 585 an arginine and a cysteine residue that are both disordered in the CBD2-BD structure. In agreement with the NMR structure, no heat pulses could be detected for NCX1 CBD2-BD in response to CaCl2 additions as high as 100 mM, thus reflecting the inability of exon B-containing NCX1 splice variants to specifically bind Ca2+.
In NCX2, for which no splice variants exist, replacement of Asp-552 by a histidine residue removes one of the two sole Ca2+-coordinating atoms (Fig. 2B) and thereby eliminates site II in this isoform, resulting in a single site of moderate affinity (Fig. 2C and Fig. S3D). Similarly, mutation of Asp-552 in NCX1 CBD2-AD (D552V mutant) (7) also results in binding of a single Ca2+ ion with comparable affinity (Fig. S3I).
For NCX3, three splice variants are reported, NCX3 CBD2-AC, CBD2-B, and CBD2-BC. NCX3 CBD2-AC has the same Ca2+-coordinating residue types as NCX1 CBD2-AD and appears to bind two Ca2+ ions, albeit one with remarkably high affinity (Fig. 2C and Fig. S3E). However, NCX3 CBD2-B has largely adopted the Ca2+-coordination scheme of CBD1 (Fig. 2B) between the CD- and EF-loops and its binding isotherm suggests coordination of three Ca2+ ions (Fig. 2C and Fig. S3F). Hence, exon B-containing variants of NCX1 and NCX3 are fundamentally different from each other whereas their exon A-containing variants apparently coordinate two Ca2+ ions in a very similar manner. Control experiments with the double mutants H552D/D585K of NCX2 CBD2 (Fig. S3G) and E578D/E585K of NCX3 CBD2-B (Fig. S3H) indeed restore two Ca2+-binding sites with macroscopic Ca2+-binding constants similar to those of NCX1 CBD2-AD (5).
To investigate a potential role of the cassette exons (Fig. 1A) on Ca2+ binding to CBD1, we also compared NCX1 CBD12-AD and CBD12-ACDEF, the brain and heart splice variant constructs comprising CBD1 and CBD2. In agreement with the expected binding of six Ca2+ ions, CBD12-AD shows both, the high- and medium-affinity Ca2+-binding isotherm characteristics of the individual domains (Fig. 2D). In strong contrast, CBD12-ACDEF entirely lacks the high-affinity component of CBD1 and displays only medium-affinity Ca2+ binding. This suggests that the additional cassette exons C, E, and F, located in proximity to the CBD1 Ca2+-binding sites, either destroy the high-affinity sites in CBD1 or increase their affinities substantially. To determine the exact stoichiometry of the Ca2+ ions present in the CBD12-AD and CBD12-ACDEF samples before the titration, we repeated the ITC measurements under sulfur- and chloride-free conditions, required for microparticle-induced X-ray emission (microPIXE) experiments (17), and confirmed the previously obtained results. The microPIXE analysis (SI Text) of the AD and ACDEF variants showed coordination of zero and four Ca2+ ions, respectively. Hence, Ca2+ appears to be trapped at the CBD1 Ca2+-binding sites in CBD12-ACDEF, suggesting that the affinity of these sites may be substantially higher than those of CBD12-AD.
Electrostatic Properties of CBDs in NCX.
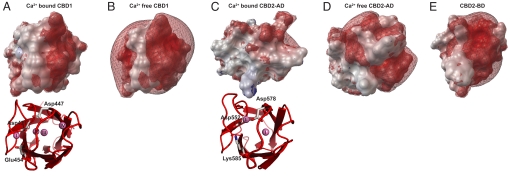
The arrangement of Ca2+ ions in CBD1 and CBD2 is reminiscent of the Ca2+-binding sites in C2-domains (6, 18) that form electrostatic switches, required for membrane association of many proteins. We calculated the electrostatic potentials of Ca2+-bound CBD1 and CBD2-AD and compared them with their corresponding Ca2+ free forms (Fig. 3 A–E). Because we lack the structure of the Ca2+-free form of CBD1, we extrapolated its electrostatic potential from the structure of the Ca2+-bound form (PDB code: 2DPK), with the four Ca2+ ions removed (Fig. 3B). In the absence of Ca2+, Ca2+-binding sites in CBD1 form a strongly negative electrostatic potential caused by the extensive cluster of aspartic and glutamic acid residues and the lack of any basic residues. Upon binding of four Ca2+ ions, the negative potential becomes drastically reduced (Fig. 3A). In comparison, the CBD2-AD electrostatic potential of the Ca2+-free form (Fig. 3D) is considerably less negative because of the reduced number of acidic residues and the presence of Arg-547, Lys-583, and Lys-585. Upon Ca2+ binding, the CBD2-AD electrostatic potential turns approximately neutral (Fig. 3C). In strong contrast, the electrostatic potential of CBD2 in exon B-containing NCX1 variants remains negative even at elevated Ca2+ concentrations because of its inability of binding Ca2+ (Fig. 3E).
Fig. 3.
Electrostatic potentials of the CBD1 and CBD2 Ca2+-binding sites in the presence and absence of Ca2+ shown in topview. (A) Ca2+-bound state of NCX1 CBD1 (PDB code: 2DPK) (6). (B) Ca2+-free state of NCX1 CBD1, calculated from the Ca2+-bound form with its four Ca2+ ions removed. (C) NCX1 CBD2-AD coordinating two Ca2+ ions (PDB code: 2QVM) (7). (D) Ca2+-free state of NCX1 CBD2-AD as determined by NMR (this work, PDB code: 2KLS) and (E) NCX1 CBD2-BD (this work, PDB code: 2KLT) incapable of Ca2+ binding despite the presence of 10 mM CaCl2. The red and blue meshes represent isocontours of the electrostatic potential at −5 and +5 kT/e that were calculated with the program apbs (29) in the presence of 0.15 M KCl and visualized using the python molecular viewer PMV (30).
Ca2+ Induces a Twist in the Hinge Between CBD1 and CBD2.
To explore effects of Ca2+ binding more globally, we used small-angle X-ray scattering (SAXS) on CBD12-AD and CBD12-ACDEF. SAXS gives orientation-averaged information about the solution structure of macromolecules by comparing calculated and observed scattering profiles (19). We derived the bimodal distributions P(r) of scattering vectors, which reveal a maximal distance (Dmax) of 120 ± 10 Å for both constructs in the Ca2+-bound form (Fig. S4 A and B). In the absence of Ca2+, Dmax values increase to 145 and 135 ± 10 Å for CBD12-AD and CBD12-ACDEF, respectively, suggesting a more extended conformation for the Ca2+-free forms.
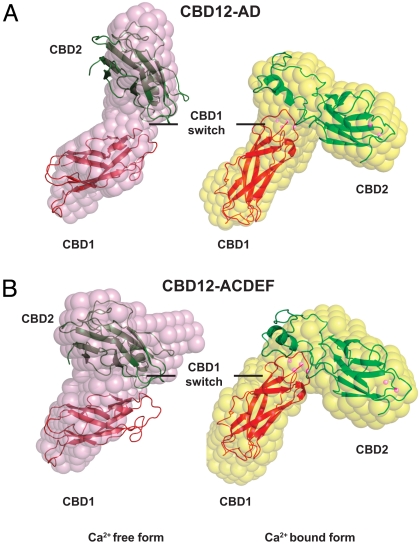
Ab initio reconstructions of the Ca2+-bound and -free forms of both constructs reveal a prominent conformational change in form of a twist in orientation between CBD1 and CBD2 (Fig. 4 A and B). Whereas ab initio reconstructions of Ca2+-bound CBD12-AD and CBD12-ACDEF are virtually identical (rmsd of 0.74 Å), CBD12-ACDEF in the Ca2+-free form displays a slightly stronger twist than CBD12-AD under the same conditions. The ab initio results were confirmed by an independent rigid-body modeling approach, where we fitted scattering profiles derived from the NMR structures of the individual domains to the SAXS data (Fig. S4 A and B). Most likely because of the large unstructured FG-loops and the lack of structural information of the Ca2+-free form of CBD1 and residues encoded by exons C, E, and F, rigid-body modeling for the Ca2+-free forms does not produce an optimal fit with the beads models. However, the substantial conformational change is supported by both modeling approaches.
Fig. 4.
SAXS shape reconstructions. Analyses of brain CBD12-AD (A) and heart CBD12-ACDEF (B) constructs in the presence and absence of Ca2+ reveal a conformational change induced by Ca2+ binding to CBD1. Pink beads reflect constructs in the absence of Ca2+ whereas yellow beads represent the Ca2+ bound forms. All beads and rigid-body models are superimposed on the CBD1 beads of the CBD12-AD construct.
Discussion
Na+-Dependent Inactivation.
Among the numerous NCX1 splice variants, the largest functional difference resides in their capability of alleviating Na+-dependent inactivation at high intracellular Ca2+ concentrations (8, 9). In NCX1, this property is determined by the type of the mutually exclusive exon that encodes the first acidic segment in CBD2 (Fig. 1A). In particular, replacement of Asp-578 by an arginine residue as in exon B removes the capability of exon A-containing NCX1 splice variants to overcome Na+-dependent inactivation (20). Strikingly, the CBD2-BD NMR structure, determined in the presence of an excessive amount of Ca2+, displays disordered Ca2+-binding sites (Fig. 1C). This suggests that Arg-578 not only prevents binding of Ca2+, but also does not form a unique salt bridge with a single surrounding acidic residue (Fig. 2B). We therefore conclude that the CBD2 Ca2+-binding sites of exon B-containing NCX1 splice variants do not specifically bind Ca2+ under physiological conditions.
Intriguingly, functional comparison of NCX1, NCX2, and NCX3 revealed very similar properties for all 3 exchanger isoforms (15), although NCX1 and NCX3 expression in skeletal muscle is reported to be developmentally regulated (21), and NCX3 distribution is apparently very tissue-specific (12). Comparison of all available NCX sequences shows strict conservation of the Ca2+-coordinating residues in CBD1 (Fig. S1); however, there is isoform and splice variant-dependent variability of three key-residues at positions 552, 578, and 585 in CBD2 (Fig. S2). Together, these residues form Ca2+-binding site II, earlier considered not to be essential for Ca2+ regulation (7). In contrast, Ca2+-binding site I, with the exception of position 578, is conserved in all NCX.
In line with the predictions of sequence analysis, ITC characterization of CBD1 from NCX1, NCX2, and NCX3 confirm binding of 4 Ca2+ ions with likely comparable affinities (Fig. 2A). In contrast, NCX2 CBD2, NCX3 CBD2-AC, and NCX3 CBD2-B display binding of one, two, and three Ca2+ ions, respectively, as well as distinct macroscopic Ca2+-binding constants ranging from 250 nM to 19 μM (Fig. 2C and Fig. S3 D–F). In view of the lack of Ca2+ binding to exon B-containing NCX1 splice variants, it is thus tempting to speculate that the number and affinity of Ca2+-binding sites in CBD2 determines the capability of overcoming Na+-dependent inactivation in the numerous NCX isoforms and splice variants. Support for this hypothesis arises from functional studies and is manifold. First of all, the decline of exchange current of guinea pig NCX1.1 (containing exons ACDEF) decreases as the level of regulatory Ca2+ is increased (9). In the presence of 1 μM Ca2+, the transient peak of exchange current decays to a relatively low steady-state value, most probably as a consequence of only partial alleviation of Na+-dependent inactivation. In comparison, at Ca2+ concentrations of 10 μM and above, Na+-dependent inactivation is fully eliminated (9, 10) as presumably the second Ca2+-binding site in CBD2 becomes occupied (5). Furthermore, the NCX1.1 K585E mutant that likely has Ca2+-binding sites comparable to NCX3 CBD2-B (Fig. S3F) is reported to be significantly less susceptible to Na+-dependent inactivation than WT NCX1 (7). Intriguingly, the NCX1.1 D552V mutant sustains an exchange current comparable to WT NCX1.1 (7), even though it only binds one Ca2+ ion at the CBD2 Ca2+-binding sites at high intracellular Ca2+ concentrations (Fig. S3I). However, the unexpectedly strong exchange current may be a consequence of the difference in Ca2+ regulation properties of NCX1.1 D552V compared with WT, rather than a difference in Ca2+ binding to the CBD2 Ca2+-binding sites (7). To restore the Ca2+-coordination scheme of CBD2-AD, we made the point-mutant R578D and the double-mutant R578D/C585K of NCX1 CBD2-BD for which functional data display modest and almost full restoration of Ca2+ relief of Na+-dependent inactivation compared with NCX1.3 (containing exons B and D) (20). ITC measurements of the R578D and R578D/C585K mutants (Fig. S5 A and B) show binding of one and two Ca2+ ions, respectively. Taken together, there are functional and thermodynamic data for exchangers that bind zero, one, two, and three Ca2+ ions at the CBD2 Ca2+-binding sites. These data strongly indicate that Ca2+ ions coordinated on the side of the CD-loop influence Na+-dependent inactivation. An increasing number of Ca2+ ions bound to CBD2 correlates with a more positive electrostatic potential and an enhanced Ca2+ relief of Na+-dependent inactivation, which has a direct impact on Na+/Ca2+ exchange. Hence, the isoform- and splice variant-specific Ca2+ relief of Na+-dependent inactivation may allow for optimal adaptation of Na+/Ca2+ exchange to special needs within the cell or a specific tissue.
Ca2+-Dependent Activation.
Ca2+-dependent exchanger activation occurs at submicromolar Ca2+ concentrations (8, 9). In view of the Ca2+ affinities determined for all CBDs, these levels of Ca2+ can generally only have an effect on the high-affinity Ca2+-binding sites in CBD1 (Fig. 2A), which is apparently sufficient to initiate Na+/Ca2+ exchange (9). As Ca2+ concentrations rise to 1 μM and above, the medium-affinity CBD1 Ca2+-binding site (Fig.S3 A–C) populates and may further stabilize the activated state of the exchanger. Conversely, a drop in Ca2+ concentration below the CBD1 Ca2+ affinities reverses the activation steps (8, 9).
As we demonstrate by ITC and microPIXE experiments, cassette exons in proximity to the CBD1 Ca2+-binding sites (Fig. 1A) may influence CBD1 Ca2+ affinities and therefore may have an impact on Ca2+-dependent exchanger activation. Biologically, increased Ca2+ affinities in the ACDEF splice variant imply that in the heart exchangers enter the activated state at considerably lower intracellular Ca2+ concentrations which results in extended Na+/Ca2+ exchange activity. Although no differences between NCX1.1 and NCX1.4 in Ca2+ sensitivity of the activation process have been reported, it has been observed that the response of the AD splice variant to changes of intracellular Ca2+ concentrations is at least 10-times faster than that of NCX1.1 (10), which would be consistent with the difference in regulatory Ca2+ affinity postulated here in the isolated CBD12 constructs.
Electrostatic Switches in CBD1 and CBD2.
Similarities with C2-domains and different numbers of Ca2+-binding sites prompted us to analyze the electrostatic potentials of CBD1 and CBD2 in the presence and absence of Ca2+. Whereas Ca2+ binding causes a drastic change in the CBD1 potential that should be similar for NCX1, NCX2, and NCX3, the electrostatic potential in CBD2 strongly depends on the isoform and splice variant. Generally, Ca2+-binding increases the electrostatic potential at the Ca2+-binding sites and amplifies attraction to negatively charged areas that in the absence of Ca2+ may cause repulsion. Such Ca2+-induced changes in the electrostatic potential are regarded as switches that can promote structural rearrangements (22). In CBD1, Ca2+-binding sites are close to a highly acidic region (residues 610–629) in the unstructured FG-loop of CBD2 for which no counterpart exists at a similar position in CBD1 (Fig. S4C). This region may be especially important as it also coincides with the location of the cassette exons and the mutation-sensitive Asp-448 (23) that is near to Arg-532 in the strictly conserved BC-loop of CBD2 (Fig. S2). In contrast, isoform- and splice variant-specific differences in the number and affinity of the Ca2+-binding sites determine the strength of the electrostatic switch in CBD2. Because exon B-containing NCX1 variants are incapable of Ca2+ binding, these splice forms are also unable to form an electrostatic switch.
To investigate the electrostatic switch in CBD1 in general and the effect of alternative splicing in particular, we analyzed CBD12-AD and CBD12-ACDEF by SAXS. Both ab initio and rigid-body SAXS models reveal a substantial conformational change between the Ca2+-free and -bound forms that is manifested by a twist in orientation between CBD1 and CBD2 (Fig. 4 A and B). Although this conformational change may be constrained by the N- and C-terminal linker regions to the CLD in the intact exchanger, the SAXS models likely reflect the effects of Ca2+ binding and release at the CBD1 Ca2+-binding sites. In terms of alternative splicing, both variants are virtually identical in the Ca2+ bound form whereas in the absence of Ca2+ the twist in the longer ACDEF variant appears to be slightly more pronounced than in the AD variant. Thus, these specific combinations of cassette exons do not appear to result in distinct activated states, but may influence the inactive state of NCX.
Potential Mechanism of Ca2+ Regulation.
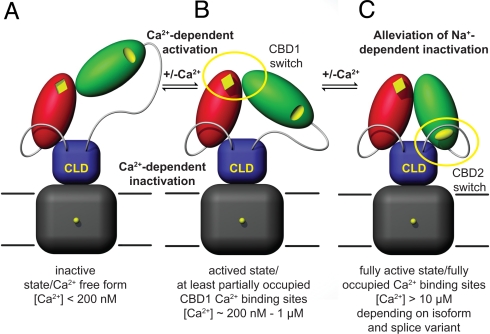
Combining our structural and biophysical data with results obtained from functional studies (7, 9, 10, 11, 20), we propose the following dual electrostatic switch mechanism for Ca2+ regulation in NCX (Fig. 5 A–C). Upon rise of intracellular Ca2+, Ca2+ ions initially bind to the CBD1 Ca2+-binding sites, thus substantially increasing their electrostatic potential. Consequently, as visualized by SAXS analysis (Fig. 4 A and B) CBD1 and CBD2 reorient and adopt a more compact conformation (Fig. 5B). This likely reduces the tension on the N- and/or C-terminal linker regions to the CLD that relays Ca2+ binding and release events to the transmembrane domain. This initial activation step (Fig. 5B) is referred to as Ca2+-dependent activation and initiates Na+/Ca2+ exchange. To alleviate counteracting Na+-dependent inactivation, Ca2+ ions must also bind to CBD2 (Fig. 5C), thereby inducing an isoform and splice variant-specific change in the electrostatic potential at the CBD2 Ca2+-binding sites. Finally, a drop in Ca2+ concentration below the CBD1 Ca2+ affinities reverses the activation steps and arrests NCX in an inactive state (Fig. 5A).
Fig. 5.
Hypothetical dual electrostatic switch mechanism in NCX regulation. (A) Inactive, Ca2+-free NCX in extended conformation. (B) Submicromolar Ca2+ concentrations induce a conformational change via the electrostatic switch in CBD1 that results in a compaction of the Ca2+-binding domains and probably reduces tension on the linker regions to the CLD. (C) Binding of Ca2+ to CBD2 allows for sustained Na+/Ca2+ exchange and removes counteracting Na+-dependent inactivation.
Materials and Methods
Expression and Purification of CBD1 and CBD2 Constructs.
NCX1 constructs (CBD1, CBD2-AD, and CBD2-BD) were derived from canine NCX1 (accession code: P23685) whereas all NCX2 and NCX3 constructs were from mouse (accession codes: Q8K596 and Q7TS90, respectively). All constructs were cloned and produced as described earlier (5). NMR samples of the apo-form of CBD2-AD and CBD2-BD contained 10 mM EDTA and 10 mM CaCl2, respectively.
NMR Spectroscopy and Structure Calculations.
NMR spectra were acquired at 306 K on Varian Inova 600 and 800 MHz spectrometers. Backbone and side chain assignments were obtained using standard protocols. Distance restraints for structure calculations were obtained from 3D-13C-NOESY-HSQC and 3D-15N-NOESY-HSQC spectra, measured with an 80-ms NOE-mixing time. All data were processed using the NMRPipe program suite (24) and analyzed using the program XEASY (25). We initially used the program RADAR (26) that automatically picks and assigns NOESY cross peaks using the module CANDID in the program CYANA 2.1 (27). NOE peak lists were then manually extended. For validation of the input data and final structures see Table S1. Figures were generated with the program PyMOL (http://pymol.sourceforge.net/)
Isothermal Titration Calorimetry.
ITC samples were produced in M9 minimal medium and concentrated in a 1 mM Hepes (Sigma, Ultrapure; pH 7.0), 2.5 mM β-mercaptoethanol buffer to minimize contaminating divalent ions. CBD1 ITC samples in addition contained 30 mM NaCl and 130 mM KCl to prevent aggregation. Protein concentrations were determined using a Nanodrop spectrophotometer ND-1000 (Isogen Life Science). Metal salts were prepared in the same buffers as the corresponding protein samples. ITC experiments were carried out with an ITC200 (Microcal) device. After thermal equilibration at 298 K, 38–114 1 μl serial injections were performed at 500 rpm stirring speed with an injection spacing of 4 min. To correct the experimental binding isotherm for background heat effects, we also titrated CaCl2 containing buffer into buffer. Information on ITC data analysis is provided in SI Text.
Small-Angle X-Ray Scattering.
CBD12-AD and CBD12-ACDEF of canine NCX1 were prepared in 20 mM Hepes (Sigma, Ultrapure; pH 7.0), 20 mM β-mercaptoethanol at three different macromolecule concentrations (1–2, 8–10, and 15–20 mg/mL) in the presence of either 10 mM CaCl2 or 10 mM EDTA. Scattering data were collected at the EMBL beamline X33 of the storage ring DORIS III (DESY) and processed with the program PRIMUS (28) using standard procedures. Further information on SAXS data analysis is provided in SI Text.
Supplementary Material
Acknowledgments.
We thank Drs. Jonathan Lytton and Oliver Weichenrieder for critical reading of the manuscript; Stephan Hilge for professional help with the figures; Drs. Luminita Damian, Adrián Velázquez-Campoy, and Manfred Roessle for fruitful discussions on ITC and SAXS, respectively; Drs. Dmitri Svergun and Petr Konarev for their support during the SAXS data collection at the beamline X33 of the EMBL outstation in Hamburg, Germany; and Drs. Melanie Bailey and Chris Jeynes for their help with the PIXE measurements. This work was supported by the Netherlands Organization for Scientific Research Grants 700.55.443 and 700.57.101. The Surrey Ion Beam Centre is supported by the Engineering and Physical Sciences Research Council under contract EP/D032210.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and chemical shift information have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2KLS and 2KLT).
This article contains supporting information online at www.pnas.org/cgi/content/full/0902171106/DCSupplemental.
References
- 1.Lytton J. Na+/Ca2+ exchangers: Three mammalian gene families control Ca2+ transport. Biochem J. 2007;406:365–382. doi: 10.1042/BJ20070619. [DOI] [PubMed] [Google Scholar]
- 2.Kang TM, Hilgemann DW. Multiple transport modes of the cardiac Na+/Ca2+ exchanger. Nature. 2004;427:544–548. doi: 10.1038/nature02271. [DOI] [PubMed] [Google Scholar]
- 3.Iwamoto T, et al. Unique topology of the internal repeats in the cardiac Na+/Ca2+ exchanger. FEBS Lett. 1999;446:264–268. doi: 10.1016/s0014-5793(99)00218-5. [DOI] [PubMed] [Google Scholar]
- 4.Nicoll DA, Ottolia M, Lu L, Lu Y, Philipson KD. A new topological model of the cardiac sarcolemmal Na+/Ca2+ exchanger. J Biol Chem. 1999;274:910–917. doi: 10.1074/jbc.274.2.910. [DOI] [PubMed] [Google Scholar]
- 5.Hilge M, Aelen J, Vuister GW. Ca2+ regulation in the Na+/Ca2+ exchanger involves two markedly different Ca2+ sensors. Mol Cell. 2006;22:15–25. doi: 10.1016/j.molcel.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Nicoll DA, et al. The crystal structure of the primary Ca2+ sensor of the Na+/Ca2+ exchanger reveals a novel Ca2+ binding motif. J Biol Chem. 2006;281:21577–21581. doi: 10.1074/jbc.C600117200. [DOI] [PubMed] [Google Scholar]
- 7.Besserer GM, et al. The second Ca2+ binding domain of the Na+/Ca2+ exchanger is essential for regulation: Crystal structures and mutational analysis. Proc Natl Acad Sci USA. 2007;104:18467–18472. doi: 10.1073/pnas.0707417104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hilgemann DW, Matsuoka S, Nagel GA, Collins A. Steady-state and dynamic properties of cardiac sodium-calcium exchange. Sodium-dependent inactivation. J Gen Physiol. 1992;100:905–932. doi: 10.1085/jgp.100.6.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilgemann DW, Collins A, Matsuoka S. Steady-state and dynamic properties of cardiac sodium-calcium exchange. Secondary modulation by cytoplasmic calcium and ATP. J Gen Physiol. 1992;100:933–961. doi: 10.1085/jgp.100.6.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyck C, et al. Ionic regulatory properties of brain and kidney splice variants of the NCX1 Na+/Ca2+ exchanger. J Gen Physiol. 1999;114:701–711. doi: 10.1085/jgp.114.5.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurtado C, et al. Cells expressing unique Na+/Ca2+ exchange (NCX1) splice variants exhibit different susceptibilities to Ca2+ overload. Am J Physiol Heart Circ Physiol. 2006;290:H2155–2162. doi: 10.1152/ajpheart.00958.2005. [DOI] [PubMed] [Google Scholar]
- 12.Quednau BD, Nicoll DA, Philipson KD. Tissue specificity and alternative splicing of the Na+/Ca2+ exchanger isoforms NCX1, NCX2, and NCX3 in rat. Am J Physiol. 1997;272:C1250–1261. doi: 10.1152/ajpcell.1997.272.4.C1250. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, et al. Cloning of the NCX2 isoform of the plasma membrane Na+/Ca2+ exchanger. J Biol Chem. 1994;269:17434–17439. [PubMed] [Google Scholar]
- 14.Nicoll DA, et al. Cloning of a third mammalian Na+/Ca2+ exchanger, NCX3. J Biol Chem. 1996;271:24914–24921. doi: 10.1074/jbc.271.40.24914. [DOI] [PubMed] [Google Scholar]
- 15.Linck B, et al. Functional comparison of the three isoforms of the Na+/Ca2+ exchanger (NCX1, NCX2, NCX3) Am J Physiol. 1998;274:C415–423. doi: 10.1152/ajpcell.1998.274.2.C415. [DOI] [PubMed] [Google Scholar]
- 16.Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 17.Garman EF, Grime GW. Elemental analysis of proteins by microPIXE. Prog Biophys Mol Biol. 2005;89:173–205. doi: 10.1016/j.pbiomolbio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Shao X, Davletov BA, Sutton RB, Südhof TC, Rizo J. Bipartite Ca2+ binding motif in C2 domains of synaptotagmin and protein kinase C. Science. 1996;273:248–251. doi: 10.1126/science.273.5272.248. [DOI] [PubMed] [Google Scholar]
- 19.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 20.Dunn J, et al. The molecular determinants of ionic regulatory differences between brain and kidney Na+/Ca2+ exchanger (NCX1) isoforms. J Biol Chem. 2002;277:33957–33962. doi: 10.1074/jbc.M206677200. [DOI] [PubMed] [Google Scholar]
- 21.Fraysse B, et al. Expression of the Na+/Ca2+ exchanger in skeletal muscle. Am J Physiol Cell Physiol. 2001;280:C146–154. doi: 10.1152/ajpcell.2001.280.1.C146. [DOI] [PubMed] [Google Scholar]
- 22.Shao X, et al. Synaptotagmin-syntaxin interaction: The C2 domain as a Ca2+-dependent electrostatic switch. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- 23.Matsuoka S, et al. Regulation of the cardiac Na+/Ca2+ exchanger by Ca2+. Mutational analysis of the Ca2+ binding domain. J Gen Physiol. 1995;105:403–420. doi: 10.1085/jgp.105.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 26.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS. J Biomol NMR. 2002;24:171–189. doi: 10.1023/a:1021614115432. [DOI] [PubMed] [Google Scholar]
- 27.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 28.Konarev PV, Volkov VV, Sokolova AV, Koch MH, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 29.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanner MF. Python: A programming language for software integration and development. J Mol Graphics Model. 1999;17:57–61. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.